Abstract
The fidelity of DNA synthesis by A-family DNA polymerases ranges from very accurate for bacterial, bacteriophage and mitochondrial family members to very low for certain eukaryotic homologues. The latter include Pol ν which, among all A-family polymerases, is uniquely prone to misincorporate dTTP opposite template G in a highly sequence-dependent manner. Here we present a kinetic analysis of this unusual error specificity, in four different sequence contexts and in comparison to Pol ν’s more accurate Family A homologue, the Klenow fragment of E. coli DNA polymerase I. The kinetic data strongly correlate with rates of stable misincorporation during gap-filling DNA synthesis. The lower fidelity of Pol ν compared to Klenow fragment can be attributed primarily to a much lower catalytic efficiency for correct dNTP incorporation, whereas both enzymes have similar kinetic parameters for G-dTTP misinsertion. The major contributor to sequence-dependent differences in Pol ν error rates is the reaction rate, kpol. In the sequence context where fidelity is highest, kpol for correct G-dCTP incorporation by Pol ν is ~ 15-fold faster than kpol for G-dTTP misinsertion. However, in sequence contexts where the error rate is higher, kpol is the same for both correct and mismatched dNTPs, implying that the transition state does not provide additional discrimination against misinsertion. The results suggest that Pol ν may be fine-tuned to function when high enzyme activity is not a priority and may even be disadvantageous, and that the relaxed active-site specificity towards the G-dTTP mispair may be associated with its cellular function(s).
Cells rely on a variety of processes to ensure faithful DNA replication and repair and DNA polymerases play key roles in these processes. The human genome encodes at least 15 DNA polymerases (reviewed in (1)). Three of these enzymes, DNA polymerases γ (Pol γ)1, θ (Pol θ) and ν (Pol ν), are members of the A-family of DNA polymerases because they share homology with Escherichia coli DNA polymerase I, encoded by the polA gene. The focus of this study is Pol ν, encoded by the human POLN gene (2). Although the biological role of Pol ν is uncertain, one clue to its possible function is its homology to Drosophila melanogaster Mus308, a Family A DNA polymerase with a helicase domain at the N-terminus (3). mus308 mutants are hypersensitive to DNA crosslinking agents (nitrogen mustard and cisplatin) but not to MMS (4), implicating Mus308 in the repair of highly toxic interstrand cross-links (2, 4, 5). POLN-knockdown cells are sensitive to various DNA-damaging agents (6, 7) including DNA cross-linking agents, indicating that Pol ν may be involved in repairing lesions generated by these agents. There is also functional and physical evidence for the interaction of Pol ν with proteins involved in the Fanconi-anemia pathway that is involved in repair of cross-linked DNA (7). Consistent with possible roles in cross-link repair, Pol ν can bypass a psoralen DNA interstrand crosslink (6) and DNA interstrand cross-links with linkages through the DNA major groove (8). It can also bypass a subclass of thymine glycol lesions (9). Pol ν is a processive enzyme and performs efficient strand-displacement synthesis, and these properties may be important for its biological function.
Several Family A DNA polymerases, including E. coli Pol I (10), T7 DNA polymerase (11, 12) and Pol γ (13-15), have intrinsic 3′ to 5′ exonuclease activity that can edit the occasional mismatches they create, thereby enhancing the fidelity with which they synthesize DNA (16, 17). Other Family A DNA polymerases, such as Taq DNA polymerase (18, 19) and Bst DNA polymerase (20), lack intrinsic 3′ to 5′ exonuclease activity and therefore cannot proofread their mistakes. Nonetheless, they are among the most accurate of the naturally exonuclease-deficient polymerases (18-23). Pol θ and Pol ν comprise yet a third subtype of Family A polymerase. They too lack 3′ to 5′ exonuclease activity, but they have low nucleotide selectivity. For example, compared to the exonuclease-deficient form of the Klenow fragment of E. coli Pol I, human Pol θ has much lower selectivity and forms a variety of different single base mismatches at high rates (24). Human Pol ν also has low nucleotide selectivity, but its specificity is much more biased, specifically for misinsertion (9) and stable misincorporation of dTTP opposite template G (25). Moreover, when stable misincorporation of dTTP was monitored opposite a large number of template guanines, the site-to-site variation in error rate for the G-dTTP mismatch was more than 30-fold (25).
Previous studies of error specificity have provided clues to the biological functions of DNA polymerases. For example, studies of the error specificity of human Pol η (26) eventually led to the conclusion that it is responsible for base substitutions at A-T base pairs during somatic hypermutation of immunoglobulin genes (27-29). Also, studies of the error specificity of yeast replicases (30-32) have led to a better understanding of leading and lagging strand DNA replication in vivo. It is our hope that the unusual error specificity of human Pol ν will likewise result in a better understanding of its biological function(s). In the present study we have focused particularly on the sequence dependence of misincorporation, a well documented phenomenon which remains poorly understood. Pol ν’s preference for forming G-dTTP mispairs in a highly sequence-dependent manner allowed us to investigate the kinetic parameters for correct and incorrect dNTP insertion and mismatch extension at hotspots and coldspots. Examining the kinetic parameters for correct and incorrect dNTP insertion by Pol ν in four different sequence contexts, we have found that the the error rate for stable misincorporation of dTTP opposite template G correlates strongly with the kinetic parameters for correct dNTP insertion, as has been previously noted for a variety of DNA polymerases (33, 34).
Experimental Procedures
Enzymes, reagents, DNA oligonucleotides
Materials for the M13mp2 fidelity assay were from sources described previously (35). Ultrapure dNTPs were purchased from Amersham Biosciences (GE Healthcare). Pol ν Fragment 1 (E175-G863, 77.1 kDa, Pol ν-77) was expressed and purified as previously described (9, 36). Exonuclease-deficient (D424A) Klenow fragment was purified according to our published procedures (37, 38). DNA oligonucleotides were synthesized by the Keck Biotechnology Resource Laboratory at Yale Medical School and were purified by denaturing gel electrophoresis as described previously (39).
