Abstract
Background
Obesity has been associated with increased mortality from hormone dependant cancers such as breast cancer which is the most prevalent cancer in women. The link between obesity and breast cancer can be attributed to excess estrogen produced through aromatization in adipose tissue. The role of steroid hormone receptors in breast cancer development is well studied but how obesity can affect the expression pattern of steroid hormones in patients with different grades of breast cancer was the aim of this study.
Methods
In this case-control study, 70 women with breast cancer participated with different grades of obesity (36 none obese, BMI < 25 kg/m2 and 34 obese, BMI ≥ 25 kg/m2). The mean age of participants was 44.53 ± 1.79 yr (21–70 yr). The serum level of estrogen, progesterone and androgen determined by ELISA. Following quantitative expression of steroid hormone receptors mRNA in tumor tissues evaluated by Real-time PCR. Patients with previous history of radiotherapy or chemotherapy were excluded. SPSS 16 was used for data analysis and P < 0.05 considered statistically significant.
Results
The difference in ERα, ERβ and PR mRNA level between normal and obese patients was significant (P < 0.001). In addition, the expression of AR mRNA was found to be higher than other steroid receptors. There was no significant relation between ERβ gene expression in two groups (P = 0.68). We observed a significant relationship between ERα and AR mRNA with tumor stage and tumor grade, respectively (P = 0.023, P = 0.015).
Conclusion
According to the obtained results, it is speculated that obesity could paly a significant role in estrogen receptors gene expression and also could affect progression and proliferation of breast cancer cells.
Keywords: obesity, breast cancer, steroid receptors, steroid hormones
Introduction
Obesity, which is the accumulation of excessive adipose tissue, is closely associated with an increased risk of cardiovascular morbidities, including hypertension, atherosclerosis and other vascular complications.1 Obese subjects have not only increased risk of developing cancer, but their mortality is also increased with increasing BMI, especially when the BMI is >40 kg m2.2,3 Adiposity or abnormality of body fat distribution is considered to be one of the major risk factors for breast cancer in postmenopausal women.4 The adipose tissue of obese women secretes more biologically active estrogen to stimulate mammary epithelial cell mitosis and to promote the development of the tumor.5 While breast cancer imposes a significant healthcare burden on women worldwide so early detection is of paramount importance in reducing mortality, yet the diagnosis of breast cancer is hampered by the lack of an adequate detection method.6 In addition, better breast cancer prognostication may improve selection of patients eligible for adjuvant therapy.7 Hence, new markers for early diagnosis, accurate prognosis and prediction of response to treatment are warranted to improve breast cancer care. The importance of Estrogen Receptor (ER) status as an independent prognostic factor in breast cancer is well established8,9 and using immunohistochemical techniques, it is now routinely determined in breast cancer prognosis, that if positive, tumor will respond better to anti-estrogen therapy.10,11 It is well established that estrogens mediate their effects through two receptors ERα and ERβ, with seemingly opposing effects in vitro, ie, ERα mediating proliferative and ERβ mediating anti-proliferative effects.12 In contrast to numerous studies that revealed undeniable impact of obesity on poor prognosis of disease in heavier subjects, the possible effect of obesity on ERs gene expression is not elucidated yet. For now a handful of studies investigated the possible impact of obesity on estrogen receptors gene expression and prognosis of diseases in patients. Interestingly, it seems that ER positive tumors are more likely to be associated with obesity than are ER negative tumors.13,14 Furthermore, obesity is associated with poor prognosis in both pre and post menopausal breast cancer.15,16 Since most breast cancers in obese patients are ER+/PR+ and these receptor positive cancers have a better prognosis, breast cancers occurring in obese patients should be expected to be less aggressive. But this is not always the case, one possible explanation is that the increased estrogen levels in obese patients may stimulate tumor growth and adversely affect the prognosis.17 The hormonal environment in obese women is consistent with the finding that obese women with breast cancer more frequently develop metastases and have a shorter survival time than non-obese breast cancer patients.18
The reasons why breast cancer in obese patients is more frequently ER+/PR+ is unknown. On the other more of these ER−/PR− tumors express androgen receptor at high levels. Obesity is reported to be associated with an increased incidence of hormone receptor positive tumors in some studies while others suggest an increase of hormone receptor negative tumors.14 This discrepancy may be explained by differences in laboratory techniques or criteria for hormone responsiveness.19
Beside ER gene expression, the role of other storied hormones such as Progesterone and androgen receptors in breast cancer development are well studied but the possible role of obesity on expression of these steroid receptors is not clear. For now only a few studies investigated the correlation between obesity and total ER at protein level. In this study we used Real Time PCR as a reliable and robust technique to evaluate the expression pattern of steroid hormone receptors gene expression in normal and obese breast cancer patients to study the possible impact of obesity on steroid hormones gene expression, and to evaluates if these findings are in concordance to the findings of protein based methods.
