Abstract
A 62-year-old presents with angina. He has been genotyped for a panel of drug metabolism enzymes through a direct-to-consumer genetics company. His results reveal a CYP2C19 *2/*2 genotype with a warning that poor metabolizers (PMs) may have “lack of therapeutic effect of clopidogrel (Plavix), resulting from failure to generate the active form of the drug.” Stress testing suggests significant ischemic burden with consequent need for percutaneous coronary intervention (PCI), possible stent placement, and antiplatelet therapy.
Clopidogrel is one of the most widely prescribed drugs in the United States, but it is associated with a great deal of interpatient variability in response (e.g., clopidogrel resistance).1 It is a prodrug that requires activation by cytochrome P450 enzymes to its active form, with only ~15% of the dose being bioactivated. This active metabolite binds irreversibly to platelet P2Y12 (ADP) receptors to inhibit aggregation (Figure 1). Cytochrome P450 2C19 (CYP2C19) is the most important enzyme in this process. The CYP2C19 gene has several common variants. The fully functional and most prevalent version of the gene is designated CYP2C19*1, and the two most common gene variants that encode enzymes with reduced activity are designated CYP2C19*2 and CYP2C19*3, with the *3 allele present primarily in Asians.2 The frequency of these alleles in various racial and ethnic groups is shown in Table 1. In addition to the reduced function alleles, there is a common gain-of-function allele designated as CYP2C19*17.
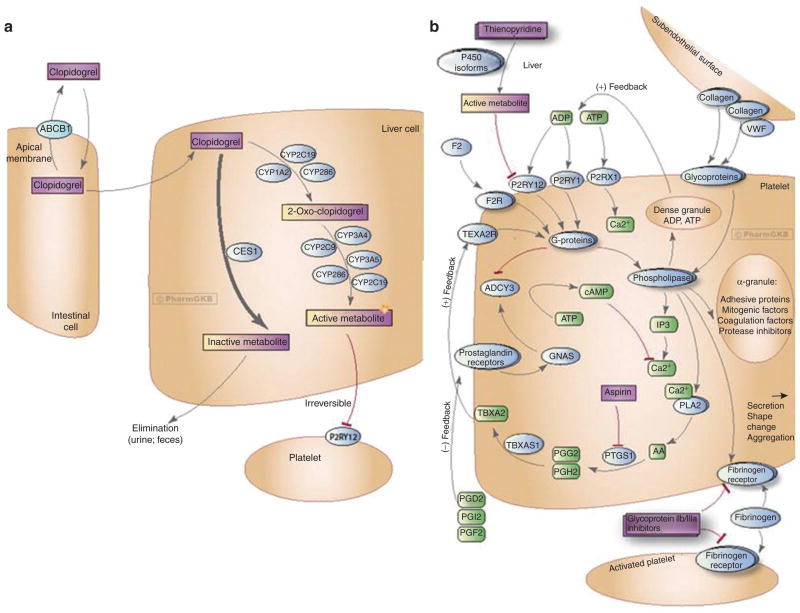
Figure 1.
Clopidogrel activation and platelet aggregation pathways. (a) Activation pathway for clopidogrel in liver.28 (b) Mechanism of clopidogrel’s antiplatelet action.29 Figure copyright PharmGKB; reprinted with permission of PharmGKB and Stanford University.
Table 1.
Frequencies of CYP2C19 *2 and*3 minor alleles and phenotype prevalence in various ethnic groups
| *2 Allele frequency | *3 Allele frequency | % IM | % PM | |
|---|---|---|---|---|
| European | 0.14 | 0.0 | 24 | 2 |
| Asian | 0.27 | 0.09 | 46 | 10 |
| African | 0.14 | 0.0 | 24 | 2 |
| African American | 0.18 | 0.008 | 30 | 3.5 |
Estimates based on HapMap and PharmGKB data (http://www.pharmgkb.org).
IM, intermediate metabolizer—*1/*2 and *1/*3 genotypes; PM, poor metabolizer—*2/*2, *3/*3, and *2/*3 genotypes.
