Summary
Here we present a unifying hypothesis about how messenger RNAs, transcribed pseudogenes, and long non-coding RNAs “talk” to each other using microRNA response elements (MREs) as letters of a new language. We propose that this “competing endogenous RNA” (ceRNA) activity forms a large-scale regulatory network across the transcriptome, greatly expanding the functional genetic information in the human genome and playing important roles in pathological conditions, such as cancer.
The Non-Coding Revolution
Lower organisms such as Caenorhabditis elegans have a comparable number of protein-coding genes as humans (Baltimore, 2001). However, the human genome is ~30 times larger than that of C. elegans, suggesting, that the non-coding portion of the genome is of crucial importance in dictating the greater complexity of higher eukaryotes (Costa, 2008; Mattick, 2009). Indeed, a significant proportion of the mammalian transcriptome does not correspond to annotated exons of protein-coding genes (Kapranov et al., 2007), implying that the fraction of the mammalian genome “carrying information” is significantly larger than previously expected. Remarkably, systematic analyses of the cancer genome and transcriptome have identified profound alterations in non-coding genes (Beroukhim et al., 2010; Futreal et al., 2004; Stratton et al., 2009). Rearrangements, such as deletion, amplification, inversion, and chromosomal translocation are observed to alter non-coding genes, as they do to coding genes.
Although recent studies have begun associating a subset of long non-coding RNAs (lncRNAs) with specific regulatory mechanisms, less is known about non-coding transcripts on a genome wide scale (Nagano and Fraser, 2011). Moreover, little is still known of the potential non-coding functions of coding genes. Recent theoretical and experimental studies have suggested that, in particular cases (Seitz, 2009; Poliseno 2010), RNAs influence each other’s levels by competing for a limited pool of microRNAs. Here we describe a unifying hypothesis that attributes this new and potentially predictable function to the coding and non-coding transcriptome. We outline this “competitive endogenous RNA” (ceRNA) hypothesis, including its logic, and then discuss recent experimental evidence for it and the consequences of altering its homeostasis. Overall, we hypothesize that all types of RNA transcripts communicate through a new “language” mediated by microRNA binding sites (MREs or “microRNA response elements”) and that recent advances in experimental techniques are finally allowing us hear and translate this language.
The ceRNA Protagonists
1. MicroRNAs
Approximately 22 nucleotides in length, microRNAs bind to sequences with partial complementarity on target RNA transcripts, called microRNA recognition elements (MRE), usually resulting in the repression of target gene expression (Bartel, 2009; Thomas et al., 2010). MicroRNAs can function in a combinatorial manner if an mRNA transcript harbors numerous MREs. Furthermore, each microRNA may repress up to hundreds of transcripts, and thus, it is estimated that microRNAs regulate a large proportion of the transcriptome (Friedman et al., 2009; Thomas et al., 2010). In fact, microRNAs have been implicated in numerous diseases (http://cmbi.bjmu.edu.cn/hmdd.), including cancer (Calin et al., 2002; Lu et al., 2008).
2. The transcriptome
A. The protein coding genes
Approximately 20,000 protein-coding genes have been identified in the human genome, many of which are densely covered in MREs (Baltimore, 2001; Friedman et al., 2009). Our increasing capacity to identify MREs on coding gene transcripts allows us to predict the extent of microRNA-dependent regulation. We believe that this predictability, coupled with appropriate validation steps, will be critical in validating the ceRNA hypothesis.
B. Pseudogenes
Pseudogenes are genomic loci that resemble known genes but are defined as “nonfunctional,” “junk,” or “evolutionary relics” because, except for a few cases, they do not encode functional proteins; their translation is interrupted by a premature stop codons, frameshift mutations, insertions, or deletions (D'Errico et al., 2004). Sequencing efforts have revealed ~19,000 pseudogenes in humans, many of which are transcribed and are often well conserved, suggesting that selective pressure to maintain pseudogenes exists (Pink et al., 2011).