Materials and Methods
Patients and samples
This study was a descriptional cross sectional study. All samples were obtained from 70 women who underwent biopsy or mastectomy surgical operation at Tabriz Emam Reza Hospital from July 2009 until May 2010. The whole project carried out in Tabriz Drug Applied Research Center. The samples were examined histologically for the presence of tumor cells by a pathologist. The patients met the following criteria: primary unilateral non metastatic breast carcinoma for which complete clinical, histological and biological data were available; and no radiotherapy or chemotherapy before surgery. After the interview, during surgery tumor tissue samples were taken into liquid nitrogen in sterile tubes, then as soon as possible aliquated and stored at −70 °C until subsequent analysis. In order to reduce the influence of treatment on measurements, questionnaire data was obtained prior surgery.
Body mass index (BMI)
Accordingly WHO Rep 2000 (World Health Organization, 2000), BMI was calculated as kg/m2 using information from clinical notes at time of diagnosis. In the analyses, BMI was divided into two categories: BMI < 25 kg/m2 (non-obese) and BMI ≥ 25 kg/m2 (obese).
Collection of questionnaire data-tumor tissues
After patients were approved to participate, written informed consent was obtained from all subjects. All participants undertook to fill in questionnaire. It took information on social demographic characteristics, age, age of menark, menstrual history, body mass index and menopause status. The study was blind and data collectors were unaware of the study hypothesis. All participants were also measured for height and weight via standardized techniques. BMI, as an indicator of generalized obesity, was calculated as weight in kilograms divided by the square of height in meter.
Sample preparation
Immediately after the interview, a 10 ml blood sample was drawn into coded EDTA-treated tubes and centrifuged at 3000 rpm for 10 min at room temperature within 1 h of collection. Plasma, Buffy coat and red blood cells were separated and stored at −70 °C until subsequent analysis. To avoid the influence of treatment on measurements, questionnaire data and blood specimens were obtained prior to initiation of definitive breast cancer therapy including surgery and/or radio- or chemotherapy.
Steroid hormones level (plasma estrogen, androgen and progesterone levels)
Plasma concentrations of estrogen, androgen and progesterone were determined by the use of commercially available quantitative sandwich enzyme-linked immunosorbant assay (ELISA) kits (DRG Estradiol ELISA, EIA 2693, DRG Progesterone ELISA, EIA1561 and DRG Androgen ELISA, EIA-1559 Germany). The sensitivity of this assay was 9.714 pg/ml for estradiol, for both progesterone and androgen was 0.083 ng/ml. Masked split specimens included within each batch were used to calculate the coefficient of variation (CV) within and between batches: the intra- and inter-assay CVs of estaradiol, progesterone and androgen were below 6.81% and 7.25%, 5.4% and 9.96% and 4.16% and 9.94%, respectively. All matched blood samples were handled identically and assayed in the same analytical run. The blood samples were labeled by number only and ordered randomly within each case–control pair.
Preparation of total RNA
Approximately 100 mg of tissue from each tumor was quick frozen in liquid nitrogen and pulverized manually with a hammer to a fine powder. The cells powder were harvested and resuspended in 1 ml of RNX plus reagent in a clean RNase-free tube. After incubation for 5 minutes at room temperature the sample was done pipetteing and subsequently treated with adding 200 μl of chloroform and was taken at room temperature for 5 minutes after shaking rigorously for 15 seconds. The mixture was centrifuged at 12,000 × g for 15 minutes and then the aqueous phase containing the RNA was transferred to a clean RNase-free tube. The total RNA was precipitated by adding 0.5 ml of isopropyl alcohol and incubated at room temperature for 15 minutes. The pellet including total RNA was washed by using 75% ethanol and centrifuge at 7,500 × g for 8 minutes. After drying ethanol, the RNA pellet re-suspended in TE buffer. The concentration of total RNA was calculated based on OD260/280 ratio measurements as a means to address purity of RNA.
cDNA synthesis
RNA was converted to cDNA after treating with DNase I. Reverse transcription of RNA was done in a final volume of 20 μl by using of cDNA first strand synthesis kit (Fermentase) random hexamere and 1 μg of total RNA. The samples were incubated at 65 °C for 10 min and 42 °C for 60 min, and reverse transcriptase was inactivated by heating at 70 °C for 5 min and cooling at 4 °C for 5 min.
Real-time RT- PCR
Principle: reactions are characterized by detection of cycling amplification of the PCR product, rather than the amount of PCR product accumulated after a fixed number of cycles. If the amount of the target molecule was larger, the earlier a significant increase in fluorescence is observed. The parameter Ct (threshold cycle) is defined as the cycle number at which the fluorescence generated by cleavage of the probe passes a fixed threshold above baseline. We used the Delta;Delta;CT method for determination of relative ERs gene expression. The Ct of each target gene compared to the Ct of it’s internal control (beta actin gene).
