Abstract
The helix-disfavoring, versus alanine, propagation values of lysine (0.8) and arginine (1.0) residues placed centrally in an (Ala)9 unit have been measured by 13C NMR.
The controversy1 concerning whether alanine is helix in-different2 or has a uniquely high helix propensity1,3 has prompted studies of the roles of the solubilizing residues (Arg, Lys, and Gln) that are typically placed in Ala-rich, (AAAXA)-repeat peptide helix models4 used to derive residue helix propensities. It has been suggested2b,5 that the substantial helicity of designed Ala-rich peptide helix models is due to the solubilizing polar residues rather than a uniquely large Ala helix propensity and experimental data of Kemp and coworkers6 have been cited as supporting this view. However, in a more recent account from the Kemp laboratory,7 the helix propensity for alanine in an (Ala)15 segment, w = 1.5 (w is the propagation constant in the Lifson–Roig helix-coil model8), is in agreement with that obtained for (AAAXA)-repeat peptides.4a,b The higher helix propensity of alanine is supported by experimental studies confirming a decrease in helicity associated with increasing Lys content in Ala/Lys copolymers9 and with replacing a central Ala residue within a long continuous stretch of Ala residues by Lys.1b Experimental evidence indicating that both Lys and Arg enhance helicity when placed near the C-terminus of designed helices (where there is a favorable Coulombic interaction with the helix macrodipole10) is clear-cut,11 but there is still some controversy concerning the role of these residues (X) at central positions in designed helices consisting of AAAXA-repeats. The argument centers on whether polar sidechain-induced backbone desolvation increases helicity.5b,12 An opposing point of view, that helix backbone solvation is enthalpically favorable and does not decrease helicity, has also been published.3c Conformer ensemble energy calculations by Scheraga5b indicate that Ac-AAAAAKAAAA-NH2 is 76% helical while Ac-(A)10-NH2 is less than 10% helical when simulated at the dielectric constant of water. A folding trajectory simulation (continuum solvent) for Ac-YGAAKA- (AAAKA)2-NH2 indicated an equilibrium helicity of 64%.13 MD simulations of Ala-rich sequences with and without added Arg residues have, depending on the sequence and whether a continuum or explicit water solvent was employed, indicated both enhanced12b and diminished14 helix stability due to the inserted Arg residues.
As a result, we saw a need for additional experimental data on the specific effects of Lys and Arg residues at the X positions in AAAXA-repeat peptides. To date, most experimental measures of helix propensities have relied on CD measurements. Since different investigators use CD values ([θ]222) for 100% helicity that can differ by nearly 50%,15 we turned to other probes to answer questions concerning helix propensities. Changes in NH exchange protection and melting data for mutations within the N-terminal helix of Trp-cage miniproteins have provided two measures of the relative helix propensities of Lys and Ala.
GAAXAAYAQWLKDGGPSSGRPPPS (X = A vs. K),
| [ref. 1a] |
DAYAQWLXDGGPSSGRPPPS (X = A vs. K),
Although this was viewed as strong evidence for Lys being significantly helix destabilizing,1a it still leaves the question about the effects of polar sidechains in Ala-rich peptides unanswered.
Recently 13C=O (13C′) chemical shifts, which move downfield upon helix formation, have emerged as residue-specific measures of fractional helicity.11b,17 We have used this method to probe the effects of Lys → Ala and Arg → Lys → Ala mutations in two designed helices: Ac-WAAAHA-(AARAA)3-NH2 and KKGG-K(AAAAK)3-GGKK-NH2.‡ The first system has been extensively examined both experimentally and in MD simulations.18 The second system included multiple Lys residues at the termini to ensure solubility. (13C′)-Alanine isotopomers were prepared in each case to provide chemical shifts for sites in each of the repeats; NMR spectra were recorded in 20 mM phosphate buffer, pH = 7.0 (10% D2O) with 13C labeled urea as an internal reference. Melting curves (δobs versus T) over the range 280–340 K were obtained for each site in each peptide. The melts (vide infra) reveal helical structuring at low temperatures which melts out (ca. 80% loss of structuring shifts) by 340 K. In the case of Ac-WAAAHA(AARAA)3-NH2, the apparent melting point (the point at which the δobs versus T slope is maximal), 305 K, is in excellent agreement with the prior study.18 Chemical shifts were converted to chemical shift deviations (CSDs)§ using the coil values and methods previously described.11b,17b The results are shown below; the 13C′-labeled sites are in bold. For the second system, CSD averages were used within the first two repeats.
The RAAAAXAAAAR system indicates that propagation values must be in the order w(Ala) > w(Arg) > w(Lys). The melting curves for X =A and K appear in Fig. 1.
Fig. 1.
