SUMMARY
Here we demonstrate that protein-coding RNA transcripts can crosstalk by competing for common microRNAs, with microRNA response elements as the foundation of this interaction. We have termed such RNA transcripts as competing endogenous RNAs (ceRNAs). We tested this hypothesis in the context of PTEN, a key tumor suppressor whose abundance determines critical outcomes in tumorigenesis. By a combined computational and experimental approach, we identified and validated endogenous protein-coding transcripts that regulate PTEN, antagonize PI3K/AKT signaling and possess growth and tumor suppressive properties. Notably, we also show that these genes display concordant expression patterns with PTEN and copy number loss in cancers. Our study presents a road map for the prediction and validation of ceRNA activity and networks, and thus imparts a trans-regulatory function to protein-coding mRNAs.
INTRODUCTION
Regulation of gene expression by small non-coding RNA molecules is ubiquitous in many eukaryotic organisms from protozoa to plants and animals. In mammals, ~22 nucleotide long RNAs termed microRNAs, guide the RNA-induced silencing complex (RISC) to microRNA response elements (MREs) on target transcripts, usually resulting in degradation of the transcript or inhibition of its translation (Bartel, 2009; Bartel and Chen, 2004). Individual genes often contain MREs for multiple distinct microRNAs, and conversely, individual microRNAs often target multiple distinct transcripts (Friedman et al., 2009).
We and others recently provided experimental support to the hypothesis that RNA molecules that share MREs can regulate each other by competing for microRNA binding (Cazalla et al., 2010; Jeyapalan et al., 2010; Kloc 2008; Lee et al., 2009; Poliseno et al., 2010b; Seitz 2009), Specifically, we reported several examples of pseudogene transcripts exerting regulatory control of their ancestral cancer gene’s expression levels by competing for microRNAs that targeted sequences common to the mRNA and the pseudo-mRNA (Poliseno et al., 2010b), in keeping with the notion that the microRNA activity should be theoretically affected by the availability of its target MRE in the cellular milieu (Arvey et al., 2010).
This in turn led us to hypothesize that the mRNA/microRNA network would operate through a reverse logic whereby protein coding and non-coding mRNAs would communicate with each other in a microRNA-dependent manner, through a MRE language (Salmena et al., 2011). We proposed that a reversed RNA → microRNA function exists, whereby RNAs actively regulate each other through direct competition for microRNA binding. In this work, we tested this hypothesis experimentally and present a comprehensive scheme for the prediction and validation of ceRNA activity and networks demonstrating that bioinformatic predictions followed by a set of stringent biological tests allow for the identification and validation of ceRNAs for mRNAs of interest. We focused our analysis on the ceRNA network encompassing PTEN, a critical tumor suppressor gene which encodes a phosphatase that converts phosphatidylinositol 3,4,5-trisphosphate to phosphatidylinositol 4,5-bisphosphate, thereby antagonizing the highly oncogenic PI3K/Akt signaling pathway (Hollander et al., 2011).
PTEN was selected as a model system for three reasons: (1) PTEN expression is frequently altered in a wide spectrum of human cancers (Hollander et al., 2011); (2) subtle changes in PTEN dose dictate critical outcomes in tumor initiation and progression in vivo (Alimonti et al., 2010; Berger et al., 2011; Trotman et al., 2003) and (3) numerous microRNAs have been validated as PTEN regulators, including the proto-oncogenic miR-106b~25 cluster that is overexpressed in prostate cancer (Huse et al., 2009; Mu et al., 2009; Olive et al., 2009; Poliseno et al., 2010a; Xiao et al., 2008). Taken together, these previous studies suggested that bona fide PTEN ceRNAs, and a broader PTEN ceRNA network, may represent a previously uncharacterized RNA-dependent tumor suppressive dimension.
RESULTS
Identification of candidate PTEN ceRNAs
To identify and characterize the PTEN ceRNA network in the human genome, we devised a multifaceted scheme involving integrated computational analysis and experimental validation (Fig. 1A), an approach that we termed mutually targeted MRE enrichment (MuTaME). Initially, we sought to identify mRNAs that are targeted by PTEN-targeting microRNAs. We focused on validated PTEN-targeting microRNAs, specifically those previously implicated in the ceRNA-mediated regulation of PTEN by its pseudogene PTENP1: miR-17-5p, miR-19a, miR-19b, miR-20a, miR-20b, miR-26a, miR-26b, miR-93, miR-106a, miR-106b and miR-214 (Poliseno et al., 2010a; Poliseno et al., 2010b), while we excluded miR-214 from subsequent analysis as it is not expressed in the cell lines utilized to validate the putative PTEN ceRNAs (Supplemental Fig. S1A).
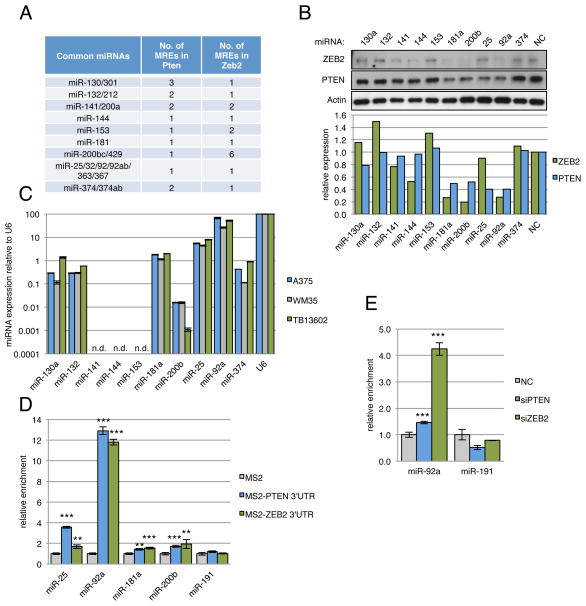
Figure 1. (related to Figure S1 and Table S1): Mutually targeted MRE enrichment (MuTaME) analysis predicts competitive endogenous RNAs for PTEN.
(A) Schematic outlining the MuTaME analysis and subsequent experimental validation strategy. Validated PTEN-targeting microRNAs were used to predict putative PTEN ceRNAs. Candidates sharing at least 7 microRNAs were considered putative PTEN ceRNAs. (B) MS2-RIP followed by microRNA RT-PCR to detect microRNAs endogenously associated with PTEN 3′UTR. Mean ± s.d., n ≥ 3, *P < 0.05, **P < 0.01. (C) Heat map showing MRE enrichment of the top 20 (upper panel) and bottom 20 (middle panel) putative PTEN ceRNAs and 25 randomly selected transcripts (lower panel). (D) Table summarizing predicted MREs in the 3′UTRs of the top 7 putative PTEN ceRNAs.