Final results, expressed as N-fold differences in tested gene (obese) relative to control gene (none obese), termed “N fold changes,” were determined as follows:
where Delta;Ct values of the sample and control are determined by subtracting the average Ct value of the target gene from the average Ct value of the beta actin gene.
qRT-PCR assay: generation of standards for target gene
To be sure of equal efficiency of PCR assay, serial dilutions of one sample that expressed target gene in high levels selected and standard curve for both ERs and control gene were prepared. PCR efficiency for both control and target gene was acceptable. Moreover, Melting curve analysis showed that there was only one segment (one pick) and this is an indication for amplification of desired specific PCR product in our study.
After determination of RNA concentration based on absorbance and cDNA synthesis, the precise amount of cDNA was added to each reaction mix. Quantitative detection of mRNA molecules was determined by quantitative real-time RT-PCR technique using the Syber Green-I (Fermentase Co. according to the instructions) by the Rotor-GeneTM 6000 system (Corbett Research, Australia) according to the manufacturer’s instructions. After cDNA synthesis, specific primers were used to amplify ERs and steroid hormone receptors progesterone and androgen receptor. (Primer sequences shown in Table 1). Human beta actin mRNA was selected as the housekeeping gene (reference gene) and amplified by using the specific forward and reverse primers. The Beta-actin mRNA measured as an internal control (a mRNA that does not vary in abundance respective to cell type) was included in each analysis. Briefly, each sample was normalized to the number of cells harvested. The protocol for detection of RNA consisted started by denatured at 95 °C for 10 minutes. This step was followed by 40 cycles of denaturing at 95 °C for 15 second; annealing at 60 °C for 30 seconds; extending at 72 °C for 30 seconds, which was then followed by melting curve analysis of 70 °C to 95 °C for conformity assessment.
Table 1.
Primer sequences for RT-real Time PCR.
| mRNA | Primer sequence | Product size | Reference |
|---|---|---|---|
| ERα | TGA TGA AAG GTG GGA TAC GA AAG GTT GGC AGC TCT CAT GT |
125 bp | Pfeiler et al34 |
| ERβ | GCT TAG TGG AGC TCA GCC TG AGGATCATGGCCTTGACACAGA |
262 bp | Pffafl et al35 |
| PR | GAACCAGATGTGATCTATGCAGGA CGAAAACCTGGCAATGATTTAGAC |
122 bp | Cremoux et al28 |
| AR | CCTGGCTTCCGCAACTTACAC GGACTTGTGCATGCGGTACTCA |
168 bp | Bieche et al27 |
| Beta actin | TCCCTGGAGAAGAGCTACG GTAGTTTCGTGGATGCCACA |
134 bp | Genbank accession: NM_001101 |
Statistical analysis
Differences between obese and non-obese groups in age at enrollment, age at menarche, age at menopause were tested using the chi-square test. In addition, Student’s t-test was used to evaluate differences in categorical breast cancer risk factors between obese and non-obese. Pearson correlation coefficients were used to examine cross-sectional relationships between steroid receptors, gene expression and BMI among subjects. Chi-square test was performed to test the relation between steroid receptors mRNA level and clinicopathological parameters (For this purpose, all steroid receptors according to their mRNA content divided in two subgroups as low and high mRNA level). All statistical analysis was conducted using SPSS statistical software (version 16). P values below the conventional level of statistical significance (P < 0.05) were considered statistically significant.
Results
In this case-control study, 70 women with breast cancer participated with different grades of obesity (36 none obese, BMI < 25 kg/m2 and 34 obese, BMI ≥ 25 kg/m2). The mean age of participants was 44.53 ± 1.79 yr (range from 21–70 years old) and half of them were under 44 years old at the time of enrollment. Patients with previous history of radiotherapy or chemotherapy were excluded from this study. Our results indicated that there is a significant relationship between family history of breast cancer and breast cancer risk in patients (P = 0.01). Baseline characteristics are summarized in Table 2. About subtypes of breast cancer samples, 71% of tumors were DCIS and 28.3% were LCIS and CIS. Although we observed no significant relationship between obesity and none of clinicopathological parameters (P > 0.05) but more of low grade tumors observed in non-obese cases (63.6%), while more of high grade tumors presented in obese ones (58.5%) (see Table 3). Moreover in case of tumor size, while more small size tumors (<2 cm) observed in none obese cases (51.9%), more of the obese ones had a tumor diameter of ≥2 cm (52.8%) (Data not shown).
Table 2.