Despite lacking canonical promoters, processed pseudogenes (i.e., ones without introns) can use proximal regulatory elements for transcription (Birney et al., 2007). Indeed, transcription of pseudogenes displays tissue-specificity and can be activated or silenced in specific pathological conditions, such as cancer (Pink et al., 2011). Importantly, the high sequence conservation between gene and associated pseudogenes implies that the same microRNAs can target them (Poliseno et al., 2010).
C. Long non-coding RNAs (lncRNAs)
LncRNAs are typically 300 to thousands of nucleotides in length. The number of reported lncRNAs is expanding, and of these, a subset have been linked to epigenetic mechanisms, including XIST (X-inactive specific transcript), which is implicated in X-chromosome inactivation (Brown et al., 1992), and the recently identified large intergenic non-coding (linc-)RNAs (Gong and Maquat, 2011; Guttman et al., 2009; Huarte et al., 2010; Khalil et al., 2009). Importantly, microRNAs also regulate lncRNAs, as shown in recent global analysis of Argonaute (Ago)-bound transcripts through the HITS-CLIP technique (Chi et al., 2009; Licatalosi et al., 2008).
The ceRNA hypothesis
RNA transcripts communicate through the ceRNA language
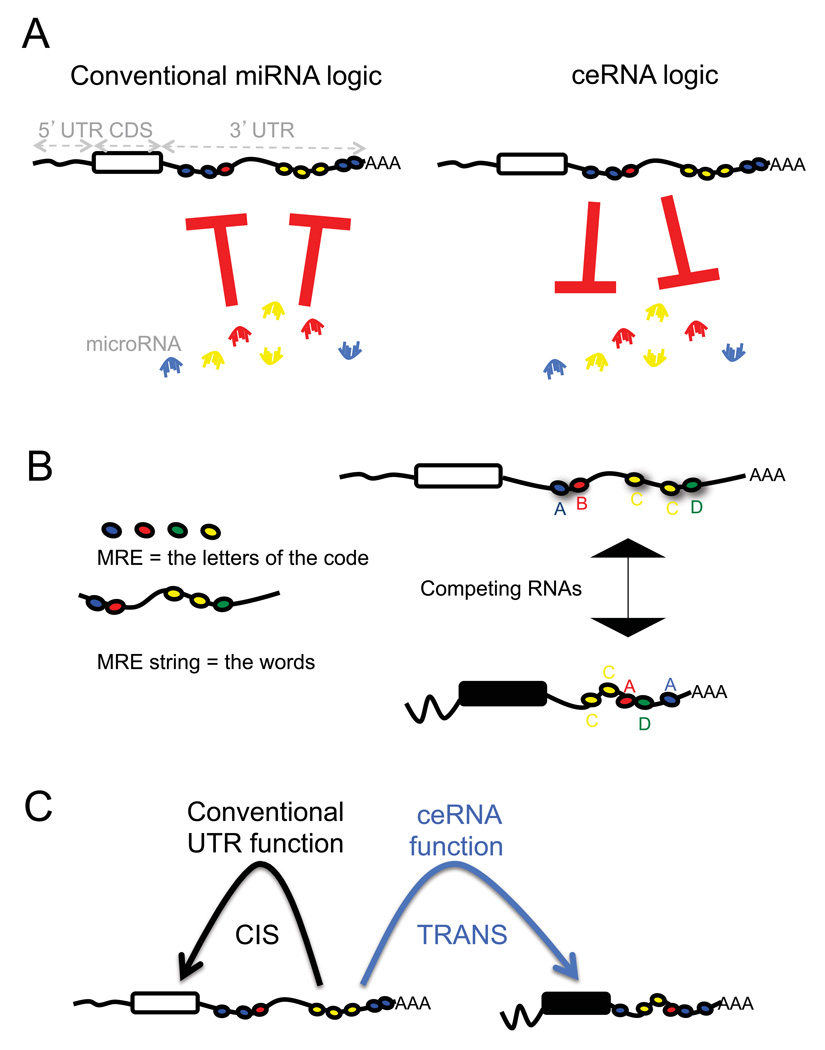
MicroRNAs are negative regulators of gene expression, decreasing the stability of target RNAs or limiting their translation (Fabian et al., 2010). Accordingly, microRNAs are commonly viewed as active regulatory elements, whereas the target mRNAs are viewed as passive targets of repression (Figure 1A, left).
