Abstract
The VH5 human antibody gene was analyzed using a computer program (mfg) which simulates transcription, to better understand transcription-driven mutagenesis events that occur during “phase 1” of somatic hypermutation. Results show that the great majority of mutations in the non-transcribed strand occur within loops of two predicted high-stability stem-loop structures, termed SLS 14.9 and SLS 13.9. In fact, 89% of the 2,505 mutations reported are within the encoded complementarity-determining region (CDR) and occur in loops of these high-stability structures. In vitro studies were also done and verified the existence of SLS 14.9. Following the formation of SLS 14.9 and SLS 13.9, a sustained period of transcriptional activity occurs within a window size of 60-70 nucleotides. During this period, the stability of these two SLSs does not change, and may provide the substrate for base exchanges and mutagenesis. The data suggest that many mutable bases are exposed simultaneously at pause sites, allowing for coordinated mutagenesis.
1. Introduction
Initiated by antigen challenge and B cell activation, somatic hypermutation (SHM) is the major source of diversification in antibody variable region (V) gene sequences, due to an approximately million-fold increase in mutation frequency (Rajewsky, 1996). SHM introduces mutations in “hotspots” of single-stranded DNA ~150 bps downstream of the V gene promoter, thereby altering the specificity of the encoded antibody. Within germinal centers of secondary lymphoid tissue, B cells which produce antibodies with higher affinity for antigen are selected, whereas B cells that generate low-affinity antibodies are deleted during the process of affinity maturation. Current research in the field is focused upon the discovery that activation-induced (cytidine) deaminase (AID) is required for various aspects of class-switch recombination, gene conversion and enzyme diversification during SHM (reviewed in Delker et al., 2009). The inter-dependence of SHM, gene conversion and class switch recombination is not yet clear, and recent evidence indicates that formation of the ssDNA substrate for SHM requires transcription but not AID (Shen et al., 2009). A central characteristic of SHM that has not yet been clarified is the mechanism underlying the direct correlation between rates of transcription and mutation, resulting in the million-fold increase in mutation frequency (Bachl et al., 2001; Fukita et al., 1998). Following the mutation of C-to-U by AID, a cascade of natural DNA repair enzymes is invoked and subverted as the theoretical “cause” of random point mutations in V genes during SHM. However, the role of repair enzymes is to chemically reverse nucleotide damage by oxidation, hydrolysis, or deamination. These enzymes do not create or insert new mutations. Moreover, the rate at which repair enzymes function is determined by the rate of mutation, i.e., by the rate of availability of their substrate. Repair enzymes are not required for SHM, and their absence in repair-deficient mice has little effect on the frequency of SHM (Li et al., 2006); therefore, they are apparently not rate-limiting for mutagenesis. Although AID activity has been demonstrated in vitro, the function and substrate of AID in vivo is under discussion (Chua et al., 2002; Franklin and Blanden, 2006; Rogozin et al., 2004), and data indicate that the activity of AID in vivo is restricted drastically as compared to cell-free assays (Shen et al., 2006). Thus, in some respects the current model of SHM may reflect in vitro rather than in vivo conditions.
Our initial dynamic model of SHM (Wright et al., 2008a, 2008b) focused on a 65-bp region and on mutation frequencies in CDR2 of the variable region heavy chain gene, VH5. A large database for this gene (Zheng et al., 2005) documents mutations which are primarily clustered in ssDNA segments of predicted Stem-Loop Structures (SLSs). The most stable and mutable of these is SLS 14.9, named for its −ΔG value. In strong support of a central role for SLS 14.9 during SHM is the fact that ex vivo experimental data of Ronai et al. (2007) were actually predicted by our in silico model of mutagenesis during SHM in VH5 (Wright et al., 2008a). The present model of SHM has now been expanded to include CDR1, thus accounting for all seven mutable sites in ssDNA loops in the variable region. Of the 2,505 total reported mutations in the CDRs of this gene, 89% are accounted for by this model (Fig. 1). All seven of the mutable sites are in loops of high-stability SLSs, and in vitro evidence, in this study, has now substantiated the existence of SLS 14.9.
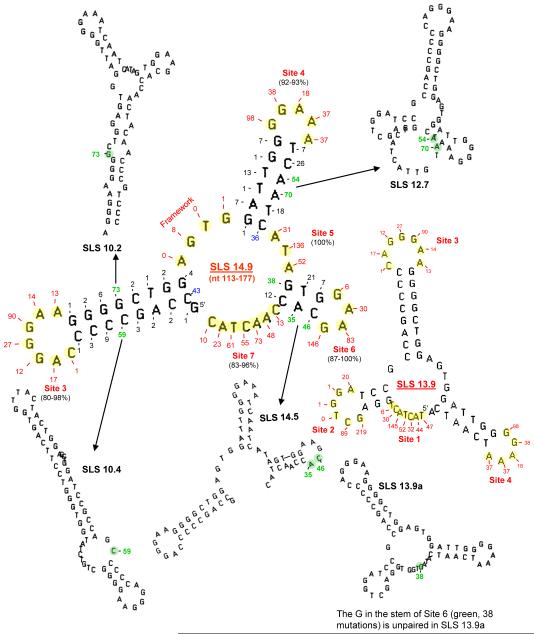
Fig. 1.
High-stability SLSs are shown in which 89% of the 2,505 mutations (Zheng et al. 2005) in CDR1 and CDR2 of VH5 are located in unpaired bases. The majority of unpaired mutable bases occur in Sites 1-7 (yellow circles) which are unpaired in the predominant 65-nt structures SLS 14.9 and SLS 13.9. Mutable bases in green are unpaired in slightly lower stability structures. The mutable G in SLS 13.9a is an unpaired C (38 mutations) in SLS 13.2 of the transcribed strand (see Fig. 6, Site 5). Only two mutable bases (shown in blue) are unpaired in low stability structures (i.e. less stable than ΔG −6.0, not shown).