Numerous high-profile studies have been published in the past 2 years that used both candidate gene and genome-wide approaches to assess variability in clopidogrel response. Most, but not all, of the studies have found a significant association between CYP2C19 reduced-function variants and clopidogrel treatment outcomes, as discussed further below. This association with outcomes was first noted in 2006 among healthy volunteers, in ex vivo platelet aggregation studies;3 this was followed, in 2009, by three candidate-gene studies showing that CYP2C19 is also associated with cardiovascular outcomes among clopidogrel- treated patients.4–6
Using a genome-wide association study, our group found CYP2C19*2 to be a major determinant of clopidogrel response as measured by ex vivo platelet reactivity in healthy Caucasian subjects.7 This association occurred in a gene–dose-dependent manner, whereby *1/*1 individuals (extensive metabolizers) had the greatest reduction in platelet aggregation in response to clopidogrel, *1/*2 individuals (intermediate metabolizers) had intermediate reduction in platelet aggregation in response to clopidogrel, and homozygous variant *2/*2 individuals (PMs) had the greatest residual platelet reactivity. The *2 variant accounted for ~12% of the total variation in platelet aggregation in response to clopidogrel. Importantly, in this genome-wide study, no other common variants with a similar effect size were discovered. Furthermore, in a separate population undergoing cardiac catheterization, carriers of the *2 allele had a 3.4-fold (95% confidence interval (CI), 1.36–8.46, P = 0.004) higher rate of occurrence of cardiovascular events while on clopidogrel therapy as compared with noncarriers.7
The majority of the candidate gene–related studies evaluating the association between CYP2C19*2 genotype and response to clopidogrel (as measured by platelet aggregation and clinical outcomes) have noted similar findings.4,5,8–11 Two meta- analyses12,13 found that, among patients on clopidogrel therapy, those carrying the CYP2C19*2 allele had a greater risk of cardiovascular events as compared with those with the *1/*1 genotype. In 9,685 patients from nine studies, 91.3% of whom underwent PCI, carriers of one CYP2C19*2 allele and two CYP2C19*2 alleles had significantly increased rates of cardiovascular events as compared with noncarriers (hazard ratio (HR) 1.55, 95% CI 1.10–2.17, P = 0.01 and HR 1.76, 95% CI 1.24–2.5, P = 0.002, respectively).12 The effect of CYP2C19*2 on stent thrombosis in clopidogrel-treated patients was even greater (HR 2.67, 95% CI 1.69–4.22, P = 0.0001 and HR 3.97, 95% CI 1.75–9.02, P = 0.001 in those with one and two copies of CYP2C19*2, respectively, relative to noncarriers).12 The findings in these studies indicate that the increase in risk for carriers of the CYP2C19*2 variants begins very early and remains throughout follow-up (1–100 months; median 12 months).
These findings have led to three US Food and Drug Administration–mandated updates of clopidogrel’s label, the most recent being in March 2010. The new boxed warning states that tests are available to determine a patient’s CYP2C19 genotype and that physicians should consider alternative treatment or treatment strategies in patients identified as PMs.
However, a growing number of publications have been reporting findings that conflict with the earlier reports. Recent large retrospective analyses of the CURE (Clopidogrel in Unstable Angina to Prevent Recurrent Events)14 and PLATO (Platelet Inhibition and Patient Outcomes)15 trials have not found an association between CYP2C19 genotype and cardiovascular outcomes. Of note, the populations in these studies differed from many of the other reported populations in that they were not necessarily undergoing PCI (with only 66% having a planned invasive strategy in PLATO and only 14.5% receiving stents in CURE), thereby suggesting that the major effect of CYP2C19*2 on clopidogrel response may be indication specific, which makes comparison with more recent studies difficult.14,15 This speculation is consistent with the observation that, among studies that have shown an association between CYP2C19*2 genotype and adverse outcomes with clopidogrel, the largest effects are seen on the outcome of stent thrombosis, with relative risks in the 3.0–6.0 range for individuals carrying a reduced-function allele as compared with those with the *1/*1 genotype.4,5,8,11 For cardiovascular outcomes other than stent thrombosis, the relative risks are lower.4,5 Interestingly, the PLATO study did see an association between CYP2C19 genotype and 30-day outcome with clopidogrel despite the association not holding true at the end of the 12-month follow-up period.15 An additional inconsistency of the clopidogrel loading dose (300 mg vs. 600 mg) exists across the cumulative experience of these studies, including those that did and those that did not demonstrate a genotype–outcome association.14,15 Particularly because increased risk linked to the CYP2C19*2 allele begins early, whether a 600-mg loading dose can to some degree overcome this PM effect remains to be elucidated.