Figure 1. Flipping the conventional logic of microRNA-mRNA interactions.
How mRNAs affect microRNAs is less well characterized than how of microRNAs affect effect mRNAs. A. The relationship between mRNAs and microRNAs could be reciprocal (Seitz 2009), causing the level of one mRNA to influence the level and activity of another mRNA. B. Thus, RNA molecules could communicate with each other through microRNA and microRNA recognition sequences (MREs). The greater the number of shared MREs, the greater the level of “communication” and thus co-regulation. C. The 3’ UTRs of RNA molecules contain MREs, which can function in cis to regulate the RNA molecule itself but also possibly in trans to regulate levels of microRNAs and consequently other RNAs.
By contrast, in 2009 Seitz hypothesized that computationally identified microRNA binding sitescan titrate miRNAs and thereby regulate the microRNA availability for (Seitz, 2009). We have more recently demonstrated experimentally that pseudogenes, due to their high sequence homology, can act as legitimate bona fide microRNA competitors thereby actively competing with their ancestral protein-coding genes for the same pool of microRNAs through sets of conserved MREs (Poliseno, 2010). The consequence of competition for microRNA is observed as a decrease in microRNA detection, and thus an impairment of microRNA activity (Cazalla et al., 2010; Lee et al., 2010; Wang et al., 2010).
Thus, we hypothesize that in addition to the conventional microRNA → RNA function, a reversed RNA → microRNA logic exists (Figure 1A, right), in which bona fide coding and non-coding RNA targets can cross-talk through their ability to compete for microRNA binding. On the basis of this hypothesis MREs can be viewed as the letters of a “RNA language” by which transcripts can actively communicate to each other to regulate their respective expression levels (Figure 1B). We hypothesize that RNAs that share multiple MREs will cross talk effectively. Importantly, we predict that this “RNA language” can be used to functionalize the entire mRNA dimension through the identification of cross-talking ceRNAs, as well as ceRNA networks.
Besides attributing a new, global function for all the non-coding RNAs, the ceRNA hypothesis challenges the notion that a protein-coding gene must be translated into a protein to exert function. We propose that mRNAs may also possess an additional and predictable function through their ability to regulate other mRNAs. Moreover, the non-coding function of mRNA may be consistent with the coding function, but the two functions could also be incoherent or even opposite in effect, thereby creating built-in regulatory loops, functional complexity, and diversification, in both physiological and pathological conditions.
Furthermore, the ceRNA hypothesis may explain the regulatory function of 3’UTRs (Rastinejad and Blau, 1993; Rastinejad et al., 1993). Besides acting as cis regulatory elements that alter the stability of their own transcripts, 3’UTRs may also act in trans to modulate gene expression through microRNA binding (Figure 1C). This is particularly relevant given the recent identification of 3’UTRs expressed separately from the associated protein-coding sequences to which they are normally linked (Mercer et al., 2010). In addition, we are proposing that all types of RNAs may compete with each other for microRNAs, generating large-scale trans-regulatory crosstalk across the transcriptome as a whole.
Logic and regulation of the ceRNA network
The ceRNA hypothesis relies on a knowledge of the precise number and location of MREs; “the letters” of the RNA code. Although several target prediction algorithms are successful in identifying some microRNA targets, they commonly fail to predict some important microRNA targets, mainly because the rules of targeting are still not understood (Bartel, 2009; Thomas et al., 2010). We expect that better target prediction algorithms and innovative biochemical techniques will contribute significantly to the definition of the ceRNA language. For example, the high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation, or HITS-CLIP allows the identification of MREs associated with the RNA-induced silencing complex (RISC)(Thomas et al., 2010).