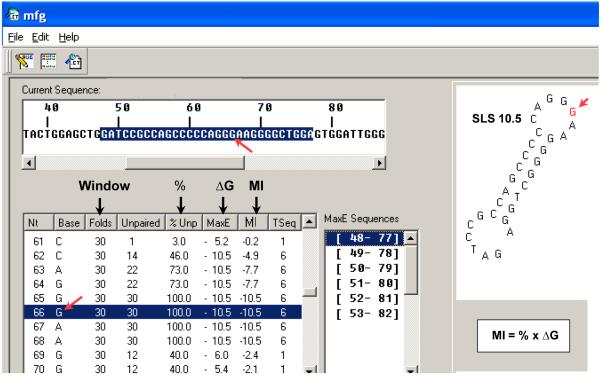
The proposed mechanism of transcription-driven mutagenesis during SHM involves selection of the most mutable bases in the most stable secondary structures associated with increasing levels of transcription, implicating a mechanism by which SHM may have evolved. In order to simulate in vivo conditions in response to increased rates of transcription, these studies have used a computer algorithm, mfg (Wright et al., 2003), that has demonstrated predictive value in both prokaryotes (Burkala et al., 2007; Reimers et al., 2004; Schmidt et al., 2006) and eukaryotes (Kim et al., 2010; Pereira et al., 2008; Wright et al., 2008a, 2008b), thereby passing the primary and essential test required of any model. The mfg computer program is used to mimic the generation of secondary structures during transcription. Fig. 2 is an example of computer output from the analysis of a given gene sequence. In this analysis, the sequence was folded in 30 nt segments or “windows”, revealing that the G at nt 66 is unpaired in 100 percent (%) of its total folds during simulated transcription of the gene segment (highlighted). Mfg interfaces with the mfold program (Markham and Zuker, 2005) which folds single-stranded segments of a specified length for any given sequence. A sequence and window size are initially chosen for an mfg analysis and mfold is used to fold the relevant successive segments. For each successive SLS in the sequence, mfg reports the stability (ΔG) of the most stable secondary structure, among many, in which that base is unpaired, and also reports the percent of total folds in which it is unpaired. The Mutability Index (MI) of each unpaired base is the product of these two variables. In Fig. 2, mfg simulates transcription in vivo by showing progressive “snap shot” views of the most stable 30 nt SLSs (e.g., SLS 10.5) in which each base is unpaired within the sequence analyzed, similar to RNA polymerase progression along the template strand during transcription. The user-determined window size relates to the rate of transcription and the amount of ssDNA available for folding. These simulations involve a running competition for shared nts between successive, inter-converting secondary structures of different stabilities (Wright et al., 2008a, 2008b). In general, the highest stability SLSs form repeatedly relative to those of lower stability. In order to find the most likely SLS(s) for coordinating mutagenesis during SHM, a series of window sizes was examined in VH5 and in two other VH genes, VH94 and VH186.2 (Wright et al., 2008 b), and the number of repeats recorded (Table 1). In all three genes, the data suggest that at a window size of 60-nt, repeats of a high stability structure are first observed. In the VH5 variable region the optimum SLS has a stability (−ΔG) of 14.9 and SLSs are formed at window sizes between ~60 and 70 nts.
Fig. 2.
An example of mfg computer output. A 30-nt segment of the sequence analyzed (bolded) is shown. The G (red arrow) at mfg nt 66 was selected as the most stable SLS (ΔG = −10.5) in which that base is unpaired (shown to the right). The output also provides the window size, percent unpaired of total SLSs in which the G nt is unpaired, the ΔG of the SLS, and the Mutability Index (MI) which is the product of % unpaired and ΔG.
Table 1.
Maximum number of SLS repeats at different window sizes as predicted by mfg during the plateau period.
| VH5a | VH94a | VH186.2a | ||||
|---|---|---|---|---|---|---|
| Window Size (nt) |
ΔGb | Number of repeats |
ΔG | Number of repeats |
ΔG | Number of repeats |
| 30 | −1.3 | 20 | −10.8 | 21 | −6.4 | 29 |
| 35 | −10.5 | 29 | −11.6 | 9 | −7.5 | 18 |
| 40 | −11.2 | 17 | −13.6 | 15 | −7.5 | 28 |
| 45 | −12.9 | 17 | −13.6 | 26 | −9.1 | 26 |
| 50 | −13.6 | 26 | −14.4 | 18 | −10.4 | 26 |
| 55 | −13.6 | 36 | −16.2 | 25 | −11.5 | 31 |
|
| ||||||
| 60 | −14.9 | 27 | −16.4 | 23 | −12.8 | 25 |
| 65 | −14.9 | 37 | −17.0 | 26 | −12.8 | 29 |
| 70 | −14.9 | 47 | −17.3 | 27 | −12.8 | 33 |
|
| ||||||
| 75 | −16.0 | 38 | −17.0 | 26 | −14.2 | 21 |
| 80 | −16.5 | 39 | −18.4 | 25 | −14.1 | 21 |
| 85 | −16.9 | 40 | −18.9 | 28 | −15.4 | 43 |
| 90 | −17.8 | 41 | −19.0 | 27 | −15.4 | 18 |
from supplement database of Zheng et al. 2005
ΔG of the SLS
2. Materials and methods
2.1 The mfg program
The program output is shown (Fig. 2) and directions for downloading and using mfg are freely available at http://www.dbs.umt.edu/research_labs/wrightlab/upload/mfg.html. Mfg is an open source desktop application that runs on Windows, Linux and Mac operating systems.
2.2 Database sorting
Data in Tables 1 and 2 pertaining to VH genes were obtained from the supplementary dataset of Zheng et al. (2005) which contains 28,307 mutations in different VH genes and referenced sequences. We examined mutations in three genes: VH5 (GenBank accession numbers X92278 and M99684), VH94 (GenBank accession number L10094), and VH186.2 (GenBank accession number L10088). The data were derived by using Excel to sort columns (See Supplementary Method).
Table 2.
Frequency and types of G and C mutations in ssDNA and antibody genes.
| G-to-A Intrinsic |
G-to-T | G-to-C | C-to-T Intrinsic |
C-to-A | C-to-G | |
|---|---|---|---|---|---|---|
| A. Thermodynamic characteristics of nucleosides leading to mutation in single-stranded DNA | ||||||
| Type of Base Substitution |
Transitionsa | Depurination (G → T) (A → T) |
Depurination (G → C) (A → C) |
Deamination (C →U→T) |
Depyrimidation (C → A) (T → A) |
Depyrimidation (C → G) (T → G) |
| Mutations/ day/ 1010 bases/cell (% of total) |
(~95%) | (na) | (na) | (95.6%) | (~2.2%) | (~2.2%) |
| B. Silent mutations in framework regions of VH genes b | ||||||
| Number of Mutations (% of total) |
1,156 (67.5%) |
274 (16.0%) |
283 (16.5%) |
1,066 (80.4%) |
103 (7.8%) |
156 (11.8%) |
| C. Missense mutations in germline light chain genes c | ||||||
| Number of Mutations (% of total) |
48 (72.7%) |
5 (7.6%) |
13 (19.7%) |
27 (81.8%) |
5 (15.2%) |
1 (3.0%) |
| D. Missense mutations in CDRs of VH5 gene b | ||||||
| Number of Mutations (% of total) |
440 (45.3%) |
85 (8.7%) |
447 (46.0%) |
149 (36.2%) |
57 (13.9%) |
205 (49.9%) |
About 5,000 purine bases turn over each day due to hydrolytic depurination, with G usually being replaced by A because of its size. (Lindahl, 1993)
Data were obtained from the VH5, VH94, and VH186.2 genes (see Materials and methods).