Characteristics of obese and non-obese breast cancer patient.
| Baseline characteristics | None obese (n = 36) Mean ± SE |
Obese (n = 34) Mean ± SE |
P value |
|---|---|---|---|
| Age at enrollment (yr) | 43.82 ± 1.78 | 45.42 ± 2.27 | 0.45 |
| BMI (kg/m2) | 23.23 ± 0.23 | 29.80 ± 0.63 | 0.000 |
| Family history of breast cancer | No. (%) | No. (%) | |
| Yes | 18 (54.5) | 6 (22.2) | |
| No | 15 (45.5) | 21 (77.8) | 0.01 |
| Family history of obesity | |||
| Yes | 23 (69.7) | 22 (81.5) | |
| No | 10 (30.3) | 5 (18.5) | 0.22 |
| Menopause status | |||
| Premenopausal | 21 (63.6) | 12 (46.2) | |
| Postmenopausal | 12 (36.4) | 14 (53.8) | 0.14 |
| Age at first live birth (yr) | |||
| 17> | 12 (37.5) | 12 (48.0) | |
| 18–23 | 11 (34.4) | 10 (40) | 0.32 |
| 24< | 9 (28.1) | 3 (12) | |
| Age at menarche (yr) | |||
| 12> | 10 (30.3) | 6 (23.1) | |
| 13 | 5 (15.2) | 10 (38.5) | 0.12 |
| 14< | 18 (54.5) | 10 (38.5) | |
Table 3.
Relation between obesity and tumor grade in breast cancer samples.
| Tumor garde BMI (kg/m2) | 1 | 2 + 3 | P |
|---|---|---|---|
| Nonobese (n = 31) | 14 (63.6) | 17 (41.5) | 0.078 |
| Obese (n = 32) | 8 (38.4) | 24 (58.5) |
Serum level of estrogen, progesterone and androgen in blood samples
We analyzed serum level of estrogen, progesterone and androgen in 70 cases with breast cancer. The mean- ± SE serum level of estrogen was 117.81 ± 17.81 in none obese (n = 33) 149.83 ± 54.75 vs. in obese cases (n = 37), and mean ± SE progesterone was 0.96 ± 0.26 vs. 1.60 ± 0.63 and also the serum level of Androgen was 1.56 ± 0.042 vs. 1.57 ± 0.02. The mean serum level of estrogen and progesterone was higher in obese cases (P = 0.07 and P = 0.03 respectively) (see Table 4).
Table 4.
Mean ± SE serum level of steroid hormones.
| Mean ± SE Sterid hormone | None obese (BMI < 25 kg/m2) (N = 36) | Obese cases (BMI ≥ 25 kg/m2) (N = 36) | P |
|---|---|---|---|
| Estrogen (pg/ml) | 117.81 ± 17.81 | 149.83 ± 54.75 | 0.07 |
| Progesterone (ng/ml) | 0.96 ± 0.26 | 1.60 ± 0.63 | 0.03 |
| Androgen (ng/ml) | 1.56 ± 0.042 | 1.57 ± 0.02 | 0.45 |
As a result, there was a significant correlation between serum level of estrogen and progesterone with ERα mRNA (Spearman 2 tailed correlation r = 0.409, P = 0.00, n = 66 and r = 0.85, P = 0.00 and n = 70 respectively) but no significant correlation observed between ERβ and serum level of steroid hormones (P > 0.05).
Determination of progesterone, androgen and estrogen receptors (ERα and ERβ) gene expression in breast cancer tissues
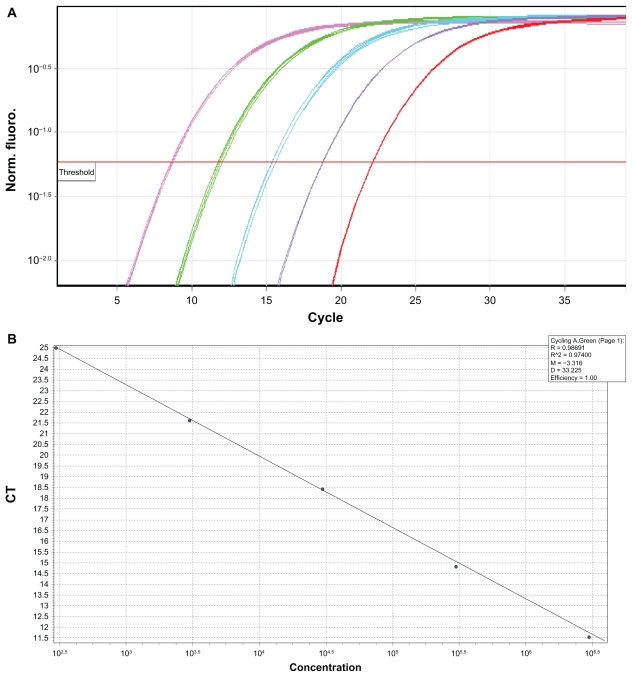
Results of comparison of steroid hormones mRNA level between obese and non-obese cases according to ΔC values are summarized in Table 5. It is obvious that as the ΔC value is high, so the expression of target gene will be less. According to this table, obese subjects expressed a higher level of ERα and also progesterone receptors in comparison to non-obese subjects and the difference in ERα and PR mRNA level between these two groups of patients was significant (both P < 0.001). In case of ERβ, the expression of ERβ was lower in obese ones and the difference in ERβ mRNA level between two groups was statistically significant (P < 0.001). There was no difference in AR mRNA between obese and none obese cases (P = 0.40). Through these steroid receptors, the ERβ mRNA show the lowest and AR mRNA show the highest expression in studied samples. 2ΔΔCt values of ERα, ERβ, PR and AR were 1–35, 1–11, 1–5 and 1–2.4 respectively. Standard curve and amplification of one of the steroid hormone receptors is (ERβ) represented in Figures 1 and 2.
