Abstract
Mouse (m) 11β-hydroxysteroid dehydrogenase type 2 (11βHSD2) was homology-modeled, and its structure and ligand-receptor interaction were analyzed. The modeled m11βHSD2 showed significant 3D similarities to the human (h) 11βHSD1 and 2 structures. The contact energy profiles of the m11βHSD2 model were in good agreement with those of the h11βHSD1 and 2 structures. The secondary structure of the m11βHSD2 model exhibited a central 6-stranded all-parallel β-sheet sandwich-like structure, flanked on both sides by 3-helices. Ramachandran plots revealed that only 1.1% of the amino acid residues were in the disfavored region for m11βHSD2. Further, the molecular surfaces and electrostatic analyses of the m11βHSD2 model at the ligand-binding site exhibited that the model was almost identical to the h11βHSD2 model. Furthermore, docking simulation and ligand-receptor interaction analyses revealed the similarity of the ligand-receptor bound conformation between the m11βHSD2 and h11βHSD2 models. These results indicate that the m11βHSD2 model was successfully evaluated and analyzed. To the best of our knowledge, this is the first report of a m11βHSD2 model with detailed analyses, and our data verify that the mouse model can be utilized for application to the human model to target 11βHSD2 for the development of anticancer drugs.
Keywords: 11βHSD2, anticancer drug, homology modeling, Molecular Operating Environment (MOE), tumor
Introduction
Hydroxysteroid dehydrogenases (HSDs) are a family of enzymes that contribute to the metabolism of steroids, and 11β-HSD catalyzes the interconversion of inactive glucocorticoids (cortisone in humans; dehydrocorticosterone in rodents) and active glucocorticoids (cortisol in humans; corticosterone in rodents).1,2 There have been several reports suggesting the association of 11β-HSD type 2 (11βHSD2) with cancers, such as colonic and pituitary adenomas3,4 and breast and colorectal cancers.5–7 Our previous studies showed that glycyrrhetinic acid (GA), a specific 11βHSD2 inhibitor, was selectively toxic toward central nervous system-derived tumor cells.8,9 These studies exhibited that 11βHSD2 could play an important role in tumor regulation and that targeting 11βHSD2 for tumor prevention and therapy could be an effective strategy with highly selective 11βHSD2 inhibitors. The underlying explanation for the aberrant 11βHSD2 expression remains unknown, but it has been postulated to control glucocorticoid regulation of cellular proliferation.10 The antiproliferative actions of glucocorticoids were demonstrated using malignant transformed cell lines, and the local inactivation of glucocorticoids, such as corticosterone, by 11βHSD2 was suggested to be an important oncogenic process promoting cellular proliferation.11 Further, 11βHSD2 inhibition by GA prevented colon cancer without triggering adverse side effects in the cardiovascular system.3 GA also had adverse effects on the proliferation of pituitary adenomas,12 and 11βHSD2 inhibition induced apoptosis of corticotroph tumor cells.4 Taken together, these reports suggest that 11βHSD2 inhibition can be utilized as a potential therapeutic option in controlling cancer.
Recently, molecular modeling has gained much importance in the field of drug discovery and development,13–15 and in silico analyses of 11βHSD2 and its interactions with possible ligands may lead to successful development of antitumor drugs targeting 11βHSD2. However, to the best of our knowledge, no X-ray crystal structure of 11βHSD2 is currently available. Further, although a few possible human (h) 11βHSD2 models have been reported,16–18 there have been no reports of mouse (m) 11βHSD2 models with detailed analyses. Large numbers of in vitro and in vivo mouse studies have been reported for human application, but those by in silico approaches are still to be explored. Therefore, in the present study, we performed modeling and structural analyses of m11βHSD2 using a highly sophisticated software package, the Molecular Operating Environment (MOE) 2009.10. (Chemical Computing Group Inc., Montreal, Canada).
Methods
Homology modeling of m11βHSD2
Homology modeling of m11βHSD2 was achieved by the same methods used for modeling h11βHSD2 in our previous study.18 In brief, the m11βHSD2 (NCBI reference sequence: NM_008289.2)19 sequence and the crystal structure coordinates of h11βHSD1 (PDB code: 3HFG)20 were loaded into the MOE. The primary structures of h11βHSD1 and m11βHSD2 were aligned, carefully checked to avoid deletions or insertions in conserved regions and corrected wherever necessary. A series of m11βHSD2 models were independently constructed with the MOE using a Boltzmann-weighted randomized procedure21 combined with specialized logic for the handling of sequence insertions and deletions.22 There were no differences in the numbers and organizations of the secondary structural elements and no significant main chain deviations among the 10 models generated for m11βHSD2. The models with the best packing quality function were selected in our study for full energy minimization and further inspection.