Biologically consistent with reports of CYP2C19 being associated with clopidogrel treatment outcomes is the finding that proton pump inhibitors, which are CYP2C19 inhibitors, reduce the efficacy of clopidogrel. However, this issue remains unresolved, with some investigators observing the interaction and others not.16,17
POSSIBLE COURSES OF ACTION
The recently published large studies with findings that differ from those of the early reports, coupled with the US Food and Drug Administration’s update to the clopidogrel label, have caused confusion among clinicians. Although we do not yet have high-quality prospective randomized studies to completely guide our decision making, there are several courses of action that physicians can utilize in the interim, and we describe them here. These approaches and their associated pros and cons are based on the recent American College of Cardiology Foundation/American Heart Association consensus statement18 and published data.
Do nothing
Given that we do not yet have high-quality prospective randomized data to guide us on how to use genotype information once obtained, one obvious course of action is to continue treating patients as usual without regard to CYP2C19 genotype. Given that we do not know whether it is best to increase the dose of clopidogrel or switch to another agent in CYP2C19 PMs and intermediate metabolizers, this is currently the most widely used approach in most patient-care settings.
Use an alternative antiplatelet agent in all patients
Prasugrel was approved by the US Food and Drug Administration in March 2010. It is a third-generation thienopyridine that avoids the problem of loss-of-function CYP2C19 genotype because it is less dependent on activation by cytochrome P450 enzymes.19,20 Other antiplatelet agents that do not require CYP2C19 for activation include dipyridamole and ticlopidine; other agents may become available in the future, e.g., ticagrelor and elinogrel.
Although the details of how to implement CYP2C19 genotyping vis-à-vis clopidogrel use are being worked out, another approach is to simply prescribe prasugrel in all patients. However, integrated data from the two TRITON-TIMI 38 (Trial to Assess Improvement in Therapeutic Outcomes by Optimizing Platelet Inhibition with Prasugrel—Thrombolysis in Myocardial Infarction) genetic substudies that evaluated clopidogrel and prasugrel separately showed no significant reduction in the primary outcome among *1/*1 individuals treated with prasugrel as compared with clopidogrel (relative risk 0.98, 95% CI 0.80–1.20) (ref. 21). However, there was a tendency to more major or minor bleeding (relative risk 1.38, 95% CI 1.00–1.93) (ref. 21). Furthermore, problems including increased bleeding risk, limited approved indications, contraindications in certain patient populations, increased malignancy rates, and the fact that clopidogrel will come off patent in 2011, seem to make the option of prescribing prasugrel to all patients imprudent at this time.19,22
Utilize genotype and available knowledge to stratify risk
Evidence from cross-sectional studies suggesting that the CYP2C19*2 allele has a role in influencing clopidogrel response creates a temptation to apply this information rapidly in order to individualize antiplatelet therapy, even as more definitive data from well-designed prospective clinical trials are forth-coming. With this approach, CYP2C19*2 homozygotes (PMs) would be considered to be at high risk for recurrent events if they are prescribed clopidogrel, and they should therefore be prescribed another agent, e.g., prasugrel. PMs constitute ~2–4% in the population, not a large proportion but not an insignificant number either. Less certain is the question of whether CYP2C19*2 heterozygotes (intermediate metabolizers), who constitute approximately one-third of the population, should also receive an alternative therapy. Perhaps in this group genotype information should be used in conjunction with other clinical factors (e.g., diabetes, disease burden, prior myocardial infarction) and/or platelet aggregation testing (see below) to determine the most appropriate therapy. Importantly, the use of such an algorithm may be wise only within a restricted indication population, namely, patients receiving stent implants.