Table 5.
Mean mRNA level of steroid hormone receptors gene expression in breast cancer tissues.
| Mean ± SE Steroid hormones | None obese (BMI < 25 kg/m2) (N = 36) | Obese (BMI ≥ 25 kg/m2) (N = 34) | P |
|---|---|---|---|
| ERα | 11.73 ± 0.41 | 7.40 ± 0.46 | <0001 |
| ERβ | 8.01 ± 0.53 | 14.15 ± 0.64 | <0001 |
| PR | 5.82 ± 0.27 | 3.79 ± 0.28 | <0001 |
| AR | 2.44 ± 0.334 | 2.08 ± 0.25 | 0.40 |
Figure 1.
(A and B) ERβ standard curve (R2 = 0.97, M = −3.3, E = 100).
Figure 2.
Amplification (A) and Melting curve (B) of ERβ mRNA in breast cancer samples, 83 °C.
Moreover, a two tailed Pearson coefficient test shows that there was a significant correlation between ERα, ERβ and PR mRNA level and BMI in samples (rs = 0.56, n = 70 and P = 0.00, rs = −0.56, n = 70 and P = 0.00 and rs = 0.49, n = 70 and P = 0.00). There was no correlation between AR and BMI in subjects (rs = −0.09, n = 70, P = 0.45). Moreover we observed a positive correlation between ERα and PR in samples (rs = 0.89, n = 70 and P = 0.00). Two tailed Pearson coefficient test shows that there was a positive correlation between AR mRNA and ERα and PR mRNA (rs = 0.23, n = 69, P = 0.04 and rs = 0.34, n = 69, P = 0.004 respectively) but no correlation observed between AR and ERβ mRNA level (P > 0.05).
Relation between steroid hormone receptors gene expression with clinicopatological parameters
According to the Bloom Richardson classification for classifying the tumors, the tumors were classified into 6 stages and 3 grades.20–22 To evaluate the possible relationships between steroid hormone receptors mRNA level and clinical characteristics of the tumor; ie, clinical staging and histopathological grading, analysis was performed using chi square test (Tables 6 and 7). For this purpose, all steroid receptors according to their mRNA content were divided in two subgroups as low and high mRNA level. We found a significant relationship between ERα and tumor stage and also a significant relation between AR gene expression and tumor grade in breast cancer tumors (P = 0.045 and P = 0.046 respectively) but there was no significant relation between ERβ and PR with clinocopathological parameters.
Table 6.
Observed frequency (%) of AR mRNA in different grades of breast cancer.
| Tumor garde AR mRNA level | 1 | 2 + 3 | P |
|---|---|---|---|
| Low (n = 33) | 10 (27.8) | 26 (72.2) | |
| High (n = 30) | 14 (51.9) | 13 (48.1) | 0.046 |
Table 7.
Observed frequency (%) of ERα mRNA in different stages of breast cancer.
| Tumor stage ERα mRNA level | 1 | 2A | 2B | 3A | 3B | 4 | P |
|---|---|---|---|---|---|---|---|
| Low (n = 36) | 0 (0.0) | 12 (36.4) | 8 (24.2) | 9 (27.3) | 4 (12.1) | 0 (0) | |
| High (n = 27) | 2 (6.7) | 13 (43.3) | 5 (16.7) | 1 (3.3) | 8 (26.7) | 1 (3.3) | 0.045 |
Conclusion
Many studies have evaluated the possible role of obesity on some steroid hormones via protein detection assays but none evaluated the possible impact of obesity on expression of other steroid receptors gene expression such as androgen and progesterone receptor and specially ERβ. So we used real-time quantitative RT-PCR to assess steroid hormone receptors gene expression in human breast tumors. This recent approach to nucleic acid quantification is suited to the development of target gene assays, having a high degree of inter-laboratory standardization and also yielding statistical confidence values.
Obesity and steroid hormone receptors gene expression in breast cancer patients
Our results show that the difference in ERα mRNA level between normal and obese patients was significant (P < 0.001) and the expression of ERα mRNA was found to be highest in obese samples. This finding is in relation to that of Dario Giuffkida 1992, Cleary MP 1997, Maehle BO 1996, and Nomura Y 1984.13,14,23,24 We confirm their finding that estrogen receptor (ER)-positive tumors are more likely to be associated with obesity than are ER-negative tumors.