M13mp2 fidelity assay
The fidelity assay scores errors generated in the lacZ α-complementation gene in M13mp2 during synthesis to fill a 407-nucleotide gap. Reaction mixtures (25 μl) to fill the gap contained 0.2 nM M13mp2 gapped DNA substrate, 20 mM Tris-HCl (pH 7.5 or 8.8), 8 mM magnesium acetate, 2 mM dithiothreitol, 80 μg of bovine serum albumin, 4% (v/v) glycerol and 1 mM each of dATP, dGTP, dCTP and dTTP. Reactions were initiated by adding Pol ν-77 (200 nM), incubated at 37°C for 30 min, and terminated by adding EDTA to 20 mM. Half of the reaction mixture was mixed with SDS buffer (20 mM Tris-HCl, pH 8.0, 5 mM EDTA, 5% SDS, 0.5% bromophenol blue and 20% glycerol) and analyzed by agarose gel electrophoresis. The results indicated that the gap was completely filled for reactions performed at both pH values (data not shown, but for a typical result, see (25)). Aliquots of the remaining DNA products were then used to determine lacZ mutant frequencies for the purpose of obtaining error rates, as described (25). Correct synthesis produces M13mp2 DNA that yields dark blue M13 plaques when introduced into an E. coli α-complementation strain and plated on indicator plates. Polymerase errors are scored as light blue or colorless M13 plaques. DNA from independent mutant clones was sequenced to define the sequence changes in lacZ, and this information was used to calculate error rates as described in (35). Error rates for individual types of mutation were calculated according to Equation: ER=((Ni/N)×MF)/(D×0.6) where Ni is the number of mutations of a particular type, N is the total number of mutants analyzed, MF is frequency of lacZ mutants, D is the number of detectable sites for the particular type of mutation, and 0.6 is the probability of expressing a mutant lacZ allele in E. coli (35).
Chemical quench kinetic measurements
Table 1 lists the sequences of the duplex DNA substrates used in this study. Primer strands, 5′-labeled with 32P, were annealed to a 1.5-fold molar excess of the appropriate template strand. Primer extension reactions were carried out at room temperature (20-22 °C) using a rapid-quench-flow instrument (KinTek Corp., Model RQF-3) for fast reactions and manual quenching when the reaction was sufficiently slow. In either case, the reaction was initiated by mixing equal volumes of a polymerase-DNA mix and a dNTP solution, both in Pol ν reaction buffer, 20 mM Tris-HCl, pH 7.5, 8 mM Mg(OAc)2, 2 mM DTT, 4 % (v/v) glycerol, and 80 μg/ml bovine serum albumin. The final reaction mixture contained 5 nM primer-template duplex, 7 nM Pol ν-77 and varying dNTP concentrations. The preparation of Pol ν-77 used in the majority of the kinetics measurements was ~ 23% active, and therefore the concentrations listed above gave burst kinetics (40, 41) with a burst amplitude of ~ 0.3. Reactions were quenched at appropriate time intervals using excess EDTA and were fractionated on denaturing polyacrylamide-urea gels and quantitated on a Fuji FLA 5100 scanner. The fraction of labeled DNA converted to product was plotted as a function of reaction time and fitted to the burst equation: A(1 – exp(−kobst)) + ksst .
Table 1. DNA duplex oligonucleotides used in kinetics experiments.
| Name | DNA sequence | |
|---|---|---|
| Misinsertion substrates | ||
| 13/19mer-G | 5′GAGTCAACAGGTC 3′CTCAGTTGTCCAGGTATGG |
|
| 13/19mer-T | 5′GAGTCAACAGGTC 3′CTCAGTTGTCCAGTGATGG |
|
| G(165) hotspot | 5′GCGATCGGTGCGGG 3′CGCTAGCCACGCCCGGAGAAGCGA |
|
| G(164) hotspot | 5′GCGATCGGTGCGGGC 3′CGCTAGCCACGCCCGGAGAAGCGA |
|
| G(−66) coldspot | 5′TGAGTGAGCTAACT 3′ACTCACTCGATTGAGTGTAATTAA |
|
| Mismatch extension substrates (X = C or T) | ||
| 14/19mer-G | 5′GAGTCAACAGGTCX 3′CTCAGTTGTCCAGGTATGG |
|
| G(165) hotspot | 5′GCGATCGGTGCGGGX 3′CGCTAGCCACGCCCGGAGAAGCGA |
|
| G(−66) coldspot | 5′TGAGTGAGCTAACTX 3′ACTCACTCGATTGAGTGTAATTAA |
Burst rate constants (kobs, corresponding to the rate constant for the first turnover) were determined at a series of dNTP concentrations and kpol and Kd were determined from a plot of kobs vs. dNTP concentration fitted to the hyperbolic equation: kobs = kpol[dNTP]/(Kd + [dNTP]). In a few instances, where lack of sufficient Pol ν precluded use of the rapid-quench instrument, rate measurements were carried out at very low dNTP concentrations using manual quenching. At low dNTP concentrations, the hyperbolic equation reduces to kobs = kpol[dNTP]/Kd, allowing the ratio kpol/Kd to be obtained directly from the slope of the plot of kobs against dNTP concentration.
Gel mobility shift studies of DNA binding
Pol ν-77, at a series of concentrations from 0.05 to 100 nM, was incubated with 32P-labeled duplex DNA in Pol ν reaction buffer, plus an additional 10% glycerol, for 10 min at 22 °C. The samples were loaded onto an 8% polyacrylamide gel, poured and run in 50 mM Tris borate, 2 mM MgCl2 and 0.1 mM EDTA. The samples were loaded with the gel running at 300 V, and then electrophoresis was continued at 150 V for 3 to 4 h at 4 °C. The gel was dried down and radioactivity was quantitated on a Fuji FLA 5100 scanner. Two types of information were obtained from plots of the fraction of bound DNA as a function of the concentration of Pol ν-77, as illustrated in Supplementary Figure S1. With DNA at 1 nM, which is > KD, the fraction of DNA bound to Pol ν-77 is given by f[Pol]/[DNA], so that the slope of the graph allowed us to determine f, the fraction of the added Pol ν-77 that is active in binding DNA. At a DNA concentration below KD, the fraction of bound DNA approximates to the hyperbolic relationship [Pol]/([Pol]+ KD) allowing calculation of KD.