Missense mutations in germline Gs and Cs in VkOx1 light chain genes (Phung et al., 1998). These values are similar in Msh2 −/− and Msh2 +/+ (Shen et al., 2006).
2.3 Construction of VH5-containing plasmids for generation of ssDNA
A 570-bp portion of the VH5 gene was amplified from chromosomal DNA of the pre-B cell line JM1 by PCR using primers (below) V1F1 and Set2R (Integrated DNA Technologies, Coralville, IA) and called VH5. Two smaller fragments were also amplified; a 370-bp fragment called VH5.1 (primers: 5′VH5hinatg and 3′VH5xma), and a 170-bp fragment called VH5.2 (primers: Set1F, and V1R2). These fragments were independently cloned into the high-copy number vector pCR2.1-TOPO (Invitrogen) in both orientations. The resulting six constructs were (individually) transformed into the XL1Blue MRF E. coli strain (Stratagene, LaJolla, CA), and selected on LB plates containing ampicillin (100 μg/ml). Each E. coli strain containing one of the plasmids was grown to an OD600 of 0.05 and ssDNA was generated by infecting with helper phage M13K07 (NEB, Ipswich, MA) or R408 (Stratagene) to capitalize on the f1 origin on the plasmid, according to manufacturer’s instructions. Phage DNA was harvested using a Qiagen (Valencia, CA) midi protocol modified for this purpose, including an incubation in a solution composed of 30% PEG and 3 M NaCl for 1 h at 4°C, followed by lysis of the phage in a solution of 1% Triton X-100, 500 mM guanidine HCl and 10 mM MOPS (pH 8.0) at 80°C before applying to the column. After harvest, the ssDNA was stored for up to two weeks at 4°C.
2.4 Polymerase pausing in VH5
Appropriate primers were hybridized to the complementary ssDNA (below). Radioactive end-labeling of primers was conducted by using T4 polynucleotide kinase (NEB) and [32Pγ]-ATP (MP Biomedicals, Solon, OH) as per standard protocols. The primer XhoMCS2rev was hybridized to single-stranded templates by heating for 2 min at 75°C and then cooling to room temperature. Following the protocol of Weaver and DePamphilis (1984), polymerase buffer and dNTP’s (NEB) were added to the hybridized templates at 37°C along with T7 DNA polymerase. The reaction was stopped by adding loading dye at various time points and immediately cooling to create labeled primer extension fragments. Polymerase products were of a size that corresponds to the length of DNA from the primer start site to the pause sites. The end-labeled products were visualized as discrete bands when separated on a large-format 7% polyacrylamide gel containing urea (7 M final concentration). Sequence ladders (Sequenase 2.0, USB/Affymetrix, Santa Clara, CA) were generated using the same template and primers and were labeled with [32Pα]-ATP (MP Biomedicals). Gels were run for 2-4 h at 1400 mV and then exposed to x-ray film overnight at −80°C with intensifier screens. Pause sites correspond to bands of the sequencing ladder which are complementary. Each pausing reaction result was confirmed by repeating the procedure at least three times using both the transcribed and the non-transcribed strands, and using at least two pools of ssDNA.
2.5 Primers
VH5: V1F1 (ATCCATACAAAGGCACCACC)
Set2R (GTGCCTCTCGCACAGTAATACACA)
VH5.1: 3′VH5xma (11 bases overlapSet2R) (GATCCCGGGCCGTGCCTCTCGCACAG)
5′VH5hinatg (CTCAAGCTTGCCGCCATGTCCCAGGGCTCACTGTGGGTTTC)
VH5.2: Set1F (TCTATGGTGGGTCCTTCAGTGGTT)
V1R2 (AGGGAGAACTGGTTCTTGGA)
Hybridization: XhoMCS2rev (CTCGAGCGGCCGCCAGTG)
2.6 Construction of VH5-containing plasmids for generation of dsDNA for S1 endonuclease cleavage assays
Two oligomers were synthesized (IDT): a 65-mer, called SLS 14.9 (GCCAGCCCCCAGGGAAGG GGCTGGAGTGGATTGGGGAAATCAATCATAGTGGAAGCACCAACTAC), corresponding to the immediate upstream region of CDR2 in the VH5 segment and the 5′ end of CDR2, and an 81-mer, called SLS 14.5 (CCGCCAGCCCCCAGGGAAGGGGCTGGAGTGGATTGGGGAAATCAATCATA GTGGAAGCACCAACTACAACCCGTCCCTCAA), corresponding to the 3′ end of CDR1 and the 5′ end of CDR2. The 5′ ends of both oligomers were end-labeled with [32Pα]-ATP using T4 poly-nucleotide kinase (NEB) and digested separately with endonuclease CviKI-1 (NEB) and T4 endonuclease VII (MCLab, S. San Francisco, CA) for 1 hr at 37°C, according to manufacturer’s instructions. Gel loading dye (NEB) was immediately added and the mixture placed on either a 7% or 20% denaturing polyacrylamide gel to separate fragments. The gel was placed at −80°C overnight with x-ray film to visualize bands. Limited Maxam-Gilbert (1980) and S1 endonuclease ladders (Wyatt et al., 1990) were generated as per published protocols.
3. Results
3.1 The relationships between window size, SLS stability, transcription and mutation frequency
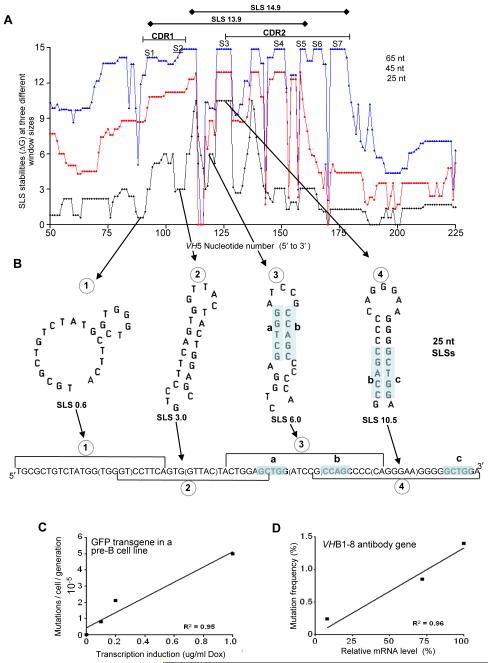
Two phases of SHM have been proposed. The first phase, called “phase 1”, is initiated at the beginning of the CDR and includes an unprecedented increase in the rates of transcription and mutation, and is targeted at Cs and Gs (reviewed in Di Noia and Neuberger, 2007). As nucleotide number increases (Fig. 3A), due to the increase in transcription frequency (Bachl et al., 2001; Fukita et al., 1998), the size and stability (ΔG) of SLSs at all window sizes (25, 45 and 65 nts) increase, and mutable Sites S1 through S7 appear successively. Mutable Sites correspond to high stability peaks. There is a strong correlation between mutation and transcription, for example, in the pre-B cell line 18-81 (Fig. 3C) and in the VHB1-8 antibody gene (Fig. 3D) using promoters of different strengths in mice. Moreover, the SLS stability at a single window size also increases (Fig. 3B). This increase involves a completely different mechanism, as illustrated by the four (encircled, 1-4) successive rearrangements in the 25-nt SLSs. These SLSs formed by segments of ssDNA have apparently been selected during evolution for their increases in stability (17.5 fold), due only to an increased ratio of double-stranded to single-stranded DNA. The blue tint areas in SLSs 6.0 and 10.5 (Fig. 3B) show that the same segment (b) is used in two SLSs; the segments involved are also bracketed in the sequence. These successive rearrangements result in the first appearance of the strongest stem in VH5 (at Site 3) which dominates in the formation of and perhaps the evolution of SLSs 14.9 and 13.9 (see Figs. 4 and 5).