Importantly, we also examined the physical association of the PTEN 3′UTR with endogenous levels of these microRNAs in the cell line used in our study. We reasoned that this represents a critical selective criterion to guide the computational component of our analysis (Fig. 1A). To do so, we performed RNA-immunoprecipitation (RIP) to pull down endogenous microRNAs associated with the PTEN 3′ UTR (Supplemental Fig. S1B) and demonstrated via real-time PCR analysis that the PTEN 3′UTR RIP in DU145 prostate cancer cells significantly enriched for miR-17-5p, 19a, 19b, 20a, 26a, 93, 106a and 106b compared to empty vector and IgG controls and a non-targeting microRNA control (miR-191) (Fig. 1B). These results further support the claim that these microRNAs, which were previously validated mainly by overexpression and/or knockdown experiments, are bona fide PTEN-targeting microRNAs in this cell line and justify their inclusion in our analyses.
We next used the rna22 microRNA target prediction algorithm (Miranda et al., 2006) available at http:://cbcsrv.watson.ibm.com/rna22.html to generate MuTaME scores for the entire human protein-coding transcriptome. The choice of rna22 was based on earlier reports supporting its low rate of false prediction (Hammell et al., 2008; Ritchie et al., 2009). A central tenet of our hypothesis is that trans-regulatory ceRNA crosstalk increases with the number of microRNAs that are shared by transcripts. This is the first consideration in deriving a MuTaMe score. The second consideration results from an often ignored dependence on the length of the candidate transcript’s 3′UTR: the expected number of spurious microRNA target predictions increases with the length of the candidate 3′UTR through a non-linear relationship (Altschul et al., 1990; Miranda et al., 2006), and this holds true independently of the algorithm used to predict microRNA targets. This dependence on length and the findings described in Miranda et al. and Ritchie et al. (Miranda et al., 2006, Ritchie et al., 2009) suggest that scenarios where there are many predicted MREs in a candidate transcript, the MREs are spread over a relatively short span and are as evenly distributed within the span as possible, and multiple MREs are predicted for each of the microRNAs under consideration, ought to be favored.
To summarize, MuTaME evaluates a candidate ceRNA X based on the following: a) how many microRNAs it shares with the mRNA M of interest, in our case this is PTEN – this is captured by the ratio #microRNAs predicted to target X over #microRNAs being considered, which increases with the number of targeting microRNAs that X shares with M; b) the number of MREs predicted in X for the i-th microRNA and the width of the span they cover – this is captured by the ratio #MREs in X for i-th microRNA over distance between leftmost and rightmost predicted MRE for the i-th microRNA, which favors situations where more MREs spanning shorter distances are predicted for the i-th microRNA and captures one of the main observations in (Ritchie et al., 2009); c) how evenly distributed the predicted MREs for the i-th microRNA are over the distance they span in X – this is captured by the ratio square of the distance between leftmost and rightmost predicted MRE for the i-th microRNA over sum of the squared distances between successive MREs of the i-th microRNA, which favors more evenly distributed MREs for each microRNA and penalizes cases where the majority, but not all, of a microRNA’s MREs aggregate in a narrow neighborhood; and, d) the relation between the total number of MREs predicted in X compared to the total number of microRNAs that give rise to these MREs – this is captured by the ratio (#MREs in X for all considered microRNAs — #microRNAs predicted to target X + 1) over #MREs in X for all considered microRNAs, which favors situations where each targeting microRNA gives rise to more than one MRE in X. A priori there is no reason to favor one type of contribution more than the rest and thus each candidate transcript X receives a combined MuTaMe score obtained simply by multiplying these four components. Furthermore, for our specific setting we set stringent criteria and required that a) a candidate ceRNA be targeted by at least 7 of the 10 validated PTEN-targeting microRNAs and b) all predicted MREs should occur only in the candidate ceRNA’s 3′UTR. One potential concern here is whether this scoring approach would be biased in favor of transcripts X with longer 3′UTRs: notably, we observed no correlation between 3′UTR length and MuTaMe score (corr=−0.13), or between 3′UTR length and the number of microRNAs predicted to target a candidate ceRNA (corr=0.07). Using MuTaMe, we identified 158 candidate protein-coding transcripts representing 136 distinct genes as putative trans-regulators of PTEN (Fig. 1C, D and Supplemental Table S1). Intriguingly, the candidates displayed a non-random enrichment in selected biological categories (Supplemental Fig. S1C).
Putative PTEN ceRNAs are co-expressed with PTEN in vivo
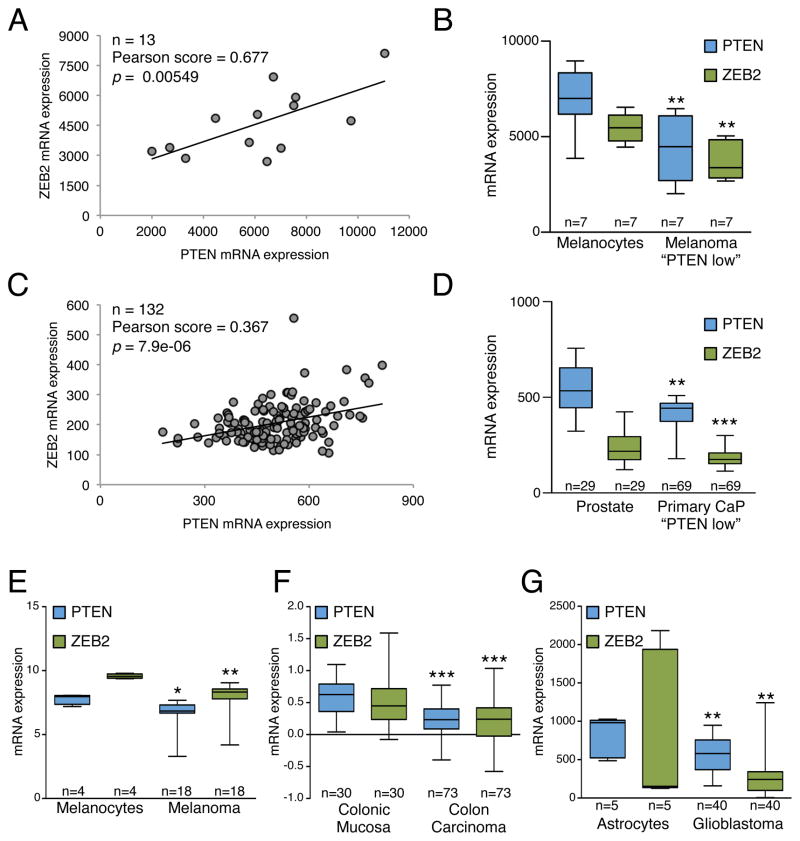
As our hypothesis predicts that transcripts within a ceRNA network are co-regulated, we first examined whether putative PTEN ceRNAs are co-expressed with PTEN in human samples. We selected the top 7 candidates from our MuTaME analysis (NCOA7, BCL11B, SERINC1, ZNF460, NUDT13, DTWD2, VAPA) (Fig. 1D) and investigated whether their expression correlated with PTEN expression in human prostate cancer (GSE21032) and glioblastoma (GSE15824), malignancies commonly defined by reduced PTEN levels. Significantly, NCOA7, SERINC1, ZNF460 and VAPA showed differential expression when samples were subdivided according to PTEN expression levels in both prostate cancer (179 total samples: 29 normal, 150 tumor) and glioblastoma (45 total samples: 5 normal, 40 tumor) (Fig. 2A, B, Supplemental Fig. S2A). Expression of these four genes was also significantly correlated with PTEN transcript levels (Supplemental Fig. S2C).