Results
Error specificity
Our earlier study highlighted the tendency of full-length human Pol ν to generate G to A substitutions via misincorporation of dTTP opposite template guanine (25). We began this study by confirming this error signature for a slightly abbreviated derivative of Pol ν (the eventual goal being structural studies), designated Pol ν-77 (36). We used the M13mp2 forward mutation assay, which detects a wide range of sequence changes in the lacZ gene in a variety of sequence contexts (35). The DNA products of gap-filling synthesis at pH 8.8 yielded an average lacZ mutant frequency of 15% (Table 2), comparable to the 18% value reported previously for the full-length protein (25, 36). In addition to similar error rates, the truncated and full-length Pol ν proteins have similar processivity (compare Figure 2 in reference (25) and Figure 4 in (36)). We also examined the fidelity at pH 7.5, the pH used in our kinetic studies below. Pol ν is more accurate at pH 7.5, as revealed by a 4-fold lower average mutant frequency of 3.5% (Table 2). Higher accuracy at lower pH is also a characteristic of other Family A DNA polymerases, including Taq polymerase (19) and the large Klenow fragment of E. coli pol I (42). When 204 independent lacZ mutants were sequenced from reactions performed at pH 7.5, a variety of single base changes were observed (Table 2). These were distributed throughout the lacZ target sequence (Figure 1), in a pattern similar to that seen earlier with full-length human Pol ν at pH 8.8 (25). Substitutions predominated, and the vast majority were G to A changes, yielding an average error rate of 22 × 10−4 for stable misincorporation of dTTP opposite the 22 template guanines where this event results in a change in plaque color (35). The second most common error was the mismatch involving the same two bases but differing with respect to the templating versus incoming base, i.e., the T-dGTP mismatch (average error rate of 2.0 × 10−4, Table 2). We have previously compared Pol ν specific error rates to the error rates of other DNA polymerases (see Table 2 in (25)).
Table 2. Summary of sequence changes generated by Pol ν-77.
| pH 7.5a | pH 8.8b | |||
|---|---|---|---|---|
| lacZ Mutant Frequency | 0.035 | 0.15 | ||
| Total mutants sequenced | 204 | 152 | ||
| Total bases sequenced | 83,028 | 61,864 | ||
| Detectable changes c | # of changes | Error Rate (×10−4) | # of changes | Error Rate (×10−4) |
| Base substitutions | 204 | 4.7 | 227 | 30 |
| Frameshifts (−1) | 16 | 0.23 | 15 | 1.2 |
| Frameshifts (+1) | 10 | 0.14 | 7 | 0.6 |
| G · dTTP | 168 | 22.0 | 190 | 150 |
| T · dGTP | 19 | 2.0 | 17 | 10 |
Error rates and mutant frequencies: Experiment 1: Mutant frequency was 2.1% (47 mutant plaques from a total of 2240) and Experiment 2: Mutant frequency was 4.8% (192 mutant plaques from a total of 3994).
Error rates and mutant frequencies were calculated using data from three independent experiments. Experiment 1: Mutant frequency was 16.0% (484 mutant plaques from a total of 2947). Experiment 2: Mutant frequency of 15% (581 mutant plaques from a total of 3771). Experiment 3: Mutant frequency was 13% (783 mutant plaques from a total of 5927).
Error rates calculated from detectable changes ((35) also see Experimental Procedures).
The total number of specific mutations reported is greater than the number of lacZ mutants sequenced due to the presence of >1 detectable errors in certain mutants.
Figure 2.
Kinetics of Pol ν-77. Panel A shows the kinetics of incorporation of dATP into the 13/19mer-T substrate (Table 1). The reactions contained 5 nM DNA primer termini, 100 μM dATP and the indicated concentrations of Pol ν-77 (measured as total protein). Because the Pol ν-77 prep was not fully active, all three concentrations of Pol ν-77 showed burst kinetics, with the burst amplitude providing a measure of the concentration of active enzyme. A plot of burst amplitude against the concentration of Pol ν-77 (Panel B) had a slope of 0.23, indicating that this Pol ν-77 preparation was 23% active. Panel C shows dTTP misinsertion by Pol ν-77 opposite template G within the G164 hotspot sequence, measured under burst conditions at a series of dTTP concentrations. The burst rate of G-dTTP misinsertion was plotted as a function of dTTP concentration and fitted to a hyperbolic equation, giving Kd(dTTP) = 35 μM, and kpol = 0.076 s−1.
Figure 4.
Regions of the A-family DNA polymerase structure where Pol ν shows interesting differences from the classical A-family conserved motifs. Panel A shows the structure of the polymerase domain of the ternary (Pol-DNA-dNTP) complex of Klentaq (3KTQ, ref. (54)). The protein structure is shown in grey, with residues 501-523 at the tip of the thumb subdomain omitted to avoid obscuring the active site region. The DNA duplex is shown in blue, with the template strand darker, and the incoming dNTP in cyan with CPK coloring. Protein sequence motifs discussed in the text are colored: the N-terminus of the N-helix (including the conserved histidine side-chain) in red, the O-helix in yellow, and the N-terminal portion of Motif 6, within the Q-helix, in green. Panel B shows the alignment of the corresponding portions of sequence from human Pol ν, compared with Klenow fragment and Klentaq, representing the classical A-family DNA polymerases. The motif designations are from Patel et al. (23). Residues highly conserved in the alignment of both classical and Pol ν-type A-family DNA polymerases (51) are highlighted with the colors of the corresponding motifs in Panel A.
Figure 1.
Spectrum of errors generated by human Pol ν-77 at pH 7.5. The 407 template nucleotides within the single-strand gap of the M13mp2 substrate are shown as 5 lines of the template sequence, bracketed by 5 base pairs at either end to indicate the boundaries of the gap. +1 represents the first transcribed nucleotide of the lacZα-complementation region. Letters above the target sequence indicate base substitutions made by Pol ν-77. Single-base deletions are represented by open triangles below the target sequence whereas single-base additions are depicted by closed inverted triangles above the target sequence. Red characters represent phenotypically detectable changes in the gap region while black characters represent phenotypically undetectable changes found in association with detectable changes. Also depicted in the figure are the error rates at positions: 169, 165, 151, 149, 148, 145, 141, 89 and 88.