Fig. 3.
The stability and rearrangements in SLSs at different window sizes during phase 1 of SHM. (A) SLS stabilities (ΔG) using three different window sizes (65 nt, blue; 45 nt, red; and 25 nt, black) are plotted against nucleotide number (presumably) during phase 1 of SHM in VH5. Seven mutable sites are shown above peaks and abbreviated S1-S7. (B) SLSs formed by four successive segments of 25 nts (1 to 4, encircled) showing increasing stability primarily due to increases in paired bases. The sequence segment below indicates corresponding segments (bracketed) that fold to form each structure. In structure 3, a short sequence in blue tint (a) forms one portion of a stem, and a different segment of the sequence (b tinted blue) forms the other half of the stem. In structure 4, sequence b pairs with a different segment (c) to create a portion of a more stable stem. Note that the most stable stem at S3 (A) first forms in this 25 nt structure (SLS 10.5) and dominates thereafter as the most stable mutable Site at all window sizes (see Fig. 5). (C) and (D) Correlations between mutation rates and transcription in a GFP transgene in a human pre-B cell line (18-81) due to the activation of transcription by doxycycline (dox) (Bachl et al., 2001), and in the VHB1-8 antibody gene (Fukita et al., 1998). Linear regression values were calculated in Excel. R2 values are statistically significant when P ≤ 0.05. For C and D, n values are 4, and 3, respectively.
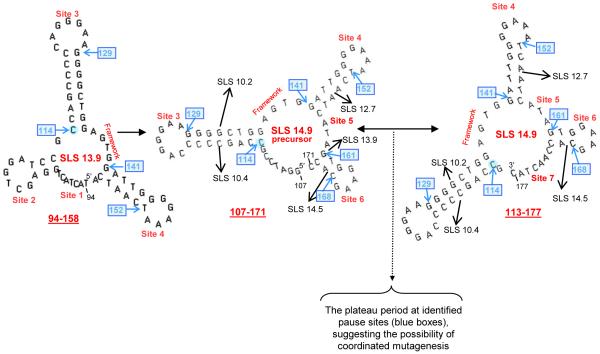
Fig 4.
Pausing and backtracking between the SLS 14.9 precursor and SLS 14.9 during the plateau period. The three dominant 65-nt SLSs containing the majority of mutable bases are SLS 13.9, SLS 14.9 precursor, and SLS 14.9. The interaction between SLS 14.9 precursor and SLS 14.9 is indicated by the double-ended black arrow; the black dotted arrow and bracket show the area of pausing and backtracking. Pause sites are indicated by blue boxes and were predicted by previous studies (Wright et al., 2008a and in Supplementary Figs. 1-3). Mutable Sites 1 to 7 are shown in red. Black arrows identify other SLSs (Fig. 1) in which bases that are paired here are unpaired and mutable.
Fig. 5.
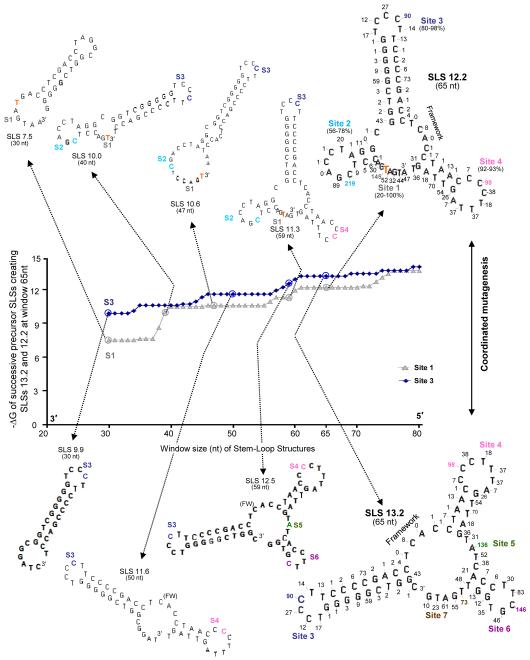
Selected high-stability SLSs in the non-transcribed strand of VH5 as window size increases from 30 to 80 nts. (A) In the plot of −ΔG versus window size solid blue diamonds indicate the stability (−ΔG) of successive SLSs leading to SLS 14.9 (lower series of SLSs) as window size increases from 30 nt to 80 nt. During the plateau period, between window sizes of 60 to 70 nts, coordinated mutagenesis and backtracking may occur (Fig. 4). Grey triangles indicate the stability (−ΔG) of successive SLSs leading to SLS 13.9 (upper series of SLSs). The predicted 30 nt precursor of Site 1 is shown (SLS 8.7), in which the most mutable base (A, orange), has 145 mutations. Blue diamonds indicate the stability (−ΔG) of successive SLSs leading to SLS 14.9 (lower series of SLSs). Each Site and the number of mutations in the two largest SLSs are color-coordinated. (B) SLSs at Site 1 for SLS 13.9 (a, b) and (C) Site 7 (c, d) for SLS 14.9. Note that during the plateau period, although mutable bases are added (small arrows), the stabilities of SLS 14.9 and SLS 13.9 do not change.
Fig. 4 shows the three key, high-stability SLSs (SLS 13.9, SLS 14.9 precursor, and SLS 14.9) which now account for 89% of the mutations in the CDR (Fig. 1 and Wright et al., 2008a). Interestingly, analyses of the these high-stability structures support the possibility that pausing (or “backtracking”) and coordinated mutagenesis may culminate during a plateau period, in which SLSs do not change in stability over several window sizes, although they can continue to acquire new mutable bases (shown in Fig. 5). Note that bases are “lost” from the 5′ end of 14.9 precursor, while a comparable number of bases are added to the 3′ end of SLS 14.9 (Fig 4). Backtracking in mfg at paired bases is the consequence of two essential and realistic assumptions made in the program: First, each fold must be initiated with an unpaired base and second, the most stable SLSs will dominate the folding pathway during transcription, i.e., each unpaired base must initiate, or remain in, the fold of the most stable SLS in which it is unpaired. The existence of pause sites at the base of stems (boxed in blue) has now been verified by in vitro analyses (see section 3.2).