Assessment of the modeled structures
Structure assessment was performed by the same methods described in our previous study.18 In brief, the overall geometric and stereochemical qualities of the final modeled structure of m11βHSD2 were examined using Ramachandran plots generated within the MOE.23,24 The qualities of the protein folds of the m11βHSD2 homology model were evaluated with the MOE by calculating the effective atomic contact energies, comprising the desolvation free energies required to transfer atoms from water to the interior of the protein.25 The contact desolvation energies were calculated for 18 different atom types of the 20 common amino acids that were resolved based on the clustering pattern of their properties. The contact potentials for each atom type were measured within a contact range of 6 Å by explicitly accounting for neighboring interactions.
Binding site selection and exploration
The binding site selection and exploration were also achieved by the same methods described in our previous study.18 In brief, the Site Finder module of the MOE was used to identify possible substrate-binding pockets within the newly generated 3D structure of m11βHSD2. Hydrophobic or hydrophilic alpha spheres served as probes denoting zones of tight atom packing. These alpha spheres were utilized to define potential ligand-binding sites (LBSs) and as centroids for the creation of dummy ligand atoms.26,27 The dummy atoms were matched to the corresponding alpha spheres during the characterization of the LBSs in m11βHSD2. This method possibly generates bound conformations that approach crystallographic resolutions.28 The LBS volume for m11βHSD2 was also analyzed with the Site Finder module.
Molecular electrostatic potential (MEP) mapping
The MEP mapping was performed as previously reported.18 In brief, electrostatic potential surfaces were calculated by solving the nonlinear Poisson-Boltzmann equation using a finite difference method as implemented in the MOE. The molecular electrostatic interactions form a crucial part of the noncovalent interaction energy between the molecules. The MEP on a molecular surface can be used to visually compare different molecules, analyze docking studies and identify sites that interact with ligands. For example, the surface MEP of h11βHSD2 was utilized to relate a nucleotide mutation with the potential values.16 In the present study, the MEP of m11βHSD2 was colored in deep blue to indicate the most positive potential and in deep red to represent the most negative potential.
Alpha sphere and excluded volume-based ligand-receptor docking (ASE-Dock)
The docking and analysis of the ligand-receptor interaction between corticosterone (or GA) and m11βHSD2 were performed with ASE-Dock in the MOE.29 In the ASE-Dock module, ligand atoms have alpha spheres within 1 Å. Based on this property, concave models are created and ligand atoms from a large number of conformations generated by superimposition with these points can be evaluated and scored by maximum overlap with alpha spheres and minimum overlap with the receptor atoms. The scoring function used by ASE-Dock is based on ligand-receptor interaction energies and the score is expressed as a Utotal value. The ligand conformations were subjected to energy minimization using the MMF94S force field,30 and 500 conformations were generated using the default systematic search parameters. Five thousand poses per conformation were randomly placed onto the alpha spheres located within the LBS in m11βHSD2. From the resulting 500,000 poses, the 200 poses with the lowest Utotal values were selected for further optimization with the MMF94S force field. During the refinement step, the ligand was free to move within the binding pocket.