What cellular conditions must exist for ceRNA network to occur? First, the relative concentration of the ceRNAs and their microRNAs is clearly important. Changes in the ceRNA expression levels need to be large enough to either overcome or relieve the microRNA repression on competing ceRNAs. This is exemplified by RNA transcripts “switched” on or off at the transcriptional level in different developmental stages or physiological/pathological conditions. Similarly, the expression levels of the sequestered microRNAs could be neither absent, nor grossly overexpressed, because either condition will abolish competition.
Second, the effectiveness of a ceRNA would depend on the number of the number of microRNAs it can “sponge”. This in turn would depend on the ceRNA’s accessibility to microRNA molecules, which is influenced by its subcellular localization and its interaction with RNA binding proteins. The specific tissue, developmental, or pathological context in which the ceRNA is expressed would also impact its overall influence because not all microRNAs are present everywhere and at all times (Venables et al., 2009). Although a ceRNA network could be built around a single microRNA, we hypothesize that the most robust ceRNA networks would contain transcripts that share multiple MREs targeted by multiple microRNAs. Thus, overall ceRNA networks would also depend on the identity, concentration, and subcellular distribution of the RNA and the microRNA species that are present in a given cell type at a given moment.
Third, not all the MREs on ceRNAs are equal. Although 2 MREs may be predicted to bind the same microRNA, their specific nucleotide composition may be partially different and the effectiveness of each MRE to bind a microRNA is critical for overall ceRNA function.
Similarily, microRNAs are predicted to target tens to hundreds of RNAs, but they do not exert the same degree of repression on all of them; the primary targets are usually few, while the rest are finely tuned (Bartel and Chen, 2004; Seitz, 2009). It is conceivable that, if a given microRNA is sequestered by a ceRNA, the primary targets of that microRNA would be preferentially affected.
Experimental evidence supporting the ceRNA hypothesis
More recently, our work demonstrated experimentally that indeed a non-coding pseudogene can bind to and compete for the same collection of microRNAs as its ancestral gene (Poliseno et al., 2010). Specifically, we found that many MREs in the tumor suppressor gene PTEN are conserved in its related pseudogene PTENP1, and overexpression of the PTENP1 3’UTR increased levels of PTEN and growth inhibition in a DICER-dependent manner. Interestingly, copy number losses at the PTENP1 locus in sporadic colon cancer suggest that PTENP1 could be considered a tumor suppressor gene (Poliseno et al. 2010).
We and other laboratories have extended this analysis to other gene-pseudogene partners (e.g. KRAS and its pseudogene KRAS1P) and protein-coding mRNAs [e.g. PTEN 3’UTR (Poliseno et al., 2010), versican 3’UTR (Lee et al., 2010; Lee et al., 2009), CD44 3’UTR (Jeyapalan et al., 2010)]. Overall, these findings suggest that 3’UTRs from both pseudogenes and coding genes may possess powerful biological activity through their ability to act as endogenous decoys for microRNAs.
Approximately three years before the identifications of these endogenous decoys, numerous studies found that exogenously expressed “microRNA sponges,” were able to inhibit microRNA function specifically and effectively (Ebert et al., 2007; Brown et al., 2007; Gentner et al., 2009). MicroRNA sponges are artificial transcripts that contain multiple copies of a single MRE in tandem. They are often cloned into viral vectors so that they can be expressed at high levels (Ebert and Sharp, 2010). The applications for sponging constructs are exciting, and perhaps the future of RNA-based therapeutic modalities (Brown et al., 2007; Gentner et al., 2009). Analogously, we propose that ceRNAs are “endogenous sponges” that are able to impact the distribution of microRNA molecules on all their targets. Unlike artificial sponges, ceRNAs contain MREs for a combination of different microRNAs, thus they can impact the multiple targets of multiple microRNAs.