Fig. 5A shows mfg-predicted increases in SLS size and stability in the non-transcribed strand of VH5 as transcription and window size increase during the formation of SLS 13.9 (first series of SLSs) and SLS 14.9 (second series of SLSs). These analyses are based on two assumptions: First, bases with a high mutation frequency will increase the complementarity of antibody for antigen during affinity maturation and be selected most frequently, while those of lower affinity will be deleted; and second, the most stable SLS harboring the mutable bases at each window size will also be selected most frequently and are the precursors of 65-nt SLS 14.9 and SLS 13.9. For example, the secondary structures in the second series of SLSs (Fig. 5A, denoted by the blue diamonds), initiated with SLS 10.5 and ending with SLS 14.9, were identified by the following procedures: An mfg window scan of the non-transcribed strand was used to identify the most mutable base at each Site and the highest ΔG SLSs at each window size, from 30-80 nts, one nt at a time. The most mutable base at Site 3 in SLS 14.9 is the G (blue) with 90 mutations and therefore SLS 10.5 was chosen as the first most stable structure found by plotting ΔG versus window size at 30 nts, while lower stability SLSs were (presumably) discarded and not identified. We next show a 44-nt SLS with Site 4 (pink); this site contains the most mutable base (G), with 98 mutations. For Site 5 (green) the T in this 50-nt SLS has 136 mutations (SLS 13.6). This SLS now has three of the most mutable Sites in SLS 14.9. As seen in SLS 14.9, the most mutable base at Site 6 (purple) is the G with 146 mutations and at Site 7 (brown), the A with 73 mutations is the most mutable. In the upper series of SLSs (grey triangles) similar analyses show selected SLSs that form as window sizes increase, revealing potential precursors of SLS 13.9. Fig. 5B and C show two successive SLSs, in SLS13.9 (a and b) and SLS14.9 (c and d), adding one base at a time during the plateau period (nt windows of ~60-70), in which these SLS do not change in stability while mutable bases are added to Site 1 of SLSs 13.9 and to Site 7 in SLS 14.9. Coordinated mutagenesis (Figs. 4 and 5) could occur during this period (Wright et al., 2008a).
Fig. 6 shows similar analyses of the transcribed strand, in which the SLSs are somewhat less stable than those in the non-transcribed strand. However, their evolution as transcription levels increase to form SLSs 13.2 and 12.2, is similar to that observed in the non-transcribed strand, and their SLSs provide some unpaired bases in which complementary bases are paired in SLSs of the non-transcribed strand (see Fig. 1). The upper set of SLSs (grey triangles) shows a series of predicted potential precursors of SLS 12.2, and the lower set (blue diamonds) shows successive precursor SLSs containing Site 3 (blue in 30 nt SLS 9.9) to Site 6 (purple in SLS 12.5) that represent precursors of SLS 13.2 (lower set of SLSs).
Fig. 6.
The mfg-predicted evolution of SLS 13.2 and SLS 12.2 initiated from the 3′ end of the transcribed strand as window size increases from 30 to 80 nts. Solid blue diamonds indicate the stability (−ΔG) of successive SLSs leading to SLS 13.2 (lower series of SLSs) as window size increases. A predicted 30-nt precursor of Site 3, SLS 9.9, has 90 mutations in the most mutable C base for that Site (dark blue). Grey triangles indicate the stability (−ΔG) of successive SLSs leading to SLS 12.2 (upper series of SLSs) as window size increases from 30 nt to 80 nt. The predicted 30 nt precursor of Site 1 shows a T (orange) as the most mutable base, and subsequent precursors follow as in Fig. 4. Although SLS 13.2 is of lower stability than the most stable SLSs in the non-transcribed strand (SLS 14.9), it accounts for the exposure of three mutable bases at Sites 5 and 6 that are paired in SLS 14.9 in the non-transcribed strand (Fig. 5A), implicating a role for the transcribed strand in mutagenesis (see section 4.2). Note that Sites 3 and 4 are seen in both 65 nt structure SLS 12.2 and SLS 13.2 and that they are not exact complements of SLS 13.9 and 14.9, respectively.
3.2 In vitro evidence for the existence of mfg-predicted secondary structures
Evidence thus far presented describes a mechanism of mutagenesis during SHM which depends upon a dominant secondary structure, SLS 14.9. In vitro analyses have confirmed that this secondary structure exists (Fig. 7).
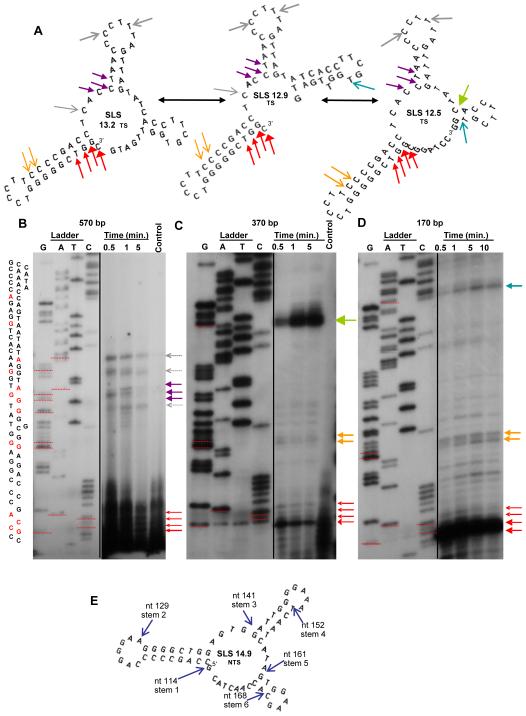
Fig. 7.
Polymerase pausing analyses of three different size segments of VH5. (A) Mfg-predicted structures are shown with pause sites occurring primarily at or in stems (colored arrows excluding grey). Color-coordinated arrows in the three SLSs correspond to locations of polymerase pausing on gels (below). Black double-headed arrows indicate dynamic interactions between the three structures. (B) Polymerase pause sites (bands) shown at 0.5, 1 and 5 min. for (B) a 570-bp and (C) a 370-bp fragment of VH5 respectively, with colored arrows corresponding to the initiation of predicted stems. (D) Pausing analysis for a 170 bp fragment of VH5. Weight of arrows corresponds to band intensity. (E) Previously predicted pause sites (Wright et al., 2008a) (blue arrows), also indicated in Fig. 4.