Results and Discussion
Evaluation of the 2D and 3D structures of the homology modeled m11βHSD2
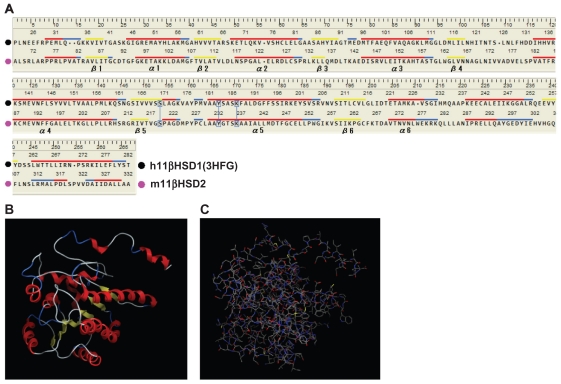
Figure 1A shows the sequence alignments of h11βHSD1 and m11βHSD2. h11βHSD1 (PDB code: 3HFG) was selected as a template for the present structure modeling because of its good crystal structure resolution (2.3 Å) and its information was the latest (from 2009) among the reported 11βHSD1 models. As shown in Figure 1A, the generated model extended from residues Ala68 to Ala332 for m11βHSD2. The LBS was characterized as non-metallo-oxidoreductase site and contained the conserved Ser219, Tyr232 and Lys236 catalytic triad. It has been proposed that Ser219 is associated with catalysis by stabilizing the reaction intermediates, that the Tyr232 hydroxyl group is the proton donor involved in the electrophilic attack on the substrate carboxyl group in a reduction reaction, and that Lys236 facilitates the proton transfer from the hydroxyl oxygen of Tyr232 to the substrate.31 The cofactor-related motif of Gly89-XXX-Gly93-X-Gly951,2,31,32 was also conserved in m11βHSD2. The secondary structure of the m11βHSD2 model exhibited a central 6-stranded all-parallel β-sheet sandwich-like structure, flanked on both sides by 3-helices (Fig. 1A and B), which are in agreement with the h11βHSD1 and 2 models.18,33 For the construction of the m11βHSD2 model, 10 independent models of the target proteins were built using a Boltzmann-weighted randomized modeling procedure in the MOE that is adapted from reports by Levitt21 and Fechteler et al.22 The intermediate models were evaluated by a residue packing quality function, which is sensitive to the degrees to which non-polar side-chain groups are buried and hydrogen bonding opportunities are satisfied. The m11βHSD2 3D model with the best packing quality function and full energy minimization is shown in Figure 1C. Root mean square deviation (RMSD) values between the main chain atoms of the h11βHSD1 vs m11βHSD2 and h11βHSD2 vs m11βHSD2 after main chain fit were 1.03 and 2.62 Å, respectively. RMSD values for each residue were also analyzed. The RMSD values for the residues located in the LBS near the cofactor-binding motif and the catalytic activity-related triad were always less than 2 Å.
Figure 1.
Evaluation of the 2D and 3D structures of the m11βHSD2 model. (A) Homology-aligned sequences of h11βHSD1 (PDB code: 3HFG; black) and m11βHSD2 (magenta). Red line: α-helix. Blue line: turn. Yellow line: β-sheet. The alignments reveal that the cofactor-related GXXXGXG and catalytic YXXXK domains are conserved in m11βHSD2. The catalytic domain is often in the vicinity of a conserved S, and this is also the case for m11βHSD2. (B) The secondary structure of the m11βHSD2 model exhibits a central 6-stranded all-parallel β-sheet sandwich-like structure, flanked on both sides by 3-helices. (C) The constructed m11βHSD2 model. The model exhibits similar 3D structure to the structures of h11βHSD2 and 2.18,33
Evaluation of the stereochemical qualities of the m11βHSD2 model
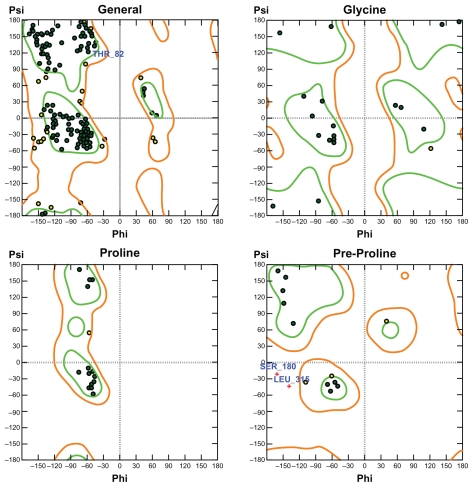
The phi and psi backbone dihedral angles for m11βHSD2 were scored using 2D probability distributions calculated on a high-resolution collection of X-ray structures containing approximately 100,000 residues from 500 protein structures.34 Each probability distribution was estimated with 2-degree spacing for each of the phi and psi backbone dihedral angles with separate histograms for pre-proline, proline, glycine and general amino acids. The stereochemical qualities of the m11βHSD2 model were assessed by Ramachandran plots (Fig. 2). 87.1% of the residues were in the favored region, 11.8% were in the allowed region and only 1.1% were in the disfavored region. The residues in the disfavored regions were located far away from the residues in the LBSs. These results indicate that the phi and psi backbone dihedral angles in the m11βHSD2 model are reasonably accurate.