In addition to pseudogenes, other examples of ceRNA have been reported recently. Franco-Zorrilla and colleagues demonstrated that the non-coding RNA IPS1 in Arabidopsis thaliana sequesters miR-399 by mimicking its target site, a phenomenon called “target mimicry” (Franco-Zorrilla et al., 2007). Analogously, a noncoding-RNA in herpesvirus saimiri RNA has been shown to bind to and cause the degradation of human miR-27 to possibly produce a permissive cellular environment for viral infection and transformation (Cazalla et al., 2010). Furthermore, highly up-regulated in liver cancer (HULC) lncRNA sequestered endogenous miR-372 to modulate its own transcriptional upregulation in HCC (Wang et al., 2010). Notably, all the endogenous sponges reported thus far do not encode for proteins.
Although our hypothesis applies to both protein-coding and non-coding RNAs, we speculate that non-coding ceRNAs may be highly effective inhibitors precisely because they are devoted to microRNA binding, without any interference from active translation (Gu et al., 2009). Further development and widespread use of HITS-CLIP and other related techniques will ultimately reveal the full extent of pseudogene and lncRNA regulation by microRNAs, and consequently their respective impact and positioning within ceRNA networks.
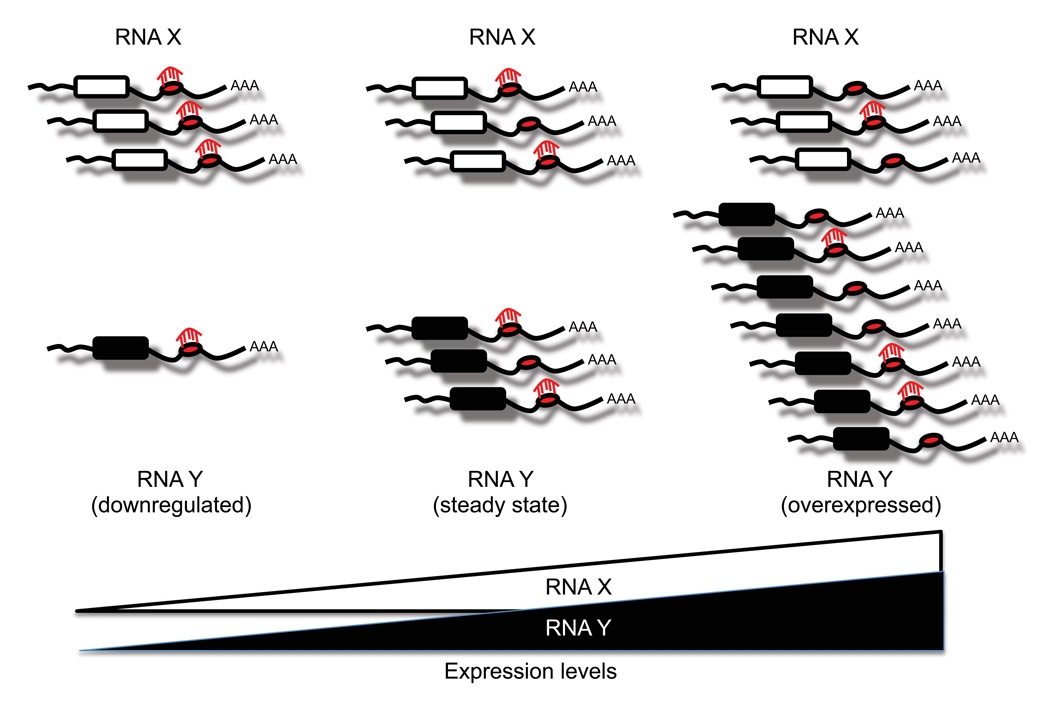
The ceRNA hypothesis is strongly supported by the fact that a single microRNA’s effectiveness is influenced by the concentration of its target mRNAs (Arvey et al., 2010). MicroRNAs that have a larger repertoire of target genes may down-regulate each individual target gene to a lesser extent than microRNAs with fewer targets. By the same token, when a given mRNA is upregulated, the repression conferred by its targeting microRNAs would be diluted because the total number of MREs exceeds that of the microRNAs themselves (Figure 2). Thus, altering the expression levels of an individual ceRNA would have repercussions on other ceRNAs with which it shares MREs.
Figure 2. MicroRNA effectiveness is influenced by the cellular concentration of its MREs.