As before (25), G to A mutation rates varied by sequence context. The highest mutation rate for a phenotypically detectable site was observed at template guanine number +165 (designated the G(165) hotspot), where 38 substitutions were observed among 204 sequenced lacZ mutants. This corresponds to an error rate of 1.1%; because of this high error rate, the G(165) hotspot sequence was chosen for our kinetic analysis. Additionally, we measured dNTP insertion kinetics at the neighboring guanine, G(164), which is also a hotspot. Although substitutions at G(164) are phenotypically silent, two G to A substitutions were observed as silent hitchhikers among 204 sequenced lacZ mutants, corresponding to an error rate of 1.6%, similar to the error rate at G(165) 2. For comparison, we analyzed Pol ν kinetics at a mutational coldspot, G(−66), selected because only three G to A changes were detected at this phenotypically-detectable position, corresponding to an error rate of 0.09%. dNTP misinsertion. The design of the kinetics experiments was influenced by the properties of the Pol ν-77 preparations. Purification of Pol ν-77 is challenging and the purified protein (36) was typically not fully active, as shown by active-site titration using the measurement of burst amplitudes in primer extension reactions (Figure 2A, B). An alternative method using DNA binding to assess the concentration of active Pol ν-77 is illustrated in Supplementary Figure S1. Measurements by the two methods were in good agreement. The Pol ν-77 preparation used in the majority of the kinetics experiments contained 23% active polymerase. As described in Methods, we measured reaction rates under burst conditions, with the concentration of active Pol ν-77 about 3-fold less than the concentration of the DNA substrate. This had the advantage of conserving our stocks of enzyme and also avoided the problem that high concentrations of some Pol ν-77 preparations inhibited the reaction.
We measured misinsertion by Pol ν-77 on several different DNA substrates (Table 1). The 13/19-mer duplex is a DNA substrate that we have used routinely in recent kinetic studies of Klenow fragment (38, 43). We compared misinsertions at a template T and a template G in this sequence. The results with Klenow fragment (Table 3, line 2 of each section) were in good agreement with data previously obtained on a different DNA sequence (reproduced in Table 3, line 1 of each section) (44). As described above, three other DNA substrates were derived from the lacZ target sequence, in order to study dTTP misinsertion opposite template guanines in three different sequence contexts: the G(164) and G(165) hotspots and the G(−66) coldspot. In each case, we measured the rates of correct and mismatched dNTP incorporation at a series of dNTP concentrations and calculated kpol and KD(dNTP). Figure 2C and D shows an example of the data obtained for T misinsertion opposite G(164). The misinsertion data obtained for Pol ν-77, with selected Klenow fragment misinsertions for comparison, are shown in Table 3. The final column of Table 3 compares the efficiencies of correct and incorrect dNTP addition on each substrate, giving the selectivity in favor of the correct complementary dNTP.
Table 3. Kinetics of dNTP insertion catalyzed by Pol ν-77 and by exonuclease-deficient Klenow fragment.
| Protein/DNAa | Correct dNTP |
Incorrect dNTP | |||||
|---|---|---|---|---|---|---|---|
| Kd(μM) | kpol(s−1) |
kpol/Kd (M−1 s−1) |
Kd(μM) | kpol(s−1) |
kpol/Kd (M−1 s−1) |
Selectivityb | |
| G·dCTP | G·dTTP | dC/dT | |||||
| Pol I(KF)c | 8.6 | 130 | 1.5 × 107 | 180 | 0.03 | 160 | 9.4 × 104 |
| Pol I(KF), 13/19mer-G | 9.4 ± 0.7 | 63 ± 7 | 6.7 × 106 | 210d | 0.042d | 200 | 3.4 × 104 |
| Pol ν, 13/19mer-G | 75 ± 7 | 0.40 ± 0.003 | 5.3 × 103 | 200 ± 15 | 0.028 ± 0.01 | 140 | 38 |
| Pol ν, G(164) hotspot | 0.93 ± 0.34 | 0.057 ± 0.016 | 6.1 × 104 | 30 ± 8 | 0.073 ± 0.005 | 2.5 × 103 | 25 |
| Pol ν, G(165) hotspot | 1.2 ± 0.3 | 0.025 ± 0.01 | 2.1 × 104 | 24 ± 8 | 0.022 ± 0.001 | 940 | 22 |
| Pol ν, G(−66) coldspot | 15 ± 7 | 0.28 ± 0.004 | 1.9 × 104 | 480 ± 140 | 0.018 ± 0.006 | 36 | 520 |
|
| |||||||
| T·dATP | T·dGTP | dA/dG | |||||
| Pol I(KF) c | 9.1 | 144 | 1.6 × 107 | 48 | 0.16 | 3.4 × 103 | 4.7 × 103 |
| Pol I(KF), 13/19mer-T | 8.6 ± 1.0 | 110 ± 2 | 1.2 × 107 | 87 ± 2 | 0.083 ± 0.01 | 950 | 1.3 × 104 |
| Pol ν, 13/19mer-Te | - | - | 1.9 × 103 | 140 ± 7 | (5.1 ± 0.2) × 10-4 | 3.7 | 510 |
DNA substrates are listed in Table 1; Pol I(KF) is exonuclease-deficient (D424A mutation); Pol ν is the Pol ν-77 fragment.
Calculated as (kpol/Kd)correct/(kpol/Kd)incorrect.
From reference (44), using a different DNA substrate.
Single determinations; all others were the average of two or more determinations.
(kpol/Kd)correct was determined from the initial slope of the plot of kobs against dATP concentration, at low concentrations of dATP. (Average of two measurements.)
The misinsertion data for the 13/19-mer substrate fit well with the overall mutational data for both Klenow fragment and Pol ν. For example, Klenow fragment has 3-fold lower discrimination against T-dGTP errors (1.3 × 10−4, bottom of Table 3) compared to G-dTTP errors (3.4 × 10−4, top of Table 3), consistent with its higher G-dTTP error rate in the M13mp2 forward mutation assay (10). For both T-dGTP and G-dTTP errors, the individual kinetic constants for Klenow fragment show that the selectivity in favor of the correct base pair is made up of ~ 10-fold discrimination in dNTP binding and ~ 103-fold discrimination in the rate of misincorporation, with the kinetic differences between correctly paired and mispaired substrates slightly larger in the case of G-dTTP errors. For Pol ν, the most frequent error in the M13mp2 assay is G-dTTP, whose rate is 11-fold higher than that of T-dGTP (Table 2). Again, this is reflected in the kinetic data, where we observe a 13-fold difference in discrimination against the two mismatches (selectivity values of 38 for G-dTTP versus 510 for T-dGTP, third lines in top and bottom sections of Table 3).