Currently, CYP2C19 genotyping is a send-out lab at most institutions and takes several days to receive results. In most situations, therefore, it is not possible to utilize genotype information to tailor acute antiplatelet therapy in PCI patients. In the future, we can anticipate more rapid point-of-care genetic testing and/or preemptive genotyping embedded in the electronic medical record for use when the indication, i.e., clopidogrel treatment, arises. In addition, as illustrated in the clinical vignette presented above, with direct-to-consumer genetic testing being available, some patients may present for care with information regarding their CYP2C19 genotype already in hand. In these cases of preemptive genotyping, or in cases of patients presenting with information about their own genotypes, physicians would need a compelling reason not to utilize this information to individualize antiplatelet therapy. This issue of implementing pharmacogenomics is further addressed in another article in this issue of CPT by the Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network.23
Platelet-responsiveness studies
Another approach that clinicians may choose is to incorporate platelet-responsiveness studies into clinical care.1,24,25 Given that platelet aggregation is “closer” to the final phenotype (i.e., cardiovascular outcomes) and takes into account environmental factors that may influence platelet aggregation in addition to “all” genetic factors, some have proposed that such testing might be more useful than CYP2C19 genotype testing for individualizing therapy. On the other hand, results of platelet-aggregation studies can have a great deal of variability due to both biological and technical factors. This renders the results difficult to interpret during periods of acute physiological stress (e.g., acute coronary syndrome and myocardial infarction) and would probably need to be repeated over time. By contrast, genotype is stable throughout a person’s lifetime. The GRAVITAS (Gauging Responsiveness With a Verify Now Assay—Impact on Thrombosis and Safety) trial, reported at the American Heart Association Scientific Sessions (November 2010, Chicago, IL), randomized post-PCI patients with high residual platelet reactivity to continue on a 75-mg regular clopidogrel dose or to receive another 600-mg loading dose and a higher maintenance dose of 150 mg daily. At the 6-month time point of follow-up, the composite end point, namely, cardiovascular death, myocardial infarction, or stent thrombosis, was identical in both groups (2.3%), with a trend to increased bleeding complications in the high-dose clopidogrel arm. Future research will likely evaluate whether a combination of platelet-responsiveness studies and genotype-guided therapy (e.g., dose initially according to genotype, then follow response by means of platelet-function testing) may be superior to either method alone.
Additional Areas of Uncertainty
Despite the well-replicated associations between CYP2C19 genotype and clopidogrel treatment outcomes, there remain several areas of uncertainty surrounding these associations. The first of these pertains to heterozygous (i.e., *1/*2 genotype) individuals (intermediate metabolizers). Most studies have noted the existence of gene–dose relationships, with *1/*2 heterozygous individuals having intermediate outcomes between those of homozygous *1/*1 and *2/*2 individuals. However, some studies have found increased risk only in *2/*2 homozygotes.6 The differences in the findings of the studies may be due to differences in patient populations, indications for clopidogrel therapy (e.g., stent placement, acute coronary syndrome without PCI, atrial fibrillation), and/or advances in the field over time, such as the use of drug-eluting stents instead of metal stents and the shift from coronary artery bypass grafting to more aggressive nonsurgical interventions. Indeed, as mentioned above, CYP2C19*2 genotype appears to have the largest effect in studies in which a greater proportion of patients received stents, especially at the early stages, when the risk of stent thrombosis is greatest. Future studies will need to address the effect of CYP2C19 genotype for specific indications (e.g., non-PCI patients).