Figure 2.
Ramachandran plots for the m11βHSD2 model. For m11βHSD2, 87.1% of the residues are in the favored region, 11.8% are in the allowed region and only 1.1% are in the disfavored region. The residues in the disfavored regions are located far away from the residues in the LBSs. Green: favored region. Light-brown: allowed region.
Analysis of the contact energies and the MEP map for the m11βHSD2 model
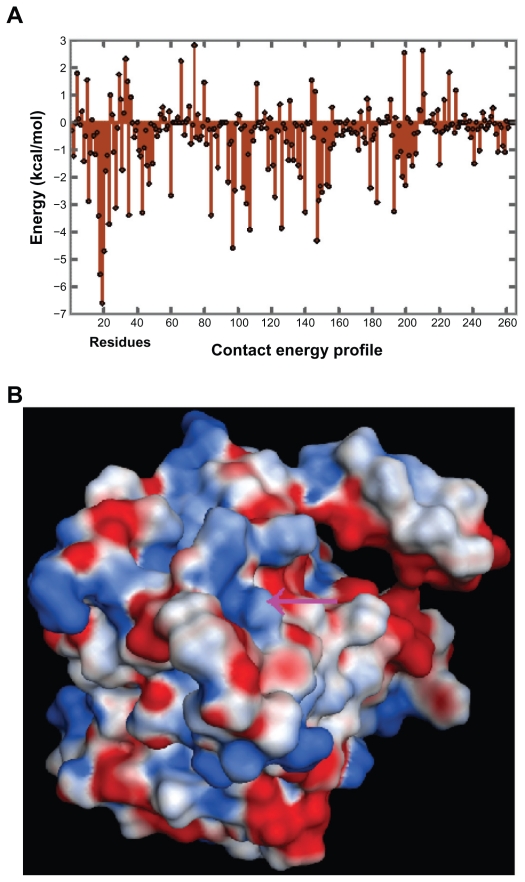
The effective atomic contact energies were calculated by the methods of Zhang et al25 using the MOE for heavy atoms of standard amino acids within a contact range of 6 Å, assigning energy terms in kcal/mol for each contact pair. These energies were summed for each residue, and in general, a large negative value indicated that the residue was predominantly in contact with hydrophobic atoms and therefore expected to be in a buried protein environment. Conversely, residues with positive energy terms indicated contacts with predominantly hydrophilic atoms, and were expected to be in more solvent-exposed regions of the proteins. The contact energy profiles of the homology-modeled m11βHSD2 (Fig. 3A) was compared with that of the X-ray structure of h11βHSD1 and the homology-modeled h11βHSD2,18 and the trends of the variation in the contact energy in most parts of the m11βHSD2 model were in good agreement with those of the h11βHSD1 and 2 structures. The binding site volume of the m11βHSD2 model was also calculated with the MOE. The calculated volume was 979 Å3, which suggests that the LBS of m11βHSD2 is sufficiently large to contain cofactors and substrates (ligands), such as NAD+ and cortisol, whose surface volumes were reported to be 420 and 297 Å3, respectively.16 During analyses and predictions of molecular interactive behaviors and properties, MEP maps can play a vital role. For example, they can be used to compare two molecules visually, which helps in identifying sites that act attractively on ligands by matching opposite electrostatics. Electrostatic interactions comprise one of the main parts of the interaction energy between ligands and receptors, and govern the strength of non-bonded interactions and molecular reactivity. In the case of a ligand-receptor interaction at the catalytic site, the ligand experiences a unique environment in terms of the electrostatic, steric and hydrophobic properties. Variations in these properties near the catalytic site of receptors can contribute to their selectivity/specificity.35 The MEP map of the m11βHSD2 model is shown in Figure 3B. The model exhibited almost identical electrostatic surfaces at the LBS in h11βHSD2,18 sharing common features such as a positively charged surface (colored in blue).
Figure 3.
(A) Contact energy profiles of m11βHSD2. The positions of the amino acid residues are shown on the x-axis, while the contact energies are shown on the y-axis. The trends in the variation of the contact energy in most parts of the m11βHSD2 model are in good agreement with those of the structures of h11βHSD1 and 2,18 indicating that the h11βHSD2 and m11βHSD2 models exhibit similar contact energy profiles. (B) The MEP map for the m11βHSD2 model at the LBS. Arrows: LBS. Deep blue: most positive potential. Deep red: most negative potential. The model exhibits an almost identical surface MEP map at the LBS in h11βHSD2,18 sharing common features such as a positively charged surface.