Multiple RNA transcripts can contain MREs for the same microRNA. Upregulation of any of these RNA transcripts increases the cellular concentration of a particular MRE, which could decrease levels of transcripts targeted by the same microRNA.
ceRNAs in the etiology of cancer
In principle, almost any RNA molecule that possesses at least one MRE accessible to microRNA binding could act as a ceRNA. Therefore, to characterize the ceRNA networks requires the accurate identification of MREs within RNA molecules. Indeed, we speculate that this type of analysis could uncover molecular interactions and gene regulatory networks that have been missed by proteomic and conventional genomic methods. In this framework, aberrant expression of coding and non-coding genes should be systematically studied in the context of human disease.
Pseudogenes are a compelling example of ceRNA because they likely posses many (if not all) of the same MREs that are harbored on their ancestral genes and thus can act as “perfect sponges.” However, the ability of pseudogenes to regulate the biology of a cell may go beyond the modulation of the levels of their ancestral genes. For instance, PTENP1 is biologically active even in a PTEN null context, as it alters the microRNA network normally regulating PTEN (Poliseno et al., 2010). Moreover, genes such as OCT4, NPM1, and many ribosomal protein pseudogenes often have numerous differentially regulated pseudogenes (Balasubramanian et al., 2009), indicating that, gene-pseudogene networks can become extensive and intricately dynamic.
In the context of cancer, a straightforward implication of our hypothesis is that pseudogenes and lncRNAs should now be systematically studied as potential tumor suppressors and oncogenes through their ceRNA function. Accordingly, the notion of endogenous lncRNA sponges was recently linked to the progression of liver cancer. It was reported that the lncRNA HULC is one of the most upregulated of all genes in hepatocellular carcinoma (Panzitt et al., 2007). Wang and colleagues identified that CREB (cAMP response element binding protein) is involved in the upregulation of HULC (Wang et al., 2010). They also demonstrated that HULC RNA inhibits miR-372 activity through a ceRNA function. This in turn leads to derepression of one of its target genes, PRKACB, which can then induce the phosphorylation and activation of CREB. Overall, HULC lncRNA is part of a self-amplifying autoregulatory loop in which it sponges miR-372 to activate CREB, and in turn upregulates its own levels.
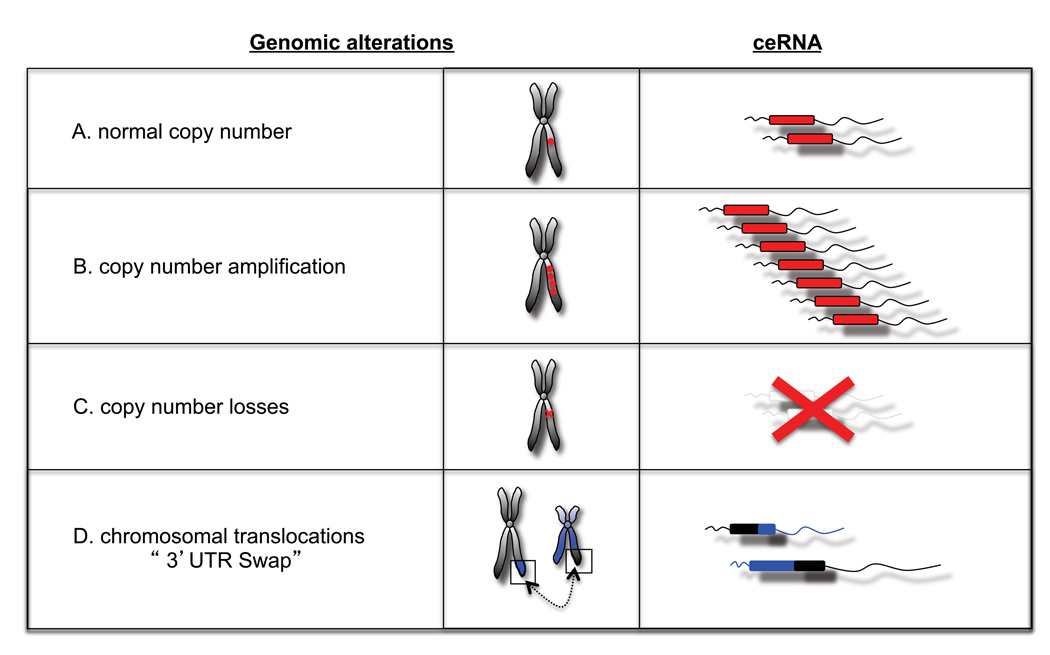
Gross genomic losses and amplifications commonly observed in cancer could have potentially dramatic consequences for the ceRNAs contained in those regions. Moreover, under the ceRNA hypothesis, gene loss events should be clearly distinguished from point mutations that abolish protein function but retain full ceRNA function.