Figure 2. (related to Figure S2): Co-expression of PTEN and PTEN ceRNAs in human cancer.
(A, B) Comparison of PTEN ceRNA expression levels in primary (A) prostate cancer and (B) glioblastoma between two subsets of samples: “PTEN high” and “PTEN low”, classified according to the average PTEN expression level. P < 0.001 except for SERINC1 in glioblastoma where P = 0.009. The ends of the whiskers represent the minimum and maximum of all the data. (C) Co-expression analysis of PTEN and PTEN ceRNAs in subsets of the human prostate cancer specimens analyzed in (A) with decreased (blue, top panels) or increased (red, bottom panels) expression of PTEN-targeting microRNAs. P < 0.001 for all graphs except for SERINC1 microRNA up (P = 0.002) and ZNF460 microRNA up (P = 0.024). See also Figure S2.
As the prostate cancer study contained integrated expression profiling of both mRNAs and microRNAs, we were also able to rank patient samples according to microRNA expression levels. This enabled us to assess whether microRNA expression levels impact the co-expression correlation of PTEN ceRNAs and PTEN. Taking into account only the expression levels of the 10 validated PTEN-targeting microRNAs used in our analysis, samples were subdivided into two groups: the first where microRNAs were expressed at a lower level compared to their expression level in all samples and the second where they were expressed at a higher level. The Pearson correlation coefficients between PTEN and its candidate ceRNAs were calculated in both groups and intriguingly, we found an increase in correlation between SERINC1, ZNF460 and VAPA expression levels and PTEN when the microRNA expression was taken into consideration (Fig. 2C). NCOA7 was not significantly correlated with PTEN in this analysis (Supplemental Fig. S2B).
The significant correlation between PTEN expression and SERINC1, ZNF460 and VAPA expression and the sensitivity of this correlation to microRNA expression levels support our hypothesis that ceRNA transcripts can regulate PTEN levels in a biologically relevant manner. We next examined the co-expression of these three genes with PTEN in a database of multi-tissue and tissue-specific conserved human-mouse gene co-expression networks (Piro et al., 2011), and found that these genes were present in the top 1% of genes co-expressed with PTEN in several human-specific co-expression networks (Supplemental Fig. S2E). Based on their consistent co-expression with PTEN, we selected SERINC1, ZNF460 and VAPA for subsequent experimental validation.
Additionally, to investigate the extent to which lower-ranking candidates from our list exerted regulatory control over PTEN, we first explored the correlation of the 20 transcripts on our list with the lowest MuTaME scores with PTEN as described above. One of the most significantly correlated transcripts in both the prostate cancer and glioblastoma datasets was CNOT6L, which was also found to be significantly correlated with PTEN in the largest number of multi-tissue and tissue-specific conserved human-mouse gene co-expression networks (Supplemental Fig. S2D). We thus selected it as another candidate for further validation analysis.
PTEN ceRNAs modulate PTEN levels
We investigated the ability of these putative PTEN ceRNAs to modulate PTEN levels by first examining the effect of depletion of these candidates on endogenous PTEN protein levels in DU145 prostate cancer cells (Fig. 3A). For this analysis, we also included two genes, ACSL4 and UNC5CL, which are not predicted targets of these PTEN-targeting microRNAs as negative controls (Fig 1C). These experiments were performed using siRNA pools (a combination of four independent siRNAs), which are designed to achieve strong on-target gene knockdown with minimal off-target effects. Real-time PCR analysis confirmed efficient siRNA-mediated knockdown of candidate PTEN ceRNAs (Supplemental Fig. S3A). Depletion of SERINC1, ZNF460, VAPA or CNOT6L transcripts did indeed result in a significant reduction in PTEN protein levels whereas depletion of ACSL4 or UNC5CL did not have a significant effect (Fig. 3A, B). Moreover, reduced PTEN protein levels were accompanied by a concomitant, albeit less significant, decrease in PTEN transcript levels for SERINC1, VAPA and CNOT6L knockdown (Fig. 3B).
Figure 3. (related to Figure S3): Putative PTEN ceRNAs modulate PTEN expression.
(A) Western blot for PTEN protein levels in DU145 cells transfected with siRNA against predicted ceRNAs SERINC1 (siSER), ZNF460 (siZNF), VAPA (siVAPA), and CNOT6L (siCNO) and selected non-targeting controls ACSL4 (siACS) and UNC5CL (siUNC). (B) Quantitation of PTEN protein shown in (A) and PTEN mRNA changes after transfection with siRNA against ceRNA as measured by RT-PCR. (C) Luciferase activity in DU145 cells co-transfected with siRNA against PTEN ceRNAs and a luciferase-PTEN 3′UTR reporter construct. (D) Luciferase activity in DU145 cells co-transfected with PTEN ceRNAs 3′UTRs and a luciferase-PTEN 3′UTR reporter construct. (E) Western blot showing PTEN protein in response to overexpression of ceRNA 3′UTRs in DU145 cells. (F) Quantitation of PTEN protein shown in (E). (G) Western blot for PTEN in HCT116 WT (top panel) and DICER−/− (bottom panel) cells transfected with siRNAs against PTEN ceRNAs. (H) Quantitation of PTEN protein shown in (G). (B,D–F,H) Mean ± s.d., n ≥ 4, *P < 0.05; **P < 0.01; ***P < 0.001.