Docking simulation and ligand-receptor interaction between corticosterone and m11βHSD2
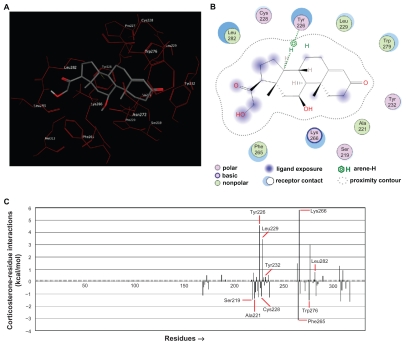
The m11βHSD2 model presented a similar structure at its LBS (Fig. 4A) to the reported LBS in h11βHSD2,16,18 and the ASE-Dock was performed to evaluate the present docking simulation. The simulation showed that the ligand (corticosterone) successfully bound to the LBS in m11βHSD2 (Fig. 4A) and had a similar binding orientation of the ligand (cortisol) to the Rossmann fold in the h11βHSD2.16 The similarity between the present docked corticosterone-m11βHSD2 pose and the h11βHSD2 model16 indicates that the present methods are capable of generating the corticosterone-m11βHSD2 model similar to the reported h11βHSD2 complex, which suggests that the mouse model in the present study can be applied to the human model. To create the ligand-receptor interaction plots for corticosteronem11 βHSD2, the Ligand Interactions module of the MOE was used, which provided a clearer arrangement of putative key intermolecular interactions that aid in interpretation of the 3D juxtaposition of the ligand and the LBS in m11βHSD2. The conserved Ser219, Tyr232 and Lys236 catalytic triad has been reported to play an important role in the binding of the ligand in h11βHSD2,31 and our present results identified that 10 residues could be of importance in the m11βHSD2 model (Fig. 4B). The bound conformation of corticosterone present in the LBS suggests that it can form a strong hydrogen bond with Tyr226. Further, the ligand-residue interaction energies were calculated by the methods of Labute36 using the MOE, assigning energy terms in kcal/mol for each residue. Generally, a negative value indicated that the residue attracted the ligand, while the residue with a positive value repelled the ligand. Among the 10 identified residues (Fig. 4B), Ser219, Ala221, Cys228, Phe265 and Trp276 appeared to attract the ligand, while Tyr226, Leu229, Tyr232, Lys266 and Leu282 could repel the ligand (Fig. 4C). Although Lys236, one of the catalytic triad, was not identified by the ligand-receptor interaction plots (Fig. 4B), the ligand-residue interaction energy profiles revealed that it would attract the ligand with a similar energy value (1.29 kcal/mol) to that (1.46 kcal/mol) of Ser219 (Fig. 4C). These results indicate that the identified 10 residues, including the catalytic triad, contribute to the stable binding of corticosterone to m11βHSD2.
Figure 4.
Ligand-receptor interaction between corticosterone and m11βHSD2. (A) Corticosterone successfully binds to the LBS in m11βHSD2. The similarity between the present docked corticosterone-m11βHSD2 pose and the h11βHSD2 model16 suggests that the present methods are capable of generating the corticosterone-m11βHSD2 model similar to the reported h11βHSD2 complex. (B) The ligand-receptor interaction plots for the corticosteronem11 βHSD2 model. Ten residues are identified as important residues in the m11βHSD2 model. The bound conformation of corticosterone present in the LBS suggests that it can form a strong hydrogen bond with Tyr226. (C) Ligand-residue interaction energies for the corticosterone-m11βHSD2 model. A negative value indicates that the residue attracts the ligand, while the residue with a positive value repels the ligand. Among the 10 identified residues (Fig. 4B), Ser219, Ala221, Cys228, Phe265 and Trp276 appear to attract the ligand, while Tyr226, Leu229, Tyr232, Lys266 and Leu282 could repel the ligand. The identified 10 residues, including the catalytic triad, would contribute to the stable binding of corticosterone to m11βHSD2.