What accounts for the low fidelity of Pol ν for G-dTTP errors? Both Klenow fragment and Pol ν have similar kinetic constants for misinsertion of dTTP opposite template G (Table 3, top right). However, correct dCTP insertion by Pol ν opposite template G is ~ 103-fold less efficient than is correct dCTP insertion catalyzed by Klenow fragment (Table 3, top left). The difference in the efficiency of correct incorporation therefore translates directly to ~ 103-fold lower selectivity of Pol ν against G-dTTP mismatches, compared with Klenow fragment. By contrast, at template T, dGTP misinsertion by Pol ν is ~ 260-fold less efficient than that of Klenow fragment (Table 3, lower right, compare 950 vs. 3.7). This difference moderates the effect of the large (6,300-fold) difference in the efficiency of correct (T-dATP) incorporation (Table 3, lower left, compare 1.2 × 107 to 1.9 × 103), resulting in ~ 24-fold lower selectivity of Pol ν against T-dGTP errors.
The kinetic data for G-dTTP misinsertion by Pol ν on the lacZ sequences also parallel the mutational data. Error rates in the M13mp2 fidelity assay are high at template G(165) and G(164) and lower at template G(−66). Likewise, selectivity against dTTP misinsertion (Table 3) is at least 20-fold higher opposite template G(−66) (selectivity factor of 520) than opposite templates G(165) and G(164) (selectivity factors of 22 and 25, respectively). The different selectivity values derive from some very interesting sequence-dependent differences in individual kinetic constants. First, the binding constants for both correct dCTP and incorrect dTTP are 15- to 20-fold weaker at the G(−66) position as compared to the G(164) and G(165) positions. At G(−66), where the error rate is lowest, incorporation of correct dCTP is ≈ 16-fold faster than misinsertion of dTTP (Table 3, kpol of 0.28 and 0.018, respectively). In contrast, at the G(164) and G(165) hotspots, the kpol values for correct dCTP incorporation are slower than at G(−66), and similar to those for misinsertion of dTTP at these same hotspot sequences. Therefore, the lower fidelity of Pol ν for G-dTTP mismatches in one sequence context as compared to another can be attributed largely to differences in the kinetics of correct G-dCTP incorporation, particularly the rate of dCTP addition, since the ratio of dCTP and dTTP binding constants is very similar at all three sequences. The kinetic constants for the template G in the 13/19-mer DNA substrate resemble to some extent those of the G(−66) sequence, with weak binding constants and a fast rate of G-dCTP incorporation.
Mismatch extension
To examine whether the mispair specificity of Pol ν operates at the level of mismatch extension, we measured the rates of extension of DNA substrates having a terminal (primer)C-G(template) basepair or a T-G mismatch (Table 4). On the 14/19-mer extension substrate, both Klenow fragment and Pol ν showed a similar discrimination between C-G and T-G termini, suggesting that mismatch extension does not account for the difference in mutational specificity of these two DNA polymerases. However, the discrimination by Pol ν against extension of the T-G mismatch was modulated by sequence context, with ~ 8-fold less discrimination at G(165) as compared to G(−66). The difference was primarily due to a faster rate of mispair extension at (G165); the rates of extension of a C-G terminus were similar. Gel mobility shift assays (Figure 3) indicated that binding of Pol ν to the T-G mismatch-extension substrates corresponding to the G(165) and G(−66) template-primers was indistinguishable (KD = 0.1 nM, in terms of fully active Pol ν). Therefore the behavior of Pol ν-77 on the two substrates does not result from a difference in binding affinity. Likewise, there was no difference in the DNA binding behavior of Pol ν when comparing the corresponding insertion substrates and C-G extension substrates at the same two sequences (data not shown).
Table 4. Kinetics of mismatch extension catalyzed by Pol ν-77 and by exonuclease-deficient Klenow fragment.
| Protein/DNAa | (primer)C-G | (primer)T-G | |||||
|---|---|---|---|---|---|---|---|
| Kd(μM) | kpol(s−1) |
kpol/Kd (M−1 s−1) |
Kd(μM) | kpol(s−1) |
kpol/Kd (M−1 s−1) |
Selectivityb | |
| Pol I(KF), 14/19mer-G (+dATP) | 5.4 + 1.8 | 68 + 14 | 1.3 × 107 | 230 + 60 | 0.43 + 0.05 | 1.8 × 103 | 6.8 × 103 |
| kobs(s−1) at 1 mM dNTP | kobs(s−1) at 1 mM dNTP | Rate(CG):(TG) | |||||
| Pol I(KF), 14/19mer-G (+dATP)c | 68 | 0.35 | 190 | ||||
| Pol ν, 14/19mer-G (+dATP) | 0.25d | (1.8 + 0.2) × 10−3 | 140 | ||||
| Pol ν, G(165) hotspot (+dCTP) | 0.16 + 0.01 | (4.4 + 0.6) × 10−3 | 36 | ||||
| Pol ν, G(-66) coldspot (+dATP) | 0.22 + 0.05 | (7.2 + 0.7) × 10−4 | 300 | ||||
Using extension substrates listed in Table 1.
Calculated as (kpol/Kd)correct/(kpol/Kd)incorrect.
Rate constants (kobs) at 1 mM dATP were calculated using the kpol and Kd parameters listed above.
Single determination; all others were the average of two or more determinations.
Figure 3.