To ascertain whether this observed effect is dependent upon regulation of the PTEN 3′UTR, we constructed a chimeric luciferase construct tagged with the PTEN 3′UTR (Luc-PTEN3′UTR). This approach allows us to uncouple regulation of PTEN via 3′UTR-targeting microRNAs from PTEN mRNA transcription and protein stability. siRNA-mediated reduction of SERINC1, VAPA and CNOT6L transcripts in Luc-PTEN3′UTR expressing cells significantly lowered luciferase activity (Fig. 3C). However, siRNA-mediated ZNF460 knockdown did not significantly reduce Luc-PTEN3′UTR activity, suggesting that the observed effect on PTEN protein is not solely mediated by the PTEN 3′UTR. We thus excluded it from subsequent analysis.
Conversely, ectopic overexpression of PTEN ceRNA 3′UTRs in DU145 cells led to a marked upregulation of both Luc-PTEN3′UTR and endogenous protein levels, similar to the effect of overexpression of the PTEN 3′UTR on PTEN protein levels (Fig. 3D–F). Due to their large size, the VAPA and CNOT6L 3′UTRs were each cloned as two separate fragments subdivided based on the location of predicted MREs. Thus, sequestration of only a fraction of PTEN-targeting microRNAs may impact PTEN expression.
MicroRNA dependency of ceRNA-mediated PTEN regulation
To investigate the microRNA dependency of ceRNA-mediated PTEN regulation, we utilized isogenic wild-type and DICER mutant HCT116 colon carcinoma cells. In the latter, gene targeting was used to disrupt a well-conserved segment of the N-terminal helicase domain in exon 5 of DICER (Cummins et al., 2006). As DICER is a critical enzyme involved in the processing of mature microRNAs, the DICEREx5 mutant cells presented an ideal system to evaluate microRNA-dependent effects. While DICER processes the vast majority of mature microRNAs, not all microRNAs exhibit DICER-dependent processing. We thus assessed whether processing of the microRNAs used in our analyses (Fig. 1A, B) is abrogated in DICEREx5 HCT116 cells. Indeed, microRNA real-time PCR analysis confirmed that these microRNAs were significantly downregulated in DICEREx5 HCT116 cells (Supplemental Fig. S3C). We also confirmed that siRNA-mediated gene silencing is independent of DICER processing and hence is fully functional in DICEREx5 HCT116 cells (Supplemental Fig. S3B).
Similar to the experiments in DU145 prostate cancer cells, siRNA-mediated depletion of SERINC1, VAPA or CNOT6L expression resulted in a significant downregulation of PTEN protein in wild-type HCT116 colon cancer cells (Fig. 3G, H). Importantly, PTEN downregulation by ceRNA loss was profoundly attenuated in DICEREx5 HCT116 cells (Fig. 3G, H), suggesting that mature microRNAs are essential for the regulation of PTEN by these three transcripts.
PTEN ceRNAs are regulated by PTEN-targeting microRNAs
After successfully validating SERINC1, VAPA and CNOT6L as bona fide PTEN ceRNAs that regulate PTEN levels in a microRNA-dependent manner, we next investigated their association with the PTEN-targeting microRNAs used in our analysis. We focused on VAPA and CNOT6L as they had the most significant effect on PTEN at the 3′UTR level (Fig. 3C–F).
We constructed chimeric luciferase constructs tagged with the respective ceRNA 3′UTR fragments: as mentioned previously, the VAPA and CNOT6L 3′UTRs were each cloned as two separate fragments subdivided based on the location of predicted MREs due to their large size (Fig. 4A, D). Expression of miR-17, 20a, 20b and 106a significantly reduced Luc-VAPA-3′UTR1 activity, and expression of miR-19a, 26b and 106b significantly reduced Luc-VAPA-3′UTR2 activity in both DU145 and wild-type HCT116 cells (Fig. 4B). The effect of miR-93 and 106b appeared to be cell-type specific: miR-93 significantly reduced Luc-VAPA-3′UTR1 activity in HCT116 wild-type cells only, whereas miR-106b significantly reduced Luc-VAPA-3′UTR1 activity in DU145 cells only (Fig. 4B).
Figure 4. (related to Figure S4): MicroRNA-dependency of PTEN ceRNA function.
(A) Schematic outlining the predicted binding sites of PTEN-targeting microRNAs to the 3′UTR of VAPA. The 2 fragments VAPA 3′U1 and VAPA 3′U2 were used for the luciferase experiments. (B) Luciferase activity in DU145 cells co-transfected with validated PTEN-targeting microRNAs predicted to target VAPA and luciferase-VAPA-3′UTR1 and 3′UTR2 reporter constructs. (C) Western blot analysis of PTEN and VAPA expression in DU145 cells transfected with validated PTEN-targeting microRNAs predicted to target VAPA. (D) Schematic outlining the predicted binding sites of PTEN-targeting microRNAs to the 3′UTR of CNOT6L. The 2 fragments CNO 3′U1 and CNO 3′U2 were used for the luciferase and RIP experiments. (E) Luciferase activity in DU145 cells co-transfected with validated PTEN-targeting microRNAs predicted to target CNOT6L and luciferase-CNOT-3′UTR1 and 3′UTR2 reporter constructs. (F) RIP followed by microRNA RT-PCR shows enrichment of PTEN-targeting microRNAs associated with CNOT6L 3′UTR. (B–C,E–F) Mean ± s.d., n ≥ 4, *P < 0.05; **P < 0.01; ***P < 0.001.
Consistent with the luciferase results, overexpression of miRs 17, 19a, 20a, 20b, 26b, 106a and 106b caused a significant downregulation of VAPA protein levels (Fig. 4C). These results confirm that these validated PTEN-targeting microRNAs also regulate VAPA protein levels via its 3′UTR. Overexpression of miR-93 also resulted in a significant reduction of VAPA protein levels, in contrast to its effect on the Luc-VAPA-3′UTR1 reporter. This suggests that miR-93 is able to regulate VAPA protein levels independently of its 3′UTR, perhaps via targeting MREs located outside the 3′UTR or modulation of upstream regulators. It is important to note that the effects of the various microRNAs on VAPA were consistently less profound than on those on PTEN. This result is in agreement with the fact that VAPA transcript is expressed at levels significantly higher than PTEN in DU145 cells (100 fold), a factor which may significantly increase its efficacy as a PTEN ceRNA (Supplemental Fig. S4A).