Docking simulation and ligand-receptor interaction between GA and m11βHSD2
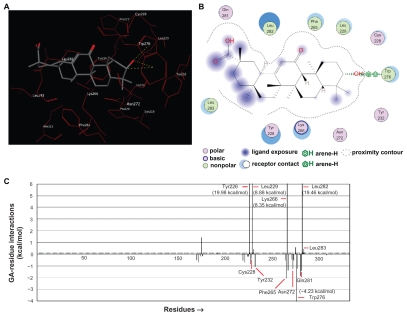
The analyses of the ligand-receptor docking and interaction between GA, the known inhibitor of 11βHSD2, and the modeled m11βHSD2 were also performed. The docking simulation showed that GA (Fig. 5A) and corticosterone (Fig. 4A) had a similar binding orientation to the LBS in the m11βHSD2. The ligand-receptor interaction plots for GA-m11βHSD2 were also created, and 11 residues were identified as important residues in the m11βHSD2 model (Fig. 5B). The bound conformation of GA present in the LBS suggests that GA can form strong hydrogen bonds with Trp276. Further, the ligand-residue interaction energies revealed that Cys228, Tyr232, Phe265, Asn272, Trp276 and Gln281 would attract the ligand, while Tyr226, Leu229, Lys266, Leu282 and Leu283 could repel the ligand among the 11 identified residues (Fig. 5C). Although the negative energy values for Ser219 and Lys236 (the two residues in the catalytic triad) were rather weak, Tyr232 (one of the residues in the catalytic triad), with a higher negative energy value, appeared to contribute to the ligand-receptor binding, attracting the ligand to m11βHSD2 (Fig. 5C). Phe265 and Trp276 were the ligand-attracting residues with rather higher negative energy values in the corticosterone-m11βHSD2 and GA-m11βHSD2 models (Fig. 4C and 5C).
Figure 5.
Ligand-receptor interaction between GA and m11βHSD2. (A) Corticosterone (Fig. 4A) and GA have a similar binding orientation to the LBS in the m11βHSD2. (B) The ligand-receptor interaction plots for the GA-m11βHSD2 model. Eleven residues are identified as important residues in the m11βHSD2 model. The bound conformation of GA present in the LBS suggests that GA can form strong hydrogen bonds with Trp276. (C) Ligand-residue interaction energies for the GA-m11βHSD2 model. The residues with the interaction energy value of less than −4 kcal/mol and more than 6 kcal/mol are shown with their interaction energy values since these values are not within the lowest and the highest graphic ranges. Among the 11 identified residues (Fig. 5B), Cys228, Tyr232, Phe265, Asn272, Trp276 and Gln281 would attract the ligand, while Tyr226, Leu229, Lys266, Leu282 and Leu283 could repel the ligand. Phe265 and Trp276 are the ligand-attracting residues with rather higher negative energy values in the corticosterone-m11βHSD2 (Fig. 4C) and GA-m11βHSD2 models.
Conclusions
The 3D model of m11βHSD2 was designed using the X-ray crystal structure of h11βHSD1 (PDB code: 3HFG) as a template. The model was successfully evaluated and analyzed in terms of its folding, stereochemical and the ligand-receptor interaction properties. Consequently, it is proposed that the m11βHSD2 model developed in the present study will be suitable for further in silico structure-based de novo drug designing. Such computer-based methodologies are now becoming an integral part of the drug discovery process, and may eventually lead to the development of potential inhibitors of 11βHSD2 in the future. To the best of our knowledge, this is the first report of a m11βHSD2 model with detailed analyses, and our present data verify that the mouse model can be utilized for application to the human model to target 11βHSD2 for the development of anticancer drugs.
Acknowledgements
This study was partially supported by a grant-in-aid from the Promotion and Mutual Aid Corporation for Private Schools of Japan.