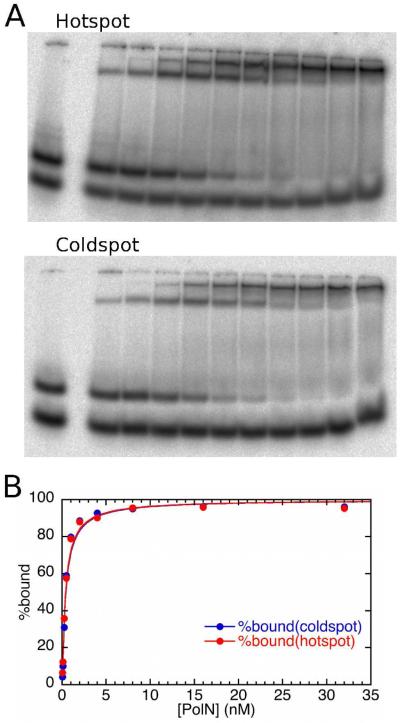
Gel mobility shift experiment comparing the binding of Pol ν-77 to a T-G mismatched DNA terminus in the context of the G(165) hotspot or the G(−66) coldspot sequence. In panel A, the labeled DNA was present at 0.025 nM; the leftmost lane of each gel shows the DNA in the absence of added protein, with excess primer strand having a faster mobility than the annealed duplex. Binding of the duplex is seen in the presence of Pol ν-77 at concentrations (from left to right) of 0.05, 0.1, 0.25, 0.5, 1, 2, 4, 8, 16, and 32 nM. A plot of bound DNA against Pol ν-77 concentration (Panel B), fitted to a quadratic equation (56), gave KD values of 0.38 and 0.40 nM for the hotspot and coldspot sequences respectively. Because only 23% of the Pol ν-77 was active, the true KD is 0.1 nM.
Discussion
Relative to many DNA polymerases that commit T-dGTP errors more frequently than any other base substitution (e.g., Klenow fragment (10), E. coli Pol III (45), Pol θ (24), Pol γ (14, 46), Pol η (26), Pol τ (47), Pol α (48), Pol ε (49), and Pol δ (50)), Pol ν is unusual in preferentially generating a mismatch with the same base composition but the opposite symmetry, i.e., dTTP opposite template G. Not only does Pol ν make this error at much higher rates than for the 11 other single base-base mismatches, it does so in a highly sequence context-dependent manner (25). The kinetic studies described here examine the binding and rate constants responsible for the unusual properties of Pol ν, in comparison with Klenow fragment, a prototypical A-family polymerase. We first consider results using a common template-primer. We then consider explanations for the site-to-site variations for Pol ν.
T-dGTP vs. G-dTTP errors
A major factor in the low fidelity of Pol ν for both errors is its > 103-fold lower efficiency of correct dNTP incorporation, compared to Klenow fragment, illustrating the general observation that differences in polymerase fidelity are frequently dominated by differences in the kinetics of correct base insertion (33, 34). The differences in selectivity of Pol ν and Klenow fragment against particular errors can be understood in terms of the kinetics of the relevant misinsertion reactions. On the 13/19-mer substrate, respresenting a generic DNA sequence not chosen for its hotspot or coldspot behavior, T-dGTP misinsertion by Pol ν is ~ 260-fold less efficient than that of Klenow fragment and this counteracts the 6,300-fold difference in the efficiency of correct (T-dATP) incorporation, resulting in ~ 24-fold lower selectivity of Pol ν against T-dGTP errors. By contrast, the kinetic parameters for G-dTTP misinsertion in the same sequence context are almost identical for the two enzymes, so that the entire 1,000-fold difference in correct (G-dCTP) incorporation is manifested in a 1,000-fold lower selectivity of Pol ν against G-dTTP errors, consistent with the abundance of these errors in synthesis by Pol ν. The similar kinetic parameters for G-dTTP misinsertion by Pol ν and Klenow fragment imply similar ground-state and transition-state energetics for this particular misinsertion in the Klenow fragment and Pol ν active sites, even though the energetics (and the corresponding kinetic parameters) of correct additions are substantially different.
Hotspot and coldspot sequences
The kinetic comparisons of the lacZ hotspot and coldspot sequences reveal that the efficiency of G-dTTP misinsertions by Pol ν varies over a range of nearly 100-fold. In contrast, the efficiency (kpol/Kd) of correct G-dCTP insertions changes very little (~ 3-fold), though the individual kpol and Kd values vary in interesting ways. Nucleotide binding is stabilized by more than 10-fold in the two hottest sequence contexts compared with the cold sequence. However, this does not contribute quantitatively to fidelity because the changes are of similar magnitude for both G-dCTP and G-dTTP insertions, indicating that ground-state binding of the ternary complex provides the same degree of selectivity against the G-dTTP nascent mispair at both hotspot and coldspot sequences. Instead, the important contributor to hotspot behavior is the rate constant, kpol. On the hotspot DNA, kpol is almost the same for correct and mismatched dNTPs, implying that the hotspot transition state fails to provide additional discrimination beyond what was already present in the ground state. In contrast, the coldspot substrate has ~ 15-fold faster kpol for correct G-dCTP incorporation compared with G-dTTP misinsertion. Thus, the tighter substrate binding at the hotspot template position appears to correlate with a less-optimal transition state for G-dCTP addition (slower reaction rate than at the coldspot). Conversely, correct G-dCTP incorporation on the coldspot template appears to use a weaker dNTP binding site and a more favorable transition state geometry, thus maintaining higher selectivity against the G-dTTP error. A similar correlation between tight dNTP binding and slow incorporation was observed in an earlier study of a Klenow fragment mutator mutant, and attributed to altered active-site geometry (44).
Sequence and structural implications
Compared to Klenow fragment, Pol ν exhibits stronger DNA binding, less efficient incorporation of correct base pairs, an active site favorable to G-dTTP nascent mispairs, and a strong influence of sequence context on the G-dTTP nascent mispair. Structural studies of Pol ν, which remain an elusive goal, will be essential for understanding differences between Klenow fragment and Pol ν and how they contribute to the kinetic properties reported here. Absent such information, sequence alignments of Pol ν with bacterial Family A DNA polymerases (51) and structural information on other Family A polymerases (23, 52-54) suggest regions of Pol ν that may contribute to its unique properties. One feature that may be relevant to the DNA binding and processivity of Pol ν is a 6-amino acid insertion predicted to be at the tip of the thumb subdomain, adjacent to conserved sequences already implicated in DNA binding in Klenow fragment (55). A strong interaction with DNA may be necessary for Pol ν’s proposed role in lesion bypass, especially when lesions compromise DNA interactions around the active site. Sequence alignments reveal that Pol ν also differs from more accurate Family A members in the O-helix (Figure 4), which makes several important interactions with the nascent basepair (Figure 4A) (9, 25). One example is Lys679 of Pol ν, which aligns with Ala759 of Klenow fragment and Thr664 of Klentaq (Figure 4B). Changing Lys679 of Pol ν to the alanine or threonine present in the more accurate polymerases did indeed increase the fidelity of Pol ν (51). Nonetheless, these changes did not alter the preferential formation of G-dTTP mismatches or the sequence dependence of Pol ν, indicating that such preferences depend on other enzyme-substrate interactions.