As predicted, miR-17, 19a, 19b, 20a, 20b and 106b significantly reduced Luc-CNO-3′UTR1 activity in both DU145 and wild-type HCT116 cells (Fig. 4E). Transfection of miR-93 significantly reduced the activity of Luc-CNO-3′UTR2 but not Luc-CNO-3′UTR1, consistent with the location of the predicted MRE (Fig. 4E). As we were unable to find a good antibody for CNOT6L for western blot analysis, we instead performed RIP to confirm the physical association of these microRNAs with the CNOT6L 3′UTR. RIP for CNOT6L 3′UTR1 in DU145 prostate cancer cells significantly enriched for miR-17-5p, 19a, 19b, 20a and 106b compared to empty vector and IgG controls, whereas RIP for CNOT6L 3′UTR2 significantly enriched for only miR-93 (Fig. 4F). Expression of miR-20b was not detected, possibly due to its low level of endogenous expression (Supplemental Fig. S1A). These results therefore confirm the microRNA:3′UTR associations predicted by our MuTaME analysis.
Reciprocal ceRNA interactions
Furthermore, we postulate that ceRNA networks will behave in a mutually reciprocal manner, i.e. ceRNAs will regulate one another bidirectionally. In an effort to study the network and reciprocal effects of ceRNA misexpression we investigated the ability of PTEN downregulation to modulate VAPA protein expression and vice-versa. We could not perform this experiment for CNOT6L due to the lack of specific antibodies as mentioned above. Interestingly, we observed that siRNA knockdown of PTEN was able to significantly reduce VAPA expression in both DU145 and HCT116 cells (Supplemental Fig. S4B, top and middle panels), thus identifying regulatory loops between ceRNAs. This reciprocal regulation was at least partially microRNA dependent, as it was significantly lost in DICEREx5 HCT116 cells (Supplemental Fig. S4B, bottom panel).
Attenuated expression of PTEN ceRNAs activates the PI3K/AKT pathway
We next investigated the biological function of CNOT6L and VAPA in accordance with defined experimental criteria (Fig. 1A). On the basis of our hypothesis, the net output of any given gene on oncogenic PI3K/AKT signaling encompasses their protein function, as well as ceRNA effect on PTEN and other transcripts. We therefore decided to determine which of the PTEN ceRNAs would not solely regulate PTEN, but also yield a robust suppressive effect on PI3K/AKT as well as on growth and tumor promoting activities. Firstly, we evaluated the consequences of ceRNA-mediated PTEN regulation on the activation of the PI3K pathway. Aberrant activation of the PI3K/AKT pathway, at least in part, accounts for the pro-tumorigenic effect of PTEN loss. DU145 cells depleted of individual PTEN ceRNAs were serum starved and re-stimulated, and AKT activation determined. Consistent with the effect on PTEN protein levels, abrogation of CNOT6L or VAPA expression significantly elevated phospho-Akt (p-Akt) levels in response to serum stimulation (Fig. 5A).
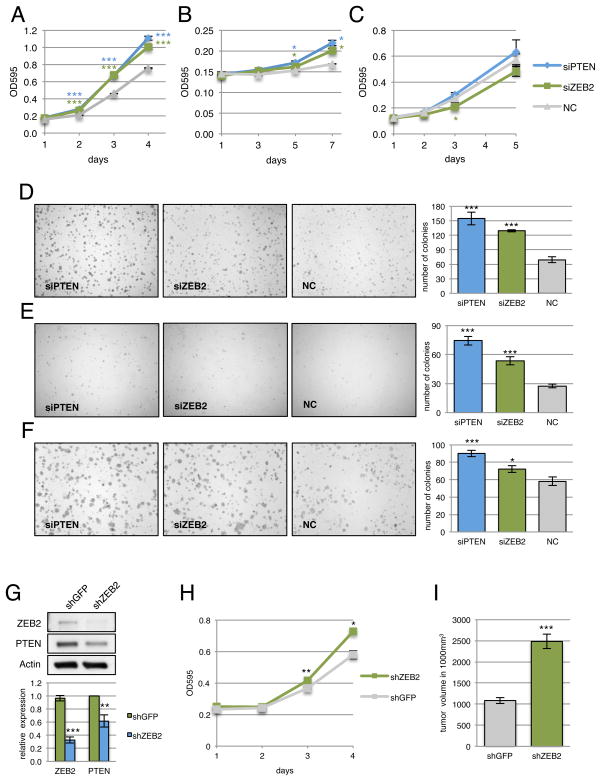
Figure 5. Depletion of PTEN ceRNAs activates the PI3K/AKT pathway and promotes growth in vitro.
(A) Western blot for phospho-AKT following serum starvation and re-stimulation of PTEN ceRNA siRNA-transfected DU145 cells (top panels), HCT116 WT (middle panels) and DICER−/− cells (lower panels). Quantitation of Western analyses is shown below the respective blots. (B) Proliferation curve of DU145 cells (top panels), HCT116 WT (middle panels) and DICER−/− cells (lower panels) transfected with siRNAs against PTEN ceRNAs. siPTEN, siCNOT6L and siVAPA result in a significant increase in growth relative to the siNC control in DU145 and HCT WT cells (P < 0.001 in DU145, P < 0.01 in HCT WT). In the HCT D−/− cells, siCNOT6L (P < 0.05) and siVAPA (P < 0.01) result in a significant decrease in growth relative to the siPTEN positive control. (C) Proliferation curve of DU145 cells transfected with plasmids overexpressing PTEN or ceRNA 3′UTRs. Relative to the empty vector control (pcDNA) transfection, CNOT 3′UTR2 (P < 0.05), VAPA 3′UTR1 (P < 0.05), VAPA 3′UTR2 (P < 0.001) and PTEN 3′UTR (P < 0.001) result in a significant decrease in growth. (B,C) Mean ± s.d., n ≥ 3.
Depletion of CNOT6L in wild-type HCT116 augmented AKT activation in response to serum starvation and re-stimulation (1.6 and 2.1 fold change at 5 and 15 min respectively relative to the negative control transfection at the same time points), (Fig. 5A) similar to the effects observed in DU145 cells. Similarly, VAPA depletion elevated p-Akt levels post-restimulation (1.8 and 2.1 fold change at 0 and 5 min respectively relative to the negative control transfection at the same time points) (Fig. 5A). Notably, the effect of CNOT6L depletion on p-AKT was completely abrogated in DICEREx5 HCT116 cells (p-Akt levels at 5 and 15 min were 0.9 and 0.4 fold respectively relative to the negative control transfection at the same time points), while the effect of VAPA depletion was significantly reduced (p-Akt levels at 0 and 5 min were 0.8 and 1.4 fold respectively relative to the negative control transfection at the same time points) (Fig. 5A). Our results therefore demonstrate that downregulation of CNOT6L and VAPA activated the PI3K/AKT pathway in a microRNA-dependent manner, consistent with their role as PTEN ceRNAs.