A second area of uncertainty is whether higher-dose clopidogrel therapy is effective in decreasing recurrent events in CYP2C19*2 carriers. Small studies in which platelet aggregation was measured indicate that some (but not all) carriers of the *2 allele may benefit from an increased dose.24–26
Less is known with regard to other CYP2C19 genotypes. Some studies have indicated that patients who carry the common CYP2C19*17 gain-of-function allele have lower ADP-induced platelet aggregation and, consequently, a higher bleeding risk as compared with *1/*1 individuals.15,27 However, whether differences exist in cardiovascular outcomes between *17 allele carriers and wild-type individuals remains unclear. Our group did not observe any difference in outcomes by *17 carrier status that was not accounted for by *2 carrier status.7 However, the recent analysis of the CURE study data found a greater reduction in adverse cardiovascular outcomes among *17 carriers than noncarriers.14
Controversy also exists regarding whether the ABCB1 3435C→T variant is associated with clopidogrel treatment outcomes. ABCB1 is an efflux pump involved in the transport of clopidogrel and may affect the bioavailability of the drug. However, recent studies have reported conflicting results. An analysis of the TRITON-TIMI 38 study found that TT individuals treated with clopidogrel were at increased risk of adverse cardiovascular outcomes as compared with CC individuals,10 whereas our study and an analysis of PLATO data found no association between ABCB1 genotype and treatment outcomes.7,15
FUTURE PERSPECTIVE
Although early epidemiologic studies demonstrated that the CYP2C19 genotype is associated with reduced clopidogrel responsiveness, and the US Food and Drug Administration–updated label reflects this information, more recent studies have called the association into question. Therefore, now more than ever, it is unclear what is to be done regarding CYP2C19 genotyping in patients who require antiplatelet therapy. Ultimately, well-powered, prospective, randomized clinical trials comparing genotype-driven clinical interventions, including comparative effectiveness studies, will be required in order to develop the evidence base for the wider adoption of more individualized antiplatelet therapy approaches. Such prospective, randomized, controlled trials are currently under way to help determine whether outcomes can be improved with genotype-guided and/or point-of-care platelet aggregometry–guided therapy. The questions are complex and will likely require several studies, both clinical and mechanistic, in order to answer all the important questions. The problem of loss of equipoise, particularly with respect to *2/*2 individuals, could hinder the completion of studies. Results from these trials must also include pharmacoeconomic analyses to evaluate whether it is cost-effective for insurance companies to cover the costs of genotyping (and/or platelet aggregation studies) and alternative drug therapy. While we await the results of these prospective studies, clinicians must use the available data to treat individual patients in the manner they deem most appropriate. However, genotype data like those presented in the case presentation cited at the beginning of this article would seem difficult to ignore. In such a case, the clinician would probably opt to treat with prasugrel.
Footnotes
Conflict of Interest
The authors declared no conflict of interest.
References
- 1.Bonello L, et al. Consensus and future directions on the definition of high on-treatment platelet reactivity to adenosine diphosphate. J Am Coll Cardiol. 2010;56:919–933. doi: 10.1016/j.jacc.2010.04.047. [DOI] [PubMed] [Google Scholar]
- 2.Xie HG, Kim RB, Wood AJ, Stein CM. Molecular basis of ethnic differences in drug disposition and response. Annu Rev Pharmacol Toxicol. 2001;41:815–850. doi: 10.1146/annurev.pharmtox.41.1.815. [DOI] [PubMed] [Google Scholar]
- 3.Hulot JS, et al. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006;108:2244–2247. doi: 10.1182/blood-2006-04-013052. [DOI] [PubMed] [Google Scholar]
- 4.Collet JP, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009;373:309–317. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 5.Mega JL, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 6.Simon T, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360:363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 7.Shuldiner AR, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giusti B, et al. Relation of cytochrome P450 2C19 loss-of-function polymorphism to occurrence of drug-eluting coronary stent thrombosis. Am J Cardiol. 2009;103:806–811. doi: 10.1016/j.amjcard.2008.11.048. [DOI] [PubMed] [Google Scholar]