If the ceRNA hypothesis proves correct, then one would need to consider the repercussions of knocking out and overexpressing ceRNAs when modeling diseases in mice. For instance, when generating knockout mice, one must consider whether only the transcript or also the protein expression is disrupted. Many experimental techniques normally neglect UTRs and limit functional studies to gene coding regions. For example, when generating transgenic mice, it has been standard to only overexpress coding sequences, but not UTRs. However, binding sites for microRNAs could occur in 3’UTRs, 5’UTRs, and coding regions (Tay et al., 2008), suggesting that the entire transcript may possess an inherent trans regulatory function. Thus, by limiting their focus or scope to coding region, many conventional tools and techniques may have been neglecting the full function of the gene.
Chromosomal translocation events and recurrent “readthrough” transcripts are common in cancers. For example, the t(15;17) translocation which generates PML-RARα and RARa-PML fusion transcripts, is often seen in Acute Promyelocytic Leukemia, whereas the or “readthrough” transcript CDK2-RAB5B is common in melanoma (Berger et al., 2010; Scaglioni and Pandolfi, 2007). Such events could be considered “UTR-swaps,” leading to perturbed MRE levels due to misplacement and consequent altered expression of UTRs (Figure 3). ceRNA perturbation could also possibly occur as a consequence of somatic genomic rearrangements affecting non-coding regions, which are emerging as hitherto unappreciated events in many cancers (Stephens et al., 2009)
Figure 3. Potential pathological alterations of cellular ceRNA.
Many types of genetic events can alter the abundance or sequence of a particular transcript. Under the ceRNA hypothesis, these events could induce ‘coding-independent’ effects, by altering the levels of microRNAs available for silencing particular transcripts.
Aberrant alternative splicing events could also introduce new RNA sequences and potentially new MREs into the cell. Because splicing can be perturbed in disease and cancer (Venables et al., 2009), the associated perturbation of the ceRNA network may also contribute to pathologies. Similarly the shortening of 3’UTRs as observed in human cancer cells (Mayr and Bartel, 2009) would not only impact microRNA-dependent mRNA regulation, but on the flipside, could also alter the capacity of a given mRNA transcript to “sponge” or titrate away microRNAs.
All these described events have a single commonality; they represent perturbations in the expression levels of a given transcript (and consequentially MREs), irrespective of whether or not the transcript is translated into a protein. Thus, it will be interesting to determine if elevated or depressed levels of a given transcript could exert oncogenic activities by altering competition for miRNAs.
Conclusions
In conclusion, we hypothesize that cross-talk between RNAs, both coding and non-coding, through MREs forms large-scale regulatory network across the transcriptome. This ceRNA activity could offer answers to evolutionary questions, as it may, in part explain the correlation of genome size and organism complexity. Moreover, perturbations of ceRNA and ceRNA networks could have consequences for diseases, but on the flip side, it may explain disease processes and present opportunities for new therapies. Although the understanding of this field and its consequences are in their infancy, we believe that experimental tools are now poised to fully identify microRNA binding sites and cataloguing the basic lexicon of the ceRNA “language.” We envision that the ceRNA language will allow us to predict and manipulate regulatory networks working through microRNA competition. Future challenges will be then to understand why such regulatory networks exist, how they may have evolved, and what the consequences are when they are perturbed. Only then, we will able to fully decipher the “Rosetta Stone” of this hidden RNA language.