Abbreviations
- 11βHSD
11β-hydroxysteroid dehydrogenase
- GA
glycyrrhetinic acid
- LBS
ligand-binding site
- MEP
molecular electrostatic potential
- MOE
Molecular Operating Environment
- RMSD
root mean square deviation
Footnotes
Disclosures
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
References
- 1.Oppermann UC, Filling C, Berndt KD, et al. Active site directed mutagenesis of 3 beta/17 beta-hydroxysteroid dehydrogenase establishes differential effects on short-chain dehydrogenase/reductase reactions. Biochemistry. 1997;36:34–40. doi: 10.1021/bi961803v. [DOI] [PubMed] [Google Scholar]
- 2.Filling C, Berndt KD, Benach J, et al. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J Biol Chem. 2002;277:25677–84. doi: 10.1074/jbc.M202160200. [DOI] [PubMed] [Google Scholar]
- 3.Zhang M, Xu J, Yao B, et al. Inhibition of 11β-hydroxysteroid dehydrogenase type II selectively blocks the tumor COX-2 pathway and suppresses colon carcinogenesis in mice and humans. J Clin Invest. 2009;119:876–85. doi: 10.1172/JCI37398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nigawara T, Iwasaki Y, Asai M, et al. Inhibition of 11β-hydroxysteroid dehydrogenase eliminates impaired glucocorticoid suppression and induces apoptosis in corticotroph tumor cells. Endocrinology. 2006;147:769–72. doi: 10.1210/en.2005-0544. [DOI] [PubMed] [Google Scholar]
- 5.Koyama K, Myles K, Smith R, Krozowski Z. Expression of the 11β-hydroxysteroid dehydrogenase type II enzyme in breast tumors and modulation of activity and cell growth in PMC42 cells. J Steroid Biochem Mol Biol. 2001;76:153–9. doi: 10.1016/s0960-0760(00)00157-6. [DOI] [PubMed] [Google Scholar]
- 6.Cohen PG. Estradiol induced inhibition of 11β-hydroxysteroid dehydrogenase 1: an explanation for the postmenopausal hormone replacement therapy effects. Med Hypotheses. 2005;64:989–91. doi: 10.1016/j.mehy.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Zbankova S, Bryndova J, Kment M, Pacha J. Expression of 11β-hydroxysteroid dehydrogenase types 1 and 2 in colorectal cancer. Cancer Lett. 2004;210:95–100. doi: 10.1016/j.canlet.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi H, Noshita T, Yu T, et al. Novel effects of glycyrrhetinic acid on the central nervous system tumorigenic progenitor cells: induction of actin disruption and tumor cell-selective toxicity. Eur J Med Chem. 2010;45:2943–8. doi: 10.1016/j.ejmech.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi H, Kidachi Y, Kamiie K, Noshita T, Umetsu H, Ryoyama K. Glycyrrhetinic acid induces anoikis-like death and cytoskeletal disruption in the central nervous system tumorigenic cells. Biol Pharm Bull. 2010;33:321–24. doi: 10.1248/bpb.33.321. [DOI] [PubMed] [Google Scholar]
- 10.Rabbitt EH, Lavery GG, Walker EA, et al. Prereceptor regulation of glucocorticoid action by 11beta-hydroxysteroid dehydrogenase: a novel determinant of cell proliferation. FASEB J. 2002;16:36–44. doi: 10.1096/fj.01-0582com. [DOI] [PubMed] [Google Scholar]
- 11.Stewart PM, Prescott SM. Can licorice lick colon cancer. J Clin Invest. 2009;119:760–3. doi: 10.1172/JCI38936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabbitt EH, Ayuk J, Boelaert K, et al. Abnormal expression of 11beta-HSD type 2 in human pituitary adenomas: a prereceptor determinant of pituitary cell proliferation. Oncogene. 2003;22:1663–7. doi: 10.1038/sj.onc.1206293. [DOI] [PubMed] [Google Scholar]
- 13.Kurogi Y, Guner OF. Pharmacophore modeling and threedimensional database searching for drug design using catalyst. Curr Med Chem. 2001;8:1035–55. doi: 10.2174/0929867013372481. [DOI] [PubMed] [Google Scholar]
- 14.Ekins S. Predicting undesirable drug interactions with promiscuous proteins in silico. Drug Discovery Today. 2004;9:276–85. doi: 10.1016/S1359-6446(03)03008-3. [DOI] [PubMed] [Google Scholar]
- 15.Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303:1813–8. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 16.Carvajal CA, Gonzalez AA, Romero DG, et al. Two homozygous mutations in the 11 beta-hydroxysteroid dehydrogenase type 2 gene in a case of apparent mineralocorticoid excess. J Clin Endocrinol Metab. 2003;88:2501–7. doi: 10.1210/jc.2002-021909. [DOI] [PubMed] [Google Scholar]