Three predicted structures are shown (Fig. 7A) in the transcribed strand (consistent with the direction of polymerase progression) with the locations of experimentally determined polymerase pauses (groups of colored arrows). Plasmids with three different inserts (B, 570 bp; C, 370 bp, and D, 170 bp) containing VH5 gene segments which were hybridized with a primer and allowed to fold after heating. T7 DNA polymerase was then added and allowed to proceed along the sequence for 0.5, 1, 5 or 10 minutes. Pausing of the polymerase was evidenced by bands on a polyacrylamide gel (Fig. 7B-D, arrows; colors correspond to locations on structures in Fig. 7A). The transcribed strand orientation of the full-length fragment (570-bp) (Fig. 7B) had pauses at sites consistent with the existence of the first two stems of the mfg-predicted structure SLS14.9 (Fig. 7E). The two smaller cloned fragments (Fig. 7C and Fig. 7D, 370 and 170 bp respectively) had pauses consistent with the first and third predicted stem. Additionally, sequence-based pauses (at G and C quartets) (Mirkin and Mirkin, 2007) were also seen. The size of each arrow head is relative to the band intensity in the gel. Black double-headed arrows indicate that the predicted structures are in dynamic equilibrium. These pauses are consistent with those previously predicted in SLS 14.9 (Fig. 7E) by backtracking analyses (Wright et al., 2008a). The process for identifying pause sites was repeated, with the DNA in the opposite orientation, using appropriate 5′ primers. Similar results were obtained (data not shown). To test the possibility that some helper phage DNA was replicated when isolating ssDNA, all primers were also annealed to either M13K07 or R408 phage DNA alone. No polymerase products were observed with phage alone for any of the primers used.
To further verify mfg-predicted structures SLS 14.9 and SLS 14.5, T4 endonuclease VII (which cleaves dsDNA) was also used. Synthesized oligonucleotide fragments of 65 nts (Supplementary Fig. 1) and 81 nts (Supplementary Fig. 2) were separated and compared to a base ladder (Maxam and Gilbert, 1980). Multiple cleavage sites were observed in each of the predicted stems, and loop structures atop all three stems were not cleaved (dotted lines on gels, and red arrows). In addition, the restriction enzyme CviKI-I was used to test for the double-stranded substrate RGCY formed by one of the predicted stems. The gels show cleavage (Supplementary Figs. 1 and 2) with CviKI-I at the predicted double stranded site (blue underlines). Supplementary Fig. 2 shows a predicted structure, designated SLS 14.5, associated with a longer 81-bp fragment that extends beyond SLS 14.9. Again, stems are cleaved in multiple locations, and loops are uncut. CviKI-I cut the predicted stem structure of SLS 14.5, as observed in SLS 14.9. Thus, the mfg predicted structures are in agreement with enzymatic cleavage analyses. Supplementary Fig. 3 shows the experimental pausing data for the sequence involved in the formation of SLS 14.9 as well as in the mfold predicted SLS 14.5. Only the mfold structure (which is less stable than SLS 14.9 and therefore not chosen by mfg) can account for both experimental pause sites, but the formation of both structures is in agreement with the mutational pattern observed in Sites 6 and 7.
4. Discussion
4.1 History of evidence that intrinsically mutable Gs and Cs are exposed in SLS loops
Since the early 1980s (Ripley and Glickman, 1983), evidence has indicated that secondary structures are associated with endogenous “background” mutations (reviewed in Wright, 2002, 2004). Hydrolytic, endogenous mutations (primarily C-to-T and G-to-A) occur at significant frequencies under physiological conditions (Amosova et al., 2006; Cooper and Krawczak, 1990; Frederico et al., 1990; Lindahl 1993; Lindahl and Nyberg, 1974; Pereira et al. 2008; Shen et al., 1994; Singer and Kusmierek, 1982; Skandalis et al., 1994; Smith, 1992; Todd and Glickman, 1982). Background maintenance levels of replication, transcription, and supercoiling allow dsDNA strands to separate sufficiently to expose segments of ssDNA which form secondary structures with loops essential for base mutability. In dsDNA, for example, N3 of cytosine is H-bonded to the N1 of guanine, and ssDNA (via transcription and/or negative supercoiling) is required for protonation at N3 and deamination to uracil. Thus, it is widely recognized that endogenous mutations of C and G are the result of their intrinsic thermodynamic instability, suggesting that ssDNA availability (during transcription) may be rate-limiting for mutation frequency under the steady-state conditions existing in vivo for SHM (Wright et al., 2008a, 2008b). This conclusion is consistent with our observation that ~89 % of a total of 2,505 mutations in the variable region of VH5 are located in loops of high stability SLSs (Fig. 1). The current study identifies successive variables that contribute to the million-fold increase in mutation frequency, especially during phase 1 of SHM, predicted to follow B cell activation (Di Noia and Neuberger 2007). At small window sizes associated with background levels of transcription just 5′ of CDR1 (Fig. 3B), 25 nt SLSs stabilities have a ΔG < −1. During the proposed initiation of phase 1 and increasing levels of transcription, sequence segment rearrangements in 25 nt structures have apparently evolved (been selected) to form higher stability SLSs that increase by as much as 17-fold, due to sequence rearrangements increasing the paired versus unpaired nt ratio. Thus window size per se is correlated with transcription levels, SLS stability, exposure of mutable bases and mutation frequency.
The overall thermodynamic characteristics of nts in ssDNA underlying the majority of background or “endogenous” mutations are shown in Table 2A. The ssDNA in which unpaired bases are exposed and mutable in vivo is presumably due to maintenance-level metabolic activities such as transcription and replication. The hydrolytic deamination of 5-methyl C and C occur ~300 times/day/human cell, which is more than sufficient to explain the observed endogenous mutation frequency (Frederico et al., 1990; Shen et al., 1994). About 5,000 purine bases turn over each day due to hydrolytic depurination, with G being replaced by A at those apurinic sites because of its size (Lindahl 1993). In the VH genes, G-to-A and C- to-T transitions in silent mutations of framework regions (Table 2B) and of missense mutations in germline light chain genes (Table 2C) predominate, presumably due to base exposure during transcription and to the intrinsic chemical instability of ssDNA. In contrast, missense mutations in CDRs of the VH5 gene (Table 2D) have apparently been selected as the fittest for binding antigen to antibody. Silent mutations (Table 2B) are perhaps the most compelling evidence for a common mechanism of mutability in unpaired bases, where the outcome of a mutation does not alter the encoded amino acid and is not selected, thus reflecting only intrinsic base instabilities.
Stems in stable SLSs are known to cause pauses that temporarily block transcription (Bagga et al., 1990; Peck and Wang, 1985), and direct correlations have been found between pause strength and the negative superhelical densities of the templates used (Krohn et al., 1992). Pausing at stems has been attributed to the slow melting of stems and associated with bursts of mRNA production and increased mutagenesis in vitro (Suo and Johnson, 1998; Weaver and DePamphilis, 1984). In mfg, the folding process results in pauses at paired bases in stems, as each successive structure must be initiated by an unpaired base. Thus, the extent of pausing correlates with stem stabilities, both in vitro and in silico.