An article published by Dario Giuffrida D and et al in 1992 is the only study which is partially similar to ours. They studied the relation between steroid receptor status and body weight in breast cancer patients via protein assay methods. They assessed the relationship between the ER and progesterone receptor (PR) status of the neoplastic tissue and obesity in a series of 615 breast cancer patients. Both ER and PR concentrations were significantly and positively correlated with obesity by multiple regression analysis. Furthermore, the estimated probability of having an ER+/PR+ carcinoma was significantly higher in obese patients (odds ratio 2.65, 95% confidence interval 1.56–4.48).13
According to Dario and our findings, it can be concluded that obese cases have a high expression of ERα and PR mRNA in comparison to their none obese counterparts.
For the first time we evaluated the possible impact of obesity on ERβ gene expression. We found that not only ERα mRNA level can be affected by excessive adipose tissue, but the mRNA level of ERβ which has a paramount importance in regulating the ERα mRNA level can be affected directly by obesity or indirectly by increased ERα expression in obese breast cancer patients. According to the results of our study, it seems that in patients with breast cancer, not only ERα but also ERβ and PR gene expression can be affected by obesity and for achieving a better result in obese subjects both estrogen receptors can be candidate as potential target therapies. In case of AR, like Bieche we find a positive correlation between AR and ERα but in contrast to Bieche in our study no correlation was observed between AR and ERβ mRNA level.25 Likewise, AR is expressed in approximately 70% to 90% of invasive breast cancers, a frequency comparable with or higher than the one reported for ER (70%–80%) and PgR (50%–70%).26 In our study, AR mRNA expression ranked the highest expression while ERβ was the lowest. Moreover there was no difference in AR mRNA between two obese and none obese cases. Although a relationship between AR and both ER and PR status has been demonstrated, a significant percentage of tumors are positive for AR and negative for ER and PR. This finding reveals the independent expression of AR in human breast cancer as we find no relation between AR and BMI in our study so it seems that AR is less affected by obesity rather than other steroid hormones.
Relation between steroid receptors gene expression with clinicopatological parameters
Studies have confirmed the relation between high grade and ER negativeness in breast cancer tissues. In one study by Bieche et al27 statistically significant links were found between ERα gene status (but not ERβ gene status) and Scarff-Bloom- Richardson (SBR) histopathological grade (P = 0.00029) and ERα and ERβ status were not significantly linked to lymph-node status or macroscopic tumor size. In another study de Cremoux found a significant correlation between ERα and tumor grade (P = 0.03) but no correlation was observed with other clinical or pathological parameters and no correlation was found between ERβ mRNA level and tumor size, nodal status, grade, histological size. In our study there was a significant relation between ER α and tumor stage (P = 0.45) and also between AR and tumor grade (P = 0.046) but there was no significant relation between the ERβ and PR with tumor stage and tumor grade. Just like many other researchers such as Cremoux Speirs V, Knoweldton JM and Cullen R it seems that ERα expression more than ERβ expression correlated with clinicopathological parameters in breast cancer tumors.8,28–30 similar to Bieche et al’s findings, it seems that AR can act as an independent factor and can affect progression of mammary cells. Several studies have reported an association between high body weight and axillary lymph node involvement.31–33 Williams et al investigated the relationship of body weight to response to endocrine therapy, steroid hormone receptors and survival of patients with advanced cancer of the breast. High body weight was correlated with advanced tumor stage (P = 0.002), but not with the presence of estrogen receptor (total ER P = 0.21).17
Giuffrida et al presented findings from prognostic parameters of tumor evaluation as tumor size, presence of metastatic lymph nodes, tumor grade and histotype were available in 538 patients, no significant correlation by an overall F test from one-way analysis of variance was found between the patient BMI and any of these parameters. As with Giuffrida, in our study also there was no significant relation between BMI and tumor stage but obese individuals were in high stages and had a larger tumor size.
Maehle and Teitli investigated 1,238 women with unilateral breast cancer treated with modified radical mastectomy living in the geographic area of Haukeland Hospital. Age- adjusted Quetelet’s index (weight/height2) showed that obese women had a 49% higher risk of dying from breast cancer than lean ones. The relative risk decreased slightly when adjusted for tumor diameter, lymph node status, and mean nuclear area of the tumor cells. They found that in patients with a hormone receptor positive tumor, obese women had a risk that was more than three times higher than lean ones. In patients with hormone receptor negative tumor, the effect of obesity was reversed, lean patients having a risk that was more than six times higher than obese ones, even after adjustment.23
It seems that in obese ER positive subjects, beside obesity, the role of other factors (eg, serum level of adipocyte-derived hormones) should be considered too.
Final conclusion
In summary, according to the obtained results, it is speculated that obesity could play a significant role in steroid receptors gene expression and also could affect progression and proliferation of breast cancer cells.
Acknowledgments
This work financially supported by Iranian National Science Foundation (INSF) with the ref code (8800060-April12/2010). We appreciate all individuals who participated in this study.