In a more global attempt to identify differences in the sequence of the O-helix of Pol ν that may affect specificity, we replaced residues in the O-helix of Klentaq with the corresponding sequence (residues 675-687: EQTKKVVYAVVYG) from Pol ν. The resulting chimera was active in gap filling synthesis, but the accuracy of this reaction was much higher than that of Pol ν and similar to that of the parental Klentaq enzyme (data not shown). Further experiments (our unpublished data) targeted two additional regions where Pol ν deviates from the A-family consensus (Figure 4). In the bacterial A-family polymerases, the N-terminus of the N-helix has the highly conserved motif D(I/V)H, where the histidine side chain interacts with the dNTP β-phosphate. The corresponding sequence is DVF in the Pol ν group. Mutation of Klenow fragment towards Pol ν consensus (H734F) caused a substantial drop in polymerase activity; however, the relative rates of T-dGTP versus G-dTTP misinsertions, and the selectivity against the G-dTTP mispair, were the same as in wild-type Klenow fragment. A similar result was obtained by changing the N-terminus of the Klenow fragment Q-helix (motif 6 of Patel et al. in (23)) towards Pol ν consensus (replacing residues 841 to 849 of Klenow fragment, RAAINAPMQ, with RQAINFVVQ). Although these experiments did not pinpoint a sequence motif responsible for the characteristic Pol ν error signature, they do suggest that the Klenow fragment and Klentaq active site sequence motifs are optimized for efficiency, so that mutations away from consensus tend to be associated with decreased polymerase activity. Pol ν, on the other hand, may have been fine-tuned by evolution for some specialized purpose where maximum enzyme activity is not a priority. Similarly, the unusual error specificity of Pol ν and its particular DNA sequence specificities may be a consequence of the relaxed active-site specificity associated with its primary function within the cell.
Supplementary Material
Acknowledgements
We thank Katarzyna Bebenek and Nigel Grindley for helpful comments on the manuscript, Komli Kofi Atsina and Ruoting (Valerie) Gong for Klenow fragment kinetic data quoted in tables, and Christina Grindley for making the Pol ν-like Klenow fragment mutants.
Footnotes
This work was supported by Project Z01 ES065070 (to T.A.K.) from the Division of Intramural Research of the NIH, NIEHS, and by NIH grant GM28550 (to C.M.J.).
- Pol
- DNA polymerase
- Taq
- Thermus aquaticus
- Bst
- Bacillus stearothermophilus
- Klentaq
- the portion of Taq DNA polymerase equivalent to Klenow fragment.
It is also possible that the hotspot for single deletions observed at G164-165 (Figure 1) results from a misinsertion-misalignment event initiated by a G-dTTP misinsertion at G(164), making the G-dTTP error rate at G(164) higher than that calculated from the observed base-substitution mutations.
Supporting Information Available
Additional gel mobility shift data for Pol ν is presented in Figure S1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 2.Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- 3.Pang M, McConnell M, Fisher PA. The Drosophila mus 308 gene product, implicated in tolerance of DNA interstrand crosslinks, is a nuclear protein found in both ovaries and embryos. DNA Repair (Amst) 2005;4:971–982. doi: 10.1016/j.dnarep.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Boyd JB, Sakaguchi K, Harris PV. mus308 mutants of Drosophila exhibit hypersensitivity to DNA cross-linking agents and are defective in a deoxyribonuclease. Genetics. 1990;125:813–819. doi: 10.1093/genetics/125.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris PV, Mazina OM, Leonhardt EA, Case RB, Boyd JB, Burtis KC. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol. Cell Biol. 1996;16:5764–5771. doi: 10.1128/mcb.16.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zietlow L, Smith LA, Bessho M, Bessho T. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 2009;48:11817–11824. doi: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D’Andrea AD. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol. Cell Biol. 2010;30:1088–1096. doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamanaka K, Minko IG, Takata K, Kolbanovskiy A, Kozekov ID, Wood RD, Rizzo CJ, Lloyd RS. Novel enzymatic function of DNA polymerase ν in translesion DNA synthesis past major groove DNA-peptide and DNA-DNA cross-links. Chem. Res. Toxicol. 2010;23:689–695. doi: 10.1021/tx900449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J. Biol. Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 10.Bebenek K, Joyce CM, Fitzgerald MP, Kunkel TA. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem. 1990;265:13878–13887. [PubMed] [Google Scholar]
- 11.Donlin MJ, Patel SS, Johnson KA. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry. 1991;30:538–546. doi: 10.1021/bi00216a031. [DOI] [PubMed] [Google Scholar]
- 12.Kunkel TA, Patel SS, Johnson KA. Error-prone replication of repeated DNA sequences by T7 DNA polymerase in the absence of its processivity subunit. Proc. Natl. Acad. Sci. U S A. 1994;91:6830–6834. doi: 10.1073/pnas.91.15.6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunkel TA, Soni A. Exonucleolytic proofreading enhances the fidelity of DNA synthesis by chick embryo DNA polymerase-γ. J. Biol. Chem. 1988;263:4450–4459. [PubMed] [Google Scholar]
- 14.Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The fidelity of human DNA polymerase γ with and without exonucleolytic proofreading and the p55 accessory subunit. J. Biol. Chem. 2001;276:38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- 15.Johnson AA, Johnson KA. Exonuclease proofreading by human mitochondrial DNA polymerase. J. Biol. Chem. 2001;276:38097–38107. doi: 10.1074/jbc.M106046200. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel TA. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 17.Kornberg A, Baker TA. DNA Replication. Second ed. University Science Books, W.H. Freeman and Company; New York: 1992. [Google Scholar]