PTEN ceRNAs display tumor suppressive properties
As CNOT6L and VAPA appeared to phenocopy PTEN-loss mediated AKT activation based on their function as PTEN ceRNAs, we next determined whether these ceRNAs possess tumor suppressive properties, as well as their status in human cancer (Fig. 1A). To address this, we first evaluated cell proliferation and transformation in response to siRNA-mediated silencing of these PTEN ceRNAs. Reduced expression of CNOT6L or VAPA in cell lines where PTEN is regulated by these ceRNAs (DU145 and HCT116) resulted in a significant increase in proliferation similar to that observed with the PTEN siRNA (Fig. 5B, top and middle panels). This effect on growth was significantly attenuated in the DICEREx5 cells, suggesting that it is partially microRNA-dependent (Fig. 5B, bottom panel). Conversely, ectopic overexpression of 3′UTRs from PTEN and its ceRNAs, CNOT6L or VAPA, in DU145 cells led to a significant reduction in proliferation, suggesting that the observed phenotype is at least partially coding-independent (Fig. 5C). Moreover, depletion of CNOT6L or VAPA in DU145 cells promoted anchorage-independent growth in semi-solid medium (Fig. 6A).
Figure 6. VAPA and CNOT6L possess tumor suppressive properties.
(A) Anchorage-independent growth of DU145 cells transfected with siRNAs against PTEN ceRNAs in semi-solid medium. Lower panel shows quantitation of colony formation after 10 days. Mean ± s.e., n ≥ 3, ***P < 0.001. (B) Heatmap depicting the genomic status of PTEN, VAPA and CNOT6L in human colon adenocarcinoma compared to normal samples. Scale: log2 copy number units, P = 1.74e−4, 1.37e−7, 3.19e−6 for PTEN, VAPA and CNOT6L respectively. (C) Model of regulation of PTEN expression. Post-transcriptional regulation via sequestration of microRNAs by ceRNAs represents a trans-regulatory dimension of PTEN regulation.
Further support for the tumor suppressive function of CNOT6L is provided in a Sleeping Beauty insertional mutagenesis screen in oncogenic BRAF-induced melanoma, which reports that CNOT6L along with other PTEN ceRNAs, are subjected to a significant enrichment of transposon insertion sites (Karreth et al, cosubmitted). PTEN itself was one of the most significant target of insertion discovered in this study, validating the ability of this approach to identify genes implicated in human melanoma. Thus, CNOT6L dysregulation may be relevant for cancer development in an in vivo mouse model.
Finally, we examined alterations of the CNOT6L and VAPA genomic loci in human cancer. Remarkably, in a large dataset of colon cancer (GSE11417, normal n=56, colon adenocarcinoma n=52) (Kurashina et al., 2008), copy number loss of both CNOT6L (P=3.19e−6, t-test = −4.926) and VAPA (P = 1.37e−7, t-test = −5.834) was present in a significant population of samples (Fig. 6B). PTEN was also significantly downregulated in this dataset (P = 1.74e−4, t-test = −3.810). This data demonstrates the existence of significant copy number losses at both the VAPA and CNOT6L genomic loci, supporting the hypothesis that they possess tumor suppressive properties and are under selective pressure to undergo copy number losses in cancer.
DISCUSSION
The findings presented herein have allowed us to reach a number of important conclusions. Firstly, we demonstrate that protein-coding mRNA transcripts can crosstalk with other mRNA transcripts by competing for common microRNAs hence attributing to protein-coding mRNA transcripts a previously unrecognized non-coding function that is encrypted in the mRNA itself. Secondly, we establish that this cross-talk occurs through a language based on MREs which allows for bioinformatic predictions towards the definition of the ceRNA network for a given mRNA. An important implication of our findings is that, in addition to their function as cis regulatory elements that regulate the expression of their own transcripts, 3′UTRs are also trans modulators of gene expression through microRNA binding. This is particularly important given the identification of 3′UTRs expressed separately from the associated protein-coding sequences to which they are normally linked (Mercer et al., 2011).
Thirdly, we propose a set of defined rules and have developed a methodology to identify, classify and validate protein-coding ceRNAs that can crosstalk by identifying shared MREs. We have demonstrated that our approach can be predictive in finding ceRNAs that can modulate PTEN through microRNA competition. More generally, we have in so doing identified a unique means to rapidly identify previously uncharacterized mRNA regulators.
Moreover, our MRE matching methodology has allowed us to identify ceRNAs with putative tumor suppressive function, which is exerted at least in part through their ability to modulate PTEN and the PI3K signaling output (Fig. 6C). This in turn functionally links a number of unexpected biological and tumor suppressive pathways to the proto-oncogenic PTEN/PI3K signaling pathway through the ceRNA non-coding language (Supplemental Fig. S1C). For example, CNOT6L is a cytoplasmic deadenylase which is involved in poly (A) tail shortening (Wang et al., 2010) and VAPA (vesicle-associated membrane protein-associated protein-A) is an integral membrane protein implicated in transport between the endoplasmic reticulum and Golgi vesicle (Prosser et al., 2008; Wyles et al., 2002).
These ceRNAs were not functionally linked to PTEN before, and have been now related to this critical tumor suppressor on the sole basis of their predicted ceRNA function. We have validated this interaction by measuring the co-expression of such ceRNAs with PTEN, determining the growth/tumor suppressive function of such ceRNAs and demonstrating that the expression of these ceRNAs and their allelic status is decreased in cancer specimens as determined by interrogating large cancer-derived gene expression datasets. Further validation stems from our identification of PTEN ceRNAs, including CNOT6L, in vivo in an oncogenic BRAF-induced mouse model of melanoma (Karreth et al., cosubmitted).
We had recently proposed that the ceRNA language would allow RNA to communicate (Salmena et al., 2011). However, it remained to be established whether this theory could be translated into useful predictions. In this study, we define a comprehensive and integrated bioinformatic and experimental approach for the prediction and validation of ceRNA activity and networks. This approach can therefore be employed to identify and validate ceRNAs for other mRNAs of interest. That the identification of ceRNAs is indeed possible through bioinformatic predictions is corroborated by an accompanying study (Sumazin et al., this issue). It is important to note that previous studies have reported a trans-regulatory modulation of gene expression through the binding of shared microRNAs, but these studies focused mainly on the regulation of a single microRNA (Cazalla et al., 2010; Lee et al., 2009) and of conserved ceRNAs such as pseudogenes (Poliseno et., 2011). This study now extends this mechanism to potentially any mRNAs that share MREs and thus, suggests the presence of a large regulatory network built from this extensive crosstalk among mRNAs.