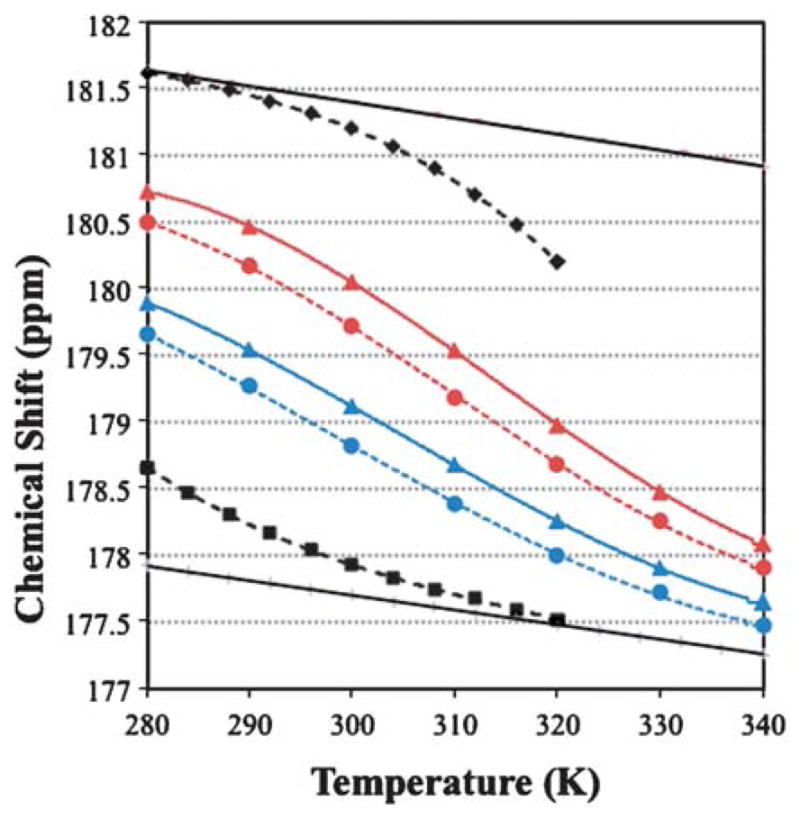
Melting curves for 13C=O chemical shifts. The data for DAYAQWLKDGGPSSGRPPPS and NAYAQWLKDGKK appear in black with the derived 100% and 0% helix lines. The data for Ac-WAAAHAAARAA
 AXAA
AXAA
 ARAA-NH2 is color coded by alanine position, with the X = A data shown as ▲ connected by a solid line and X = K as ● connected by a dashed line. The 100% and 0% helix lines for –RAA
ARAA-NH2 is color coded by alanine position, with the X = A data shown as ▲ connected by a solid line and X = K as ● connected by a dashed line. The 100% and 0% helix lines for –RAA
 AXAA
AXAA
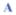 AR– appear in the Electronic Supplementary Information together with the method employed to convert CSDs to helix propagation values.†
AR– appear in the Electronic Supplementary Information together with the method employed to convert CSDs to helix propagation values.†
| -AARAAAAXAAAARAA- | ||
|---|---|---|
| X = A | 3.04 | 2.21 |
| X = R | 2.90 | 2.05 |
| X = K | 2.83 | 1.97 |
| (CSDs in ppm) | ||
The CSD data for the KAAAAXAAAAX′AAAAK system (below) indicate that the mutational effects can be observed throughout the helical span. The diminished structuring shifts in the final repeat reflect C-terminal fraying of the helix.
| -GKAAAAXAAAAX′AAAAKG- | |||
|---|---|---|---|
| X, X′= K, K | 1.92 | 2.02 | 1.28 |
| X, X′= A, K | 2.39 | 2.48 | 1.62 |
| X, X′= K, A | 2.11 | 2.35 | 1.66 |
This result is in accord with expectations based on the cooperativity inherent in helix–coil transition models. Assuming that the value of w(Ala) is not position dependent over the helical span examined, the observation of larger structuring shifts for –AAAAAAKA– versus –AKAAAAAA– suggests, as expected for a sidechain-charge–helix-macrodipole interaction, that w(Lys) increases as the Lys residue moves closer to the C-terminus of the helix.
To obtain propagation values, the changes in CSDs must be converted to fractional helicity (fH) changes, and thus, local ΔΔGH values. This requires a value for the CSD expected for 100% local helicity. The Trp-cage provides a means for obtaining such an estimate. A large body of evidence exists1a,16 to support the assertion that a Trp-cage species of the DAYAQWLKDGGPSSGRPPPS sequence is >98% folded at 280 K. The temperature dependence of the 13C′ CSDs for the sites in bold was measured. The corresponding coil values were obtained from the chemical shift melting behavior of a truncated species (NAYAQWLKDGKK),‡ which lacks the features required for Trp-cage formation and displays fH = 0.18 at the central sites at 280 K with melting complete by 320 K. For the intact Trp-cage system, a small degree of melting is observed from 300–320 K with the plot of δ13C′ vs. T for each site over the 280–300 K range suggesting an approach to a linear, pre-melting plot with essentially the same slope as the δcoil line. Together, the two melting studies afforded the following 100% CSD estimates at 280 K: A2 (3.35), A4 (3.71), and L7 (3.25 ppm). The melting curves for A4 of the Trp-cage and the truncated species appear in Fig. 1.
Prior studies, with the Δδ(13C′) for helix formation calibrated against CD estimates of fH, had placed the 100% CSD for central residue in a helix at 3.04 ppm.17b Therefore, we designed and prepared an N-capped Ala-rich peptide (GASEDE(AAAAK)3GY-NH2)‡ that was expected to have a very high fractional helicity with 13C=O units at specific alanine sites. The largest 13C′ CSD (3.41 ppm) was observed in the first repeat, immediately after the SEDE capping box, with a 3.14 ppm CSD observed in the central repeat. Kemp has also reported data indicating that Δδ(