- 17.Koch MA, Wittenberg LO, Basu S, et al. Compound library development guided by protein structure similarity clustering and natural product structure. Proc Natl Acad Sci U S A. 2004;101:16721–6. doi: 10.1073/pnas.0404719101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaguchi H, Akitaya T, Yu T, et al. Homology modeling and structural analysis of 11β-hydroxysteroid dehydrogenase type 2. Eur J Med Chem. 2011;46:1325–30. doi: 10.1016/j.ejmech.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 19.Huang Y, Li X, Lin H, et al. Regulation of 11beta-hydroxysteroid dehydrogenase 1 and 2 by IGF-1 in mice. Biochem Biophys Res Commun. 2010;391:1752–6. doi: 10.1016/j.bbrc.2009.12.148. [DOI] [PubMed] [Google Scholar]
- 20.Wan ZK, Chenail E, Xiang J, et al. Efficacious 11beta-hydroxysteroid dehydrogenase type I inhibitors in the diet-induced obesity mouse model. J Med Chem. 2009;52:5449–61. doi: 10.1021/jm900639u. [DOI] [PubMed] [Google Scholar]
- 21.Levitt M. Accurate modeling of protein conformation by automatic segment matching. J Mol Biol. 1992;226:507–33. doi: 10.1016/0022-2836(92)90964-l. [DOI] [PubMed] [Google Scholar]
- 22.Fechteler T, Dengler U, Schomberg D. Prediction of protein three-dimensional structures in insertion and deletion regions: a procedure for searching data bases of representative protein fragments using geometric scoring criteria. J Mol Biol. 1995;253:114–31. doi: 10.1006/jmbi.1995.0540. [DOI] [PubMed] [Google Scholar]
- 23.Bowie JU, Lüthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–70. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 24.Lüthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–5. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, Vasmatizis G, Cornette JL, DeLisi C. Determination of atomic desolvation energies from the structures of crystallized proteins. J Mol Biol. 1997;267:707–26. doi: 10.1006/jmbi.1996.0859. [DOI] [PubMed] [Google Scholar]
- 26.Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S. Analytical shape computation of macromolecules: I. molecular area and volume through alpha shape. Proteins. 1998;33:1–17. [PubMed] [Google Scholar]
- 27.Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S. Analytical shape computation of macromolecules: II. Inaccessible cavities in proteins. Proteins. 1998;33:18–29. [PubMed] [Google Scholar]
- 28.Goto J, Kataoka R, Hirayama N. Ph4Dock: pharmacophore-based protein-ligand docking. J Med Chem. 2004;47:6804–11. doi: 10.1021/jm0493818. [DOI] [PubMed] [Google Scholar]
- 29.Goto J, Kataoka R, Muta H, Hirayama N. ASEDock-Docking based on alpha spheres and excluded volumes. J Chem Inf Model. 2008;48:583–90. doi: 10.1021/ci700352q. [DOI] [PubMed] [Google Scholar]
- 30.Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17:490–519. [Google Scholar]
- 31.Hoffmann F, Maser E. Carbonyl reductases and pluripotent hydroxysteroid dehydrogenases of the short-chain dehydrogenase/reductase superfamily. Drug Metab Rev. 2007;39:87–144. doi: 10.1080/03602530600969440. [DOI] [PubMed] [Google Scholar]
- 32.Obeid J, White PC. Tyr-179 and Lys-183 are essential for enzymatic activity of 11 beta-hydroxysteroid dehydrogenase. Biochem Biophys Res Commun. 1992;188:222–7. doi: 10.1016/0006-291x(92)92373-6. [DOI] [PubMed] [Google Scholar]
- 33.Miguet L, Zhang Z, Barbier M, Grigorov MG. Comparison of a homology model and the crystallographic structure of human 11beta-hydroxysteroid dehydrogenase type 1 (11betaHSD1) in a structure-based identification of inhibitors. J Comput Aided Mol Des. 2006;20:67–81. doi: 10.1007/s10822-006-9037-3. [DOI] [PubMed] [Google Scholar]
- 34.Lovell SC, Davis IW, Arendall WB, III, et al. Structure validation by Cα geometry: φ, Ψ and Cβ deviation. Proteins. 2003;50:437–50. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 35.Awale M, Kumar V, Saravanan P, Mohan CG. Homology modeling and atomic level binding study of Leishmania MAPK with inhibitors. J Mol Model. 2010;16:475–88. doi: 10.1007/s00894-009-0565-3. [DOI] [PubMed] [Google Scholar]
- 36.Labute P. The generalized Born/volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area. J Comput Chem. 2008;29:1693–8. doi: 10.1002/jcc.20933. [DOI] [PubMed] [Google Scholar]