Acknowledgements
We thank all Pandolfi lab members for critical discussions. L.S. was supported by fellowships from the Canadian Institutes of Health Research and the Human Frontier Science Program. L.P. was supported by fellowships from the Istituto Toscano Tumori and the American Italian Cancer Foundation. Y.T. is supported by a Special Fellow Award from The Leukemia & Lymphoma Society. L.K. is supported by a NHMRC Overseas Biomedical Postdoctoral Fellowship. This work was supported by NIH grant R01 CA-82328-09 awarded to P.P.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Zheng D, Liu YJ, Fang G, Frankish A, Carriero N, Robilotto R, Cayting P, Gerstein M. Comparative analysis of processed ribosomal protein pseudogenes in four mammalian genomes. Genome Biol. 2009;10:R2. doi: 10.1186/gb-2009-10-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. Our genome unveiled. Nature. 2001;409:814–816. doi: 10.1038/35057267. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Berger MF, Levin JZ, Vijayendran K, Sivachenko A, Adiconis X, Maguire J, Johnson LA, Robinson J, Verhaak RG, Sougnez C, et al. Integrative analysis of the melanoma transcriptome. Genome Res. 2010;20:413–427. doi: 10.1101/gr.103697.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BD, Cantore A, Annoni A, Sergi LS, Lombardo A, Della Valle P, D'Angelo A, Naldini L. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood. 2007;110:4144–4152. doi: 10.1182/blood-2007-03-078493. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz J. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FF. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- D'Errico I, Gadaleta G, Saccone C. Pseudogenes in metazoa: origin and features. Brief Funct Genomic Proteomic. 2004;3:157–167. doi: 10.1093/bfgp/3.2.157. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annual review of biochemistry. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, Naldini L. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3' untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyapalan Z, Deng Z, Shatseva T, Fang L, He C, Yang BB. Expression of CD44 3'-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Jeyapalan Z, Fang L, Yang J, Zhang Y, Yee AY, Li M, Du WW, Shatseva T, Yang BB. Expression of versican 3'-untranslated region modulates endogenous microRNA functions. PloS one. 2010;5:e13599. doi: 10.1371/journal.pone.0013599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Shatseva T, Jeyapalan Z, Du WW, Deng Z, Yang BB. A 3'-untranslated region (3'UTR) induces organ adhesion by regulating miR-199a* functions. PLoS One. 2009;4:e4527. doi: 10.1371/journal.pone.0004527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q. An analysis of human microRNA and disease associations. PLoS One. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Wilhelm D, Dinger ME, Solda G, Korbie DJ, Glazov EA, Truong V, Schwenke M, Simons C, Matthaei KI, et al. Expression of distinct RNAs from 3' untranslated regions. Nucleic Acids Res. 2010;39:2393–2403. doi: 10.1093/nar/gkq1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Francisco Carter DR. Pseudogenes: Pseudo-functional or key regulators in health and disease? RNA. 2011 doi: 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastinejad F, Blau HM. Genetic complementation reveals a novel regulatory role for 3' untranslated regions in growth and differentiation. Cell. 1993;72:903–917. doi: 10.1016/0092-8674(93)90579-f. [DOI] [PubMed] [Google Scholar]
- Rastinejad F, Conboy MJ, Rando TA, Blau HM. Tumor suppression by RNA from the 3' untranslated region of alpha-tropomyosin. Cell. 1993;75:1107–1117. doi: 10.1016/0092-8674(93)90320-p. [DOI] [PubMed] [Google Scholar]
- Scaglioni PP, Pandolfi PP. The theory of APL revisited. Curr Top Microbiol Immunol. 2007;313:85–100. doi: 10.1007/978-3-540-34594-7_6. [DOI] [PubMed] [Google Scholar]
- Seitz H. Redefining microRNA targets. Curr Biol. 2009;19:870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, et al. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16:670–676. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]