4.2 The role of dynamic models in understanding the mechanism of mutagenesis in vivo
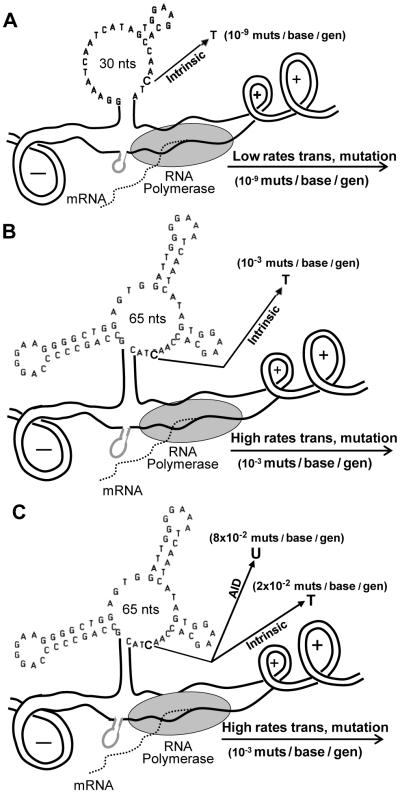
Kinetic models organize our data and serve to provide conceptual insights in realistic, dynamic terms related to the steady-state conditions of the living cell. Our model of mutagenesis revealed circumstances that were not intuitively obvious and made specific predictions that could be verified in the available data. Fig. 8 shows three successive dynamic models of events prior to (A), during (B), and immediately following (C) phase 1 of SHM, at peak levels of transcription and frequency of C mutations. At that point, AID activation could usurp (split) the fate of high frequency C mutations, initiating high frequency C-to-U mutations (at the expense of C-to-T mutations), resulting in high frequency enzyme diversification. These models elucidate mechanisms by which transcription-driven single-stranded segments of DNA are exposed and become mutable. The data presented also suggest mechanisms by which selection of the most mutable bases and of the most stable secondary structures during affinity maturation may underlie both the evolution and mechanism of SHM.
Fig. 8.
Three dynamic models of events before, during, and immediately after phase 1. (A) Circumstances resulting in background mutation frequencies of about 10−9 mutations / base/generation. (B) Consequences of a ~10,000-fold increase in transcription frequency during antigen challenge targeted primarily to Cs and Gs in ssDNA. (C) Predicted events occurring at the peak of phase 1 as a result of AID activation, resulting in a switch from high frequency C-to-T mutations to high frequency C-to-U mutations, resulting in enzyme diversification.
5. Conclusion
Mutagenesis in SHM is characterized by high levels of transcription and mutation, stable SLSs, and the exposure of intrinsically mutable bases in loops of these structures during transcription. As demonstrated by The Unity of Biochemistry (Kluyver, 1926 and see Wright, 2004), evolution has bestowed a fundamental unity of biochemical behavior on all forms of life: the same substrates, enzymes, regulatory mechanisms, biosynthetic and catabolic pathways. It is therefore not surprising that the mechanisms underlying mutagenesis in several systems, including the p53 tumor suppressor gene (Wright et al. 2011), are similar to those found in SHM.
Supplementary Material
Highlights.
The mfg computer program mimics the dynamics of transcription in vivo.
Intrinsically mutable Cs and Gs are rate-limiting for mutation frequency.
89% of identified mutable bases in VH5 are in loops of high stability DNA structures.
Intrinsic G and C mutation frequencies correlate with transcription frequency.
Acknowledgments
We thank Steve Lodmell for excellent advice regarding technical aspects of pausing and cleavage analysis. This work was supported by grants from the Stella Duncan Memorial Research Institute, and NIH grant R01CA099242.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary data associated with this article can be found in the online version.
References
- Amosova O, Coulter R, Fresco JR. Self-catalyzed site-specific depurination of guanine residues within gene sequences. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4392–4397. doi: 10.1073/pnas.0508499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachl J, Carlson C, Gray-Schopfer V, Dessing M, Olsson C. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J. Immunol. 2001;166:5051–7. doi: 10.4049/jimmunol.166.8.5051. [DOI] [PubMed] [Google Scholar]
- Bagga R, Ramesh N, Brahmachari SK. Supercoil-induced unusual DNA structures as transcriptional block. Nucleic Acids Res. 1990;18:3363–9. doi: 10.1093/nar/18.11.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkala E, Reimers JM, Schmidt KH, Davis N, Wei P, Wright BE. Secondary structures as predictors of mutation potential in the lacZ gene of Escherichia coli. Microbiology. 2007;153:2180–2189. doi: 10.1099/mic.0.2007/005470-0. [DOI] [PubMed] [Google Scholar]
- Chua KF, Alt FW, Manis JP. The function of AID in somatic mutation and class switch recombination: upstream or downstream of DNA breaks. J. Exp. Med. 2002;195:F37–41. doi: 10.1084/jem.20020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M. The mutational spectrum of single base-pair substitutions causing human genetic disease: patterns and predictions. Hum. Genet. 1990;85:55–74. doi: 10.1007/BF00276326. [DOI] [PubMed] [Google Scholar]
- Dayn A, Malkhosyan S, Mirkin SM. Transcriptionally driven cruciform formation in vivo. Nucleic Acids Res. 1992;20:5991–5997. doi: 10.1093/nar/20.22.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker R, Fuggmann SD, Papavasiliou N. A coming-of-age story: activation-induced cytidine deaminase turns 10. Nat. Immunol. 2009;10:1147–53. doi: 10.1038/ni.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Franklin A, Blanden RV. A/T-targeted somatic hypermutation: critique of the mainstream model. Trends Biochem. Sci. 2006;31:252–258. doi: 10.1016/j.tibs.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Frederico L, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- Fukita Y, Jacobs H, Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9:105–14. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- Kim H, Lee BS, Tomita M, Kanai A. Transcription-associated mutagenesis increases protein sequence diversity more effectively than does random mutagenesis in Escherichia coli. PLoS One. 2010;5:e10567. doi: 10.1371/journal.pone.0010567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluyver AJ. Die Einheit in der Biochemie in der Biochie. Chem. D. Zelle u Gew. 1926;13:134–190. [Google Scholar]
- Krasilnikov AS, Podtelezhnikov A, Vologodskii A, Mirkin SM. Large-scale effects of transcriptional DNA supercoiling in vivo. J. Mol. Biol. 1999;292:1149–1160. doi: 10.1006/jmbi.1999.3117. [DOI] [PubMed] [Google Scholar]