- 9.Lee JM, et al. Relation of genetic polymorphisms in the cytochrome P450 gene with clopidogrel resistance after drug-eluting stent implantation in Koreans. Am J Cardiol. 2009;104:46–51. doi: 10.1016/j.amjcard.2009.02.045. [DOI] [PubMed] [Google Scholar]
- 10.Mega JL, et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet. 2010;376:1312–1319. doi: 10.1016/S0140-6736(10)61273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sibbing D, et al. Cytochrome P450 2C19 loss-of-function polymorphism and stent thrombosis following percutaneous coronary intervention. Eur Heart J. 2009;30:916–922. doi: 10.1093/eurheartj/ehp041. [DOI] [PubMed] [Google Scholar]
- 12.Mega JL, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sofi F, Giusti B, Marcucci R, Gori AM, Abbate R, Gensini GF. Cytochrome P450 2C19(*)2 polymorphism and cardiovascular recurrences in patients taking clopidogrel: a meta-analysis. Pharmacogenomics. 2010 doi: 10.1038/tpj.2010.21. e-pub ahead of print 30 March 2010. [DOI] [PubMed] [Google Scholar]
- 14.Paré G, et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010;363:1704–1714. doi: 10.1056/NEJMoa1008410. [DOI] [PubMed] [Google Scholar]
- 15.Wallentin L, et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet. 2010;376:1320–1328. doi: 10.1016/S0140-6736(10)61274-3. [DOI] [PubMed] [Google Scholar]
- 16.Bhatt DL, et al. Clopidogrel with or without omeprazole in coronary artery disease. N Engl J Med. 2010;363:1909–1917. doi: 10.1056/NEJMoa1007964. [DOI] [PubMed] [Google Scholar]
- 17.Siller-Matula JM, Jilma B, Schrör K, Christ G, Huber K. Effect of proton pump inhibitors on clinical outcome in patients treated with clopidogrel: a systematic review and meta-analysis. J Thromb Haemost. 2010;8:2624–2641. doi: 10.1111/j.1538-7836.2010.04049.x. [DOI] [PubMed] [Google Scholar]
- 18.Holmes DR, Jr, Dehmer GJ, Kaul S, Leifer D, O’Gara PT, Stein CM. Society for Cardiovascular Angiography and Interventions; Society of Thoracic Surgeons; Writing Committee Members. ACC F/AHA Clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association. Circulation. 2010;122:537–557. doi: 10.1161/CIR.0b013e3181ee08ed. [DOI] [PubMed] [Google Scholar]
- 19.Effient (prasugrel) prescribing information. Eli Lilly; 2010. < http://pi.lilly.com/us/effient.pdf>. [Google Scholar]
- 20.Mega JL, et al. Cytochrome P450 genetic polymorphisms and the response to prasugrel: relationship to pharmacokinetic, pharmacodynamic, and clinical outcomes. Circulation. 2009;119:2553–2560. doi: 10.1161/CIRCULATIONAHA.109.851949. [DOI] [PubMed] [Google Scholar]
- 21.Sorich MJ, Vitry A, Ward MB, Horowitz JD, McKinnon RA. Prasugrel vs. clopidogrel for cytochrome P450 2C19-genotyped subgroups: integration of the TRITON-TIMI 38 trial data. J Thromb Haemost. 2010;8:1678–1684. doi: 10.1111/j.1538-7836.2010.03923.x. [DOI] [PubMed] [Google Scholar]
- 22.Floyd JS, Serebruany VL. Prasugrel as a potential cancer promoter: review of the unpublished data. Arch Intern Med. 2010;170:1078–1080. doi: 10.1001/archinternmed.2010.154. [DOI] [PubMed] [Google Scholar]
- 23.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin Pharmacol Ther. 89:474–477. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barker CM, Murray SS, Teirstein PS, Kandzari DE, Topol EJ, Price MJ. Pilot study of the antiplatelet effect of increased clopidogrel maintenance dosing and its relationship to CYP2C19 genotype in patients with high on-treatment reactivity. JACC Cardiovasc Interv. 2010;3:1001–1007. doi: 10.1016/j.jcin.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Bonello L, et al. Clopidogrel loading dose adjustment according to platelet reactivity monitoring in patients carrying the 2C19 2* loss of function polymorphism. J Am Coll Cardiol. 2010;56:1630–1636. doi: 10.1016/j.jacc.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Gladding P, et al. Pharmacogenetic testing for clopidogrel using the rapid INFINITI analyzer: a dose-escalation study. JACC Cardiovasc Interv. 2009;2:1095–1101. doi: 10.1016/j.jcin.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 27.Sibbing D, et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121:512–518. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 28.Sangkuhl K, Klein TE, Altman RB. Clopidogrel pathway. Pharmacogenet Genomics. 2010;20:463–465. doi: 10.1097/FPC.0b013e3283385420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein TE, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J. 2001;1:167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]