- 18.Tindall KR, Kunkel TA. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- 19.Eckert KA, Kunkel TA. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1990;18:3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiefer JR, Mao C, Hansen CJ, Basehore SL, Hogrefe HH, Braman JC, Beese LS. Crystal structure of a thermostable Bacillus DNA polymerase I large fragment at 2.1 Å resolution. Structure. 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 21.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 22.Beard WA, Wilson SH. DNA polymerases lose their grip. Nat. Struct. Biol. 2001;8:915–917. doi: 10.1038/nsb1101-915. [DOI] [PubMed] [Google Scholar]
- 23.Patel PH, Suzuki M, Adman E, Shinkai A, Loeb LA. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. J. Mol. Biol. 2001;308:823–837. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- 24.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arana ME, Takata K, Garcia-Diaz M, Wood RD, Kunkel TA. A unique error signature for human DNA polymerase ν. DNA Repair (Amst) 2007;6:213–223. doi: 10.1016/j.dnarep.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase η. J. Mol. Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 27.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase η error spectrum. Nat. Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 28.Gearhart PJ, Wood RD. Emerging links between hypermutation of antibody genes and DNA polymerases. Nat. Rev. Immunol. 2001;1:187–192. doi: 10.1038/35105009. [DOI] [PubMed] [Google Scholar]
- 29.Pavlov YI, Rogozin IB, Galkin AP, Aksenova AY, Hanaoka F, Rada C, Kunkel TA. Correlation of somatic hypermutation specificity and A-T base pair substitution errors by DNA polymerase η during copying of a mouse immunoglobulin κ light chain transgene. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9954–9959. doi: 10.1073/pnas.152126799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. Yeast DNA polymerase ε participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nick McElhinny SA, Stith CM, Burgers PM, Kunkel TA. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 2007;282:2324–2332. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beard WA, Shock DD, Vande Berg BJ, Wilson SH. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J. Biol. Chem. 2002;277:47393–47398. doi: 10.1074/jbc.M210036200. [DOI] [PubMed] [Google Scholar]
- 34.Beard WA, Wilson SH. Structural insights into the origins of DNA polymerase fidelity. Structure. 2003;11:489–496. doi: 10.1016/s0969-2126(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 35.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 36.Arana ME, Powell GK, Edwards LL, Kunkel TA, Petrovich RM. Refolding active human DNA polymerase ν from inclusion bodies. Protein Expr. Purif. 2010;70:163–171. doi: 10.1016/j.pep.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joyce CM, Derbyshire V. Purification of Escherichia coli DNA polymerase I and Klenow fragment. Methods Enzymol. 1995;262:3–13. doi: 10.1016/0076-6879(95)62003-6. [DOI] [PubMed] [Google Scholar]
- 38.Joyce CM, Potapova O, Delucia AM, Huang X, Basu VP, Grindley NDF. Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry. 2008;47:6103–6116. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- 39.Astatke M, Grindley NDF, Joyce CM. How E. coli DNA polymerase I (Klenow fragment) distinguishes between deoxy- and dideoxynucleotides. J. Mol. Biol. 1998;278:147–165. doi: 10.1006/jmbi.1998.1672. [DOI] [PubMed] [Google Scholar]
- 40.Johnson KA. Rapid quench kinetic analysis of polymerases, adenosinetriphosphatases, and enzyme intermediates. Methods Enzymol. 1995;249:38–61. doi: 10.1016/0076-6879(95)49030-2. [DOI] [PubMed] [Google Scholar]
- 41.Joyce CM. Techniques used to study the DNA polymerase reaction pathway. Biochim. Biophys. Acta. 2010;1804:1032–1040. doi: 10.1016/j.bbapap.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckert KA, Kunkel TA. Effect of reaction pH on the fidelity and processivity of exonuclease-deficient Klenow polymerase. J. Biol. Chem. 1993;268:13462–13471. [PubMed] [Google Scholar]
- 43.Bermek O, Grindley NDF, Joyce CM. Distinct roles of the active-site Mg2+ ligands, Asp882 and Asp705, of DNA polymerase I (Klenow fragment) during the prechemistry conformational transitions. J. Biol. Chem. 2011;286:3755–3766. doi: 10.1074/jbc.M110.167593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minnick DT, Liu L, Grindley NDF, Kunkel TA, Joyce CM. Discrimination against purine-pyrimidine mispairs in the polymerase active site of DNA polymerase I: a structural explanation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1194–1199. doi: 10.1073/pnas.032457899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaaper RM. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J Biol Chem. 1993;268:23762–23765. [PubMed] [Google Scholar]
- 46.Lee HR, Johnson KA. Fidelity of the human mitochondrial DNA polymerase. J. Biol. Chem. 2006;281:36236–36240. doi: 10.1074/jbc.M607964200. [DOI] [PubMed] [Google Scholar]
- 47.Bebenek K, Tissier A, Frank EG, McDonald JP, Prasad R, Wilson SH, Woodgate R, Kunkel TA. 5′-Deoxyribose phosphate lyase activity of human DNA polymerase ι in vitro. Science. 2001;291:2156–2159. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- 48.Kunkel TA, Hamatake RK, Motto-Fox J, Fitzgerald MP, Sugino A. Fidelity of DNA polymerase I and the DNA polymerase I-DNA primase complex from Saccharomyces cerevisiae. Mol. Cell Biol. 1989;9:4447–4458. doi: 10.1128/mcb.9.10.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. Unique error signature of the four-subunit yeast DNA polymerase ε. J. Biol. Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 50.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 51.Takata KI, Arana ME, Seki M, Kunkel TA, Wood RD. Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity. Nucleic Acids Res. 2010:3233–3244. doi: 10.1093/nar/gkq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 53.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minnick DT, Astatke M, Joyce CM, Kunkel TA. A thumb subdomain mutant of the large fragment of Escherichia coli DNA polymerase I with reduced DNA binding affinity, processivity, and frameshift fidelity. J. Biol. Chem. 1996;271:24954–24961. doi: 10.1074/jbc.271.40.24954. [DOI] [PubMed] [Google Scholar]
- 56.Patel SS, Bandwar RP, Levin MK. Transient-state kinetics and computational analysis of transcription initiation. In: Johnson KA, editor. Kinetic Analysis of Macromolecules. Oxford Univeristy Press; Oxford, UK: 2003. pp. 87–129. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.