- Krohn M, Pardon B, Wagner R. Effects of template topology on RNA polymerase pausing during in vitro transcription of the Escherichia coli rrnB leader region. Mol. Microbiol. 1992;6:581–9. doi: 10.1111/j.1365-2958.1992.tb01504.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Peled JU, Zhao C, Svetlanov A, Ronai D, Cohen PE, Scharff MD. A role for Mlh3 in somatic hypermutation. DNA Repair (Amst.) 2006;5:675–682. doi: 10.1016/j.dnarep.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. In: Methods in Enzymology. Moldave K, editor. Vol. 65. Academic Press; New York: 1980. pp. 499–560. Nucleic Acids, Pt. I. [DOI] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck LJ, Wang JC. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985;4:129–37. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- Pereira F, Soares P, Carneiro J, Pereira L, Richards MB, Samuels DC, Amorim A. Evidence for variable selective pressures at a large secondary structure of the human mitochondrial DNA control region. Mol. Biol. Evol. 2008;25:2759–2770. doi: 10.1093/molbev/msn225. [DOI] [PubMed] [Google Scholar]
- Phung QH, Winter DB, Cranston A, Tarone RE, Bohr VA, Fishel R, Gearhart PJ. Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J. Exp. Med. 1998;187:1745–1751. doi: 10.1084/jem.187.11.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–8. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Reimers JM, Schmidt KH, Longacre A, Reschke DK, Wright BE. Increased transcription rates correlate with increased reversion rates in leuB and argH Escherichia coli auxotrophs. Microbiology. 2004;150:1457–1466. doi: 10.1099/mic.0.26954-0. [DOI] [PubMed] [Google Scholar]
- Ripley LS, Glickman BW. Unique self-complementarity of palindromic sequences provides DNA structural intermediates for mutation. Cold Spring Harb. Symp. Quant. Biol. 1983;47(Pt 2):851–861. doi: 10.1101/sqb.1983.047.01.097. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Diaz M. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 2004;172:3382–4. doi: 10.4049/jimmunol.172.6.3382. [DOI] [PubMed] [Google Scholar]
- Ronai D, Iglesias-Ussel MD, Fan M, Li Z, Martin A, Scharff MD. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J. Exp. Med. 2007;204:181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KH, Reimers JM, Wright BE. The effect of promoter strength, supercoiling and secondary structure on mutation rates in Escherichia coli. Mol. Microbiol. 2006;60:1251–1261. doi: 10.1111/j.1365-2958.2006.05166.x. [DOI] [PubMed] [Google Scholar]
- Shen HM, Poirier MG, Allen MJ, North J, Lal R, Widom J, Storb U. The activation-induced cytidine deaminase (AID) efficiently targets DNA in nucleosomes but only during transcription. J. Exp. Med. 2009;206:1057–71. doi: 10.1084/jem.20082678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Tanaka A, Bozek G, Nicolae D, Storb U. Somatic hypermutation and class switch recombination in Msh6(-/-)Ung(-/-) double-knockout mice. J. Immunol. 2006;177:5386–92. doi: 10.4049/jimmunol.177.8.5386. [DOI] [PubMed] [Google Scholar]
- Shen JC, Rideout WM, 3rd, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 1994;22:972–976. doi: 10.1093/nar/22.6.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B, Kusmierek JT. Chemical mutagenesis. Annu. Rev. Biochem. 1982;51:655–693. doi: 10.1146/annurev.bi.51.070182.003255. [DOI] [PubMed] [Google Scholar]
- Skandalis A, Ford BN, Glickman BW. Strand bias in mutation involving 5-methylcytosine deamination in the human hprt gene. Mutat. Res. 1994;314:21–26. doi: 10.1016/0921-8777(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Smith KC. Spontaneous mutagenesis: Experimental, genetic and other factors. Mutat. Res. 1992;277:139–162. doi: 10.1016/0165-1110(92)90002-q. [DOI] [PubMed] [Google Scholar]
- Suo Z, Johnson KA. DNA secondary structure effects on DNA synthesis catalyzed by HIV-1 reverse transcriptase. J. Biol. Chem. 1998;273:27259–67. doi: 10.1074/jbc.273.42.27259. [DOI] [PubMed] [Google Scholar]
- Todd PA, Glickman BW. Mutational specificity of UV light in Escherichia coli: indications for a role of DNA secondary structure. Proc. Natl. Acad. Sci. U. S. A. 1982;79:4123–4127. doi: 10.1073/pnas.79.13.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver DT, DePamphilis ML. The role of palindromic and non-palindromic sequences in arresting DNA synthesis in vitro and in vivo. J. Mol. Biol. 1984;180:961–986. doi: 10.1016/0022-2836(84)90266-3. [DOI] [PubMed] [Google Scholar]
- Wright BE. Mini Review, A biochemical mechanism for nonrandom mutations and evolution. J. Bacteriol. 2000;182:2993–3001. doi: 10.1128/jb.182.11.2993-3001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BE. Micro Review, Stress-directed adaptive mutations and evolution. Mol. Microbiol. 2004;52:643–650. doi: 10.1111/j.1365-2958.2004.04012.x. [DOI] [PubMed] [Google Scholar]
- Wright BE, Reimers JM, Schmidt KH, Reschke DK. Hypermutable bases in the p53 cancer gene are at vulnerable positions in DNA secondary structures. Cancer Res. 2002;62:5641–5644. [PubMed] [Google Scholar]
- Wright BE, Reschke DK, Schmidt KH, Reimers JM, Knight W. Predicting mutation frequencies in stem-loop structures of derepressed genes: implications for evolution. Mol. Microbiol. 2003;48:429–441. doi: 10.1046/j.1365-2958.2003.t01-1-03436.x. [DOI] [PubMed] [Google Scholar]
- Wright BE, Schmidt KH, Minnick MF, Davis N. I. VH gene transcription creates stabilized secondary structures for coordinated mutagenesis during somatic hypermutation. Mol. Immunol. 2008;45:3589–99. doi: 10.1016/j.molimm.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BE, Schmidt KH, Davis N, Hunt AT, Minnick MF. II. Correlations between secondary structure stability and mutation frequency during somatic hypermutation. Mol. Immunol. 2008;45:3600–8. doi: 10.1016/j.molimm.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BE, Schmidt KH, Hunt AT, Lodmell JS, Minnick MF, Reschke D. The roles of transcription and genotoxins underlying p53 mutagenesis in vivo. Carcinogenesis. 2011 doi: 10.1093/carcin/bgr177. doi: 10.1093/carcin/bgr177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt JR, Puglist JD, Tinoco I., Jr. RNA pseudoknots: Stability and loop size requirements. J. Mol. Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- Zheng NY, Wilson K, Jared M, Wilson PC. Intricate targeting of immunoglobulin somatic hypermutation maximizes the efficiency of affinity maturation. J. Exp. Med. 2005;201:1467–78. doi: 10.1084/jem.20042483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.