Although we have focused only on protein-coding RNA, the fact that ceRNA function does not rely on the protein-encoding genetic blueprint that mRNA harbors within its nucleotide sequence suggests that all MRE-containing components of the transcriptome including mRNAs, transcribed pseudogenes and long non-coding RNAs (lncRNA), are capable of regulating each other in this manner. This is supported by an accompanying study where the long non-coding RNA linc-MD1 is shown to regulate muscle differentiation by acting as a ceRNA for muscle differentiation factors MEF2C and MAML1 (Cesana et al., cosubmitted).
We therefore propose that the ceRNA language expands the total collection of functional genetic information in our genome by orders of magnitude and attributes a non-coding function to protein-coding transcripts, which may play an important role in physiological processes and pathological conditions (Salmena et al., 2011). In this study, we have therefore identified an unanticipated dimension by which RNA molecules can communicate and exert their functions, and a mean to predict these functions through the annotation of the ceRNA language.
EXPERIMENTAL PROCEDURES
MicroRNA target prediction
MicroRNA target prediction was performed using Rna22, which is available through a graphical user interface or a batch utility at http://cbcsrv.watson.ibm.com/rna22.html (Miranda et al., 2006).
Reagents
Anti-HSP90 antibody 61041 (Becton Dickinson); anti-PTEN antibody 9559 (Cell Signaling); anti-VAPA antibody sc-98890 (Santa Cruz); siGENOME siRNA reagents for non-targeting 2 (siNC), PTEN (siPTEN), ZNF460 (siZNF), CNOT6L (siCNO), SERINC1 (siSER), VAPA (siVAPA), Dharmafect 1 (Dharmacon); Lipofectamine 2000, Trizol reagent, Dulbecco’s Modified Eagle Medium (DMEM), Opti-MEM reduced serum media, fetal bovine serum (FBS) (Invitrogen); psiCHECK-2 vector, dual-luciferase reporter assay (Promega); RNeasy mini kit, DNeasy blood and tissue kit, Qiaprep spin miniprep kit (Qiagen).
Plasmids
The 3′UTRs of PTEN, CNOT6L, SERINC1 and VAPA were amplified by PCR from the genomic DNA of DU145 cells and cloned into pcDNA3.1+ according to standard protocols. Due to size constraints, CNOT6L and VAPA 3′UTRs were each cloned as two separate fragments. The PTEN 3′UTR was then subcloned into psiCHECK-2 using XhoI and NotI restriction sites. PTEN 3′UTR was also subcloned into the pMS2 vector for RIP analysis. Primer sequences are as follows: PTEN3′UTR-F TAGAGGAGCCGTCAAATCCA, PTEN3′UTR-R CCCCCACTTTAGTGCACAGT, CNOT6L3′UTR1-F AAGACGGGGATCTGTTGCTA, CNOT6L3′UTR1-R GGGCTCCTCTGTGGTTCATA, CNOT6L3′UTR2-F CCACCTACTGCTCCTTGCTC, CNOT6L3′UTR2-R CTTTTGCACGCACACTTTGT, SERINC13′UTR-F AGACTTCTAGCATGAAAGTCCCACT, SERINC13′UTR-R TTCATTTATTTAGAGGTAAAACACAGC, VAPA3′UTR1-F CCTTGTGAGGCAGTTGTTGA, VAPA3′UTR1-R TTTTGCACACAAGCAAGAGG, VAPA3′UTR2-F TCACCTCACTGCAGCTTCC, VAPA3′UTR2-R TGCAAAACTTTATTTTGATTCTCG.
Cell culture and transfection
DU145, HCT116 wild-type and DICEREx5 cells were grown in DMEM plus 10% FBS, penicillin/streptomycin and glutamine at 37°C in a humidified atmosphere with 5% CO2. For the transfection of siRNAs, DU145 or HCT116 were transfected with 100nM siRNAs in 12-well dishes at a density of 100,000 or 130,000 cells per well respectively. Transfection was performed with Dharmafect 1 according to the manufacturer’s recommendations. With this protocol, more than 90% of cells were positive for the fluorescent siGLO RISC-free control siRNA (data not shown). For plasmid transfection, DU145 were seeded in 12-well dishes at a density of 120,000 cells per well. Transfection was performed 24 hours later with Lipofectamine 2000 according to the manufacturer’s recommendations. Cells were trypsinized and seeded for the various assays 8 hours post-transfection.
Protein extraction and western blot analysis
Cells were washed in chilled PBS and lysed directly in the wells by incubating on ice for 20 min with RIPA lysis buffer containing protease inhibitors. Lysates were cleared by centrifugation at 4°C for 15 min at 12,100g, and protein concentrations were determined using Bradford dye (Bio-Rad). For western blot analysis, 10 μg of total protein was size-fractionated by SDS–PAGE on 4–12% Bis-Tris acrylamide NuPAGE gels in MOPS SDS running buffer (Invitrogen), transferred to nitrocellulose membranes (Whatman) in NuPage transfer buffer (Invitrogen) containing 10% methanol. The membranes were then probed with specific primary antibodies (see Reagents section).
RNA extraction and real-time PCR
For real-time PCR analyses, total RNA was extracted from cells using Trizol reagent as per the manufacturer’s instructions and subsequently column-purified with RNeasy kits (Qiagen). cDNA synthesis was performed using the High Capacity cDNA Archive kit according to the manufacturer’s instructions (Applied Biosystems). MicroRNA reverse transcription was performed with the microRNA reverse transcription kit according to manufacturer’s instructions (Applied Biosystems). Real-time PCR was subsequently performed using the LightCycler 480 System (Roche Applied Science).
Luciferase assays
MicroRNA target validation assays were performed as described previously (Tay et al., 2008). DU145 cells were seeded 24 h before transfection at a density of 120,000 cells/well in 12-well plates. 100ng of empty psiCHECK-2 vector or psiCHECK-2+PTEN3′UTR was co-transfected with 100nM siRNA or 1μg vector constructs with Lipofectamine 2000 according to manufacturer’s instructions. In all cases, a constitutively expressed firefly luciferase gene in psiCHECK-2 was used as a normalization control for transfection efficiency. 72 hours after transfection, firefly and Renilla luciferase activities were measured consecutively with the dual-luciferase reporter system (Promega) using a luminometer (Promega).
Cell proliferation
8 hours post-transfection, DU145 cells were trypsinized, resuspended and seeded in 4 separate 12-well plates at a final density of 20,000/well. Starting from the following day (d0), one plate per day was washed once with PBS, fixed in 10% formalin solution for 10 min at room temperature and then kept in PBS at 4°C. On the last day, all the wells were stained with crystal violet. After lysis with 10% acetic acid, optical density was read at 595 nm.
Growth in semisolid medium
The bottom layer was obtained by covering 6-well dishes with 3 ml of 0.6% agar in DMEM. The day after, transfected DU145 cells were seeded on top in triplicate in 2 ml of 0.3% agar in DMEM plus 10% FBS. Colonies were counted after 10 days at 40× magnification.