Footnotes
Disclosures
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
References
- 1.Zhu W, Cheng KK, Vanhoutte PM, Lam KS, Xu A. Vascular effects of adiponectin: molecular mechanisms and potential therapeutic intervention. Clin Sci (Lond) 2008;114(5):361–74. doi: 10.1042/CS20070347. [DOI] [PubMed] [Google Scholar]
- 2.Kelesidis I, Kelesidis T, Mantzoros CS. Adiponectin and cancer: a systematic review. Br J Cancer. 2006;94(9):1221–5. doi: 10.1038/sj.bjc.6603051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou WK, Xu YX, Yu T, Zhang L, Zhang WW, Fu CL, et al. Adipocytokines and breast cancer risk. Chin Med J (Engl) 2007;120(18):1592–6. [PubMed] [Google Scholar]
- 4.Sellers TA, Kushi LH, Potter JD, et al. Effect of family history, body-fat distribution, and reproductive factors on the risk of postmenopausal breast cancer. N Engl J Med. 1992;326(20):1323–9. doi: 10.1056/NEJM199205143262004. [DOI] [PubMed] [Google Scholar]
- 5.Edman CD, Aiman EJ, Porter JC, MacDonald PC. Identification of the estrogen product of extraglandular aromatization of plasma androstenedione. Am J Obstet Gynecol. 1978;130:439–47. doi: 10.1016/0002-9378(78)90286-7. [DOI] [PubMed] [Google Scholar]
- 6.Speirs V, Walker RA. New perspectives into the biological and clinical relevance of oestrogen receptors in the human breast. J Pathol. 2007;211(5):499–506. doi: 10.1002/path.2130. [DOI] [PubMed] [Google Scholar]
- 7.Loi S, Milne RL, Friedlander ML, et al. Obesity and outcomes in premenopausal and postmenopausal breast cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(7):1686–91. doi: 10.1158/1055-9965.EPI-05-0042. [DOI] [PubMed] [Google Scholar]
- 8.Cullen R, Maguire TM, McDermott EW, Hill AD, O’Higgins NJ, Duffy MJ. Studies on oestrogen receptor-alpha and-beta mRNA in breast cancer. Eur J Cancer. 2001;37(9):1118–22. doi: 10.1016/s0959-8049(01)00088-0. [DOI] [PubMed] [Google Scholar]
- 9.Matthews J, Gustafsson JA. Estrogen signaling: a subtle balance between ER alpha and ER beta. Mol Interv. 2003;3(5):281–92. doi: 10.1124/mi.3.5.281. [DOI] [PubMed] [Google Scholar]
- 10.Markey GC, Cullen R, Diggin P, Hill AD, Mc Dermott EW, O’Higgins NJ, et al. Estrogen receptor-beta mRNA is associated with adverse outcome in patients with breast cancer. Tumour Biol. 2009;30(4):171–5. doi: 10.1159/000236409. [DOI] [PubMed] [Google Scholar]
- 11.Skliris GP, Carder PJ, Lansdown MR, Speirs V. Immunohistochemical detection of ERbeta in breast cancer: towards more detailed receptor profiling? Br J Cancer. 2001;84(8):1095–8. doi: 10.1054/bjoc.2001.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao C, hlman-Wright K, Gustafsson JA. Estrogen receptor beta: an over-view and update. Nucl Recept Signal. 2008;6:e003. doi: 10.1621/nrs.06003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giuffrida D, Lupo L, La Porta GA, et al. Relation between steroid receptor status and body weight in breast cancer patients. Eur J Cancer. 1992;28(1):112–5. doi: 10.1016/0959-8049(92)90397-k. [DOI] [PubMed] [Google Scholar]
- 14.Cleary MP, Maihle NJ. The role of body mass index in the relative risk of developing premenopausal versus postmenopausal breast cancer. Proc Soc Exp Biol Med. 1997;216(1):28–43. doi: 10.3181/00379727-216-44153b. [DOI] [PubMed] [Google Scholar]
- 15.Folsom AR, Kaye SA, Prineas RJ, et al. Increased incidence of carcinoma of the breast associated with abdominal adiposity in postmenopausal women. Am J Epidemiol. 1990;131(5):794–803. doi: 10.1093/oxfordjournals.aje.a115570. [DOI] [PubMed] [Google Scholar]
- 16.Verla-Tebit E, Chang-Claude J. Anthropometric factors and the risk of pre-menopausal breast cancer in Germany. Eur J Cancer Prev. 2005;14:419–26. doi: 10.1097/00008469-200508000-00016. [DOI] [PubMed] [Google Scholar]