RNA-immunoprecipitation and analysis
Cells were lysed on ice 48 hours after transfection with MS2 plasmids (Kim et al., 2009), and RIPs were performed as previously described (Keene et al., 2006). Briefly, cell lysates were incubated overnight at 4°C with control Rabbit IgG (Jackson ImmunoResearch) or anti-YFP antibody (Santa Cruz) that were bound to Protein A-Sepharose beads (Sigma). Beads were then washed 5 times with NT2 buffer, and RNA was eluted by incubation with SDS-TE buffer by heating at 55°C for 30 minutes. RNA was then precipitated using the standard Trizol (Invitrogen) protocol and analyzed via real-time PCR.
PTEN and ceRNA co-expression analysis and genomic status assessment
Co-expression in prostate cancer was evaluated using GEO super-series GSE21032 (Taylor et al., 2010). From this data set, whole-transcript expression data for human primary and metastatic prostate cancer samples (GSE21034) and microRNA expression data for human primary and metastatic prostate cancer samples (GSE21036) were analyzed. Processed and normalized expression data was downloaded from NCBI. To study differential expression, samples were subdivided into two subsets according to PTEN expression: a PTEN-down subset with samples showing a PTEN expression level lower than the PTEN average value calculated among all samples and a PTEN-up subset characterized by a PTEN expression level higher than this average value. To select subsets of samples with lower or higher microRNAs expression level, at least one microRNA, out of 10 microRNAs targeting PTEN, was required to show an expression level lower (or higher) than a standard deviation from its average level; samples where the different levels of expression among microRNAs were not consistent were discarded. 33 samples were thus obtained for the subset with higher expression and 26 for that with lower expression. Differential expression in glioblastoma was assessed by the analysis of GEO series GSE15824. Processed and normalized expression data was downloaded from NCBI, entrez gene ID were linked to Affymetrix probesets by Affymetrix annotation na31. Co-expression between PTEN and the ceRNA genes were analyzed in multi-tissues and normal-tissue-specific human-mouse conserved and human-specific co-expression networks generated from a specific annotated database (http://www.cbu.mbcunito.it/ts-coexp) (Piro et al., 2011); the co-expression was considered significant if it was found in the top 1% of the ranked list of genes co-expressed with PTEN. Genomic status was assessed using the GEO series GSE11417 (Kurashina et al., 2008). Oncomine™ (Compendia Bioscience, Ann Arbor, MI) was used for analysis and visualization. (www.oncomine.org)
Statistical analysis
In vitro data were analyzed using unpaired Student’s t-test. Values of P < 0.05 were considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001. The mean ± s.d. of three or more independent experiments is reported.
Supplementary Material
Figure 7.
Acknowledgments
We thank Pandolfi laboratory members for critical discussions. Real-time PCR analysis was conducted through the Harvard Catalyst Laboratory for Innovative Translational Technologies with support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (NIH Award #UL1 RR 025758 and financial contributions from Harvard University and its affiliated academic health care centers). The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, the National Center for Research Resources, or the National Institutes of Health. We thank B. Vogelstein for DICEREx5 cells, M. Gorospe for the pMS2 plasmids and I. Legnini for experimental support. Y.T. was supported by a Special Fellow Award from The Leukemia & Lymphoma Society. L.K. was supported by an NHMRC Overseas Postdoctoral Fellowship. L.S. was supported by fellowships from the Human Frontier Science Program and the Canadian Institutes of Health Research. U.A. was supported by a fellowship from the Fondazione per la Ricerca Biomedica ONLUS of Torino. S.M.T. was supported by a Department of Defense (DOD) Breast Cancer Research Program (BCRP) postdoctoral fellowship award. I.R. was supported by funding from the Jefferson Medical College of Thomas Jefferson University. This work was supported in part by NIH grant R01 CA-82328-09 to P.P.P.
Footnotes
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, Salmena L, Sampieri K, Haveman WJ, Brogi E, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Gen. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumor suppression. Nature. 2011 doi: 10.1038/nature10275. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, Ie Sage C, Agami R, Tuschl T, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyapalan Z, Deng Z, Shatseva T, Fang L, He C, Yang BB. Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2011;39:3026–3041. doi: 10.1093/nar/gkq1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M. Emerging novel functions of RNAs, and binary phenotype? Dev Biol. 2008;317:401–404. doi: 10.1016/j.ydbio.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Kurashina K, Yamashita Y, Ueno T, Koinuma K, Ohashi J, Horie H, Miyakura Y, Hamada T, Haruta H, Hatanaka H, et al. Chromosome copy number analysis in screening for prognosis-related genomic regions in colorectal carcinoma. Cancer Sci. 2008;99:1835–1840. doi: 10.1111/j.1349-7006.2008.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Shatseva T, Jeyapalan Z, Du WW, Deng Z, Yang BB. A 3′-untranslated region (3′UTR) induces organ adhesion by regulating miR-199a* functions. PLoS One. 2009;4:e4527. doi: 10.1371/journal.pone.0004527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Wilhelm D, Dinger ME, Solda G, Korbie DJ, Glazov EA, Truong V, Schwenke M, Simons C, Matthaei KI, et al. Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res. 2011;39:2393–2403. doi: 10.1093/nar/gkq1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D’Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro RM, Ala U, Molineris I, Grassi E, Bracco C, Perego GP, Provero P, Di Cunto F. An atlas of tissue-specific conserved coexpression for functional annotation and disease gene prediction. Eur J Hum Genet. 2011 Jun 8; doi: 10.1038/ejhg.2011.96. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010a;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010b;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser DC, Tran D, Gougeon PY, Verly C, Ngsee JK. FFAT rescues VAPA-mediated inhibition of ER-to-Golgi transport and VAPB-mediated ER aggregation. J Cell Sci. 2008;121:3052–3061. doi: 10.1242/jcs.028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie W, Flamant S, Rasko JE. Predicting microRNA targets and functions: traps for the unwary. Nat Methods. 2009;6:397–398. doi: 10.1038/nmeth0609-397. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H. Redefining microRNA targets. Curr Biol. 2009;19:870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, Khoo AS, Roy-Burman P, Greenberg NM, Van Dyke T, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Morita M, Yang X, Suzuku T, Yang W, Wang J, Ito K, Wang Q, Zhao C, Bartlam M, et al. Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J. 2010;29:2566–2576. doi: 10.1038/emboj.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyles JP, McMaster CR, Ridgway ND. Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J Biol Chem. 2002;277:29908–29918. doi: 10.1074/jbc.M201191200. [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.