- 17.Williams G, Howell A, Jones M. The relationship of body weight to response to endocrine therapy, steroid hormone receptors and survival of patients with advanced cancer of the breast. Br J Cancer. 1988;58(5):631–4. doi: 10.1038/bjc.1988.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmichael AR, Bendall S, Lockerbie L, Prescott RJ, Bates T. Does obesity compromise survival in women with breast cancer? Breast. 2004;13(2):93–6. doi: 10.1016/j.breast.2003.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Carmichael AR, Bates T. Obesity and breast cancer: a review of the literature. Breast. 2004;13(2):85–92. doi: 10.1016/j.breast.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Tough IC, Carter DC, Fraser J, Bruce J. Histological grading in breast cancer. Br J Cancer. 1969;23(2):294–301. doi: 10.1038/bjc.1969.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.WHO. Histological typing of breast tumors. Geneva: 1981. Ref Type: Internet Communication. [Google Scholar]
- 22.Thoresen S. Histological grading and clinical stage at presentation in breast carcinoma. Br J Cancer. 1982;46(3):457–8. doi: 10.1038/bjc.1982.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maehle BO, Tretli S, Skjaerven R, Thorsen T. Premorbid body weight and its relations to primary tumour diameter in breast cancer patients; its dependence on estrogen and progesteron receptor status. Breast Cancer Res Treat. 2001;68(2):159–69. doi: 10.1023/a:1011977118921. [DOI] [PubMed] [Google Scholar]
- 24.Nomura Y, Tashiro H, Hamada Y, Shigematsu T. Relationship between estrogen receptors and risk factors of breast cancer in Japanese pre- and postmenopausal patients. Breast Cancer Res Treat. 1984;4(1):37–43. doi: 10.1007/BF01806986. [DOI] [PubMed] [Google Scholar]
- 25.Bieche I, Parfait B, Tozlu S, Lidereau R, Vidaud M. Quantitation of androgen receptor gene expression in sporadic breast tumors by real-time RT-PCR: evidence that MYC is an AR-regulated gene. Carcinogenesis. 2001;22(9):1521–6. doi: 10.1093/carcin/22.9.1521. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez LO, Corte MD, Vazquez J, Junquera S, Sanchez R, Alvarez AC, et al. Androgen receptor expresion in breast cancer: relationship with clinicopathological characteristics of the tumors, prognosis, and expression of metalloproteases and their inhibitors. BMC Cancer. 2008;8:149. doi: 10.1186/1471-2407-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bieche I, Parfait B, Laurendeau I, Girault I, Vidaud M, Lidereau R. Quantification of estrogen receptor alpha and beta expression in sporadic breast cancer. Oncogene. 2001;20(56):8109–15. doi: 10.1038/sj.onc.1204917. [DOI] [PubMed] [Google Scholar]
- 28.de CP, Tran-Perennou C, Elie C, Boudou E, Barbaroux C, Poupon MF, et al. Quantitation of estradiol receptors alpha and beta and progesterone receptors in human breast tumors by real-time reverse transcription-polymerase chain reaction. Correlation with protein assays. Biochem Pharmacol. 2002;64(3):507–15. doi: 10.1016/s0006-2952(02)01187-5. [DOI] [PubMed] [Google Scholar]
- 29.Valerie S, Alicia T, Michael J, et al. Coexpression of Estrogen Receptor a and β Poor Prognostic Factors in Human Breast Cancer? Cancer Res. 1999;595:25–528. [PubMed] [Google Scholar]
- 30.Knowlden JM, Gee JM, Robertson JF, Ellis IO, Nicholson RI. A possible divergent role for the oestrogen receptor alpha and beta subtypes in clinical breast cancer. Int J Cancer. 2000;89(2):209–12. doi: 10.1002/(sici)1097-0215(20000320)89:2<209::aid-ijc17>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 31.Greenberg ER, Vessey MP, McPherson K, Doll R, Yeates D. Body size and survival in premenopausal breast cancer. Br J Cancer. 1985;51(5):691–7. doi: 10.1038/bjc.1985.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abe R, Kumagai N, Kimura M, Hirosaki A, Nakamura T. Biological characteristics of breast cancer in obesity. Tohoku J Exp Med. 1976;120(4):351–9. doi: 10.1620/tjem.120.351. [DOI] [PubMed] [Google Scholar]
- 33.Eberlein T, Simon R, Fisher S, Lippman ME. Height, weight, and risk of breast cancer relapse. Breast Cancer Res Treat. 1985;5(1):81–6. doi: 10.1007/BF01807654. [DOI] [PubMed] [Google Scholar]
- 34.Pfeiler GH, Buechler C, Neumeier M, et al. Adiponectin effects on human breast cancer cells are dependent on 17-beta estradiol. Oncol Rep. 2008;19(3):787–93. [PubMed] [Google Scholar]
- 35.Pfaffl MW, Lange IG, Daxenberger A, Meyer HH. Tissue-specific expression pattern of estrogen receptors (ER): quantification of ER alpha and ER beta mRNA with real-time RT-PCR. APMIS. 2001;109(5):345–55. doi: 10.1034/j.1600-0463.2001.090503.x. [DOI] [PubMed] [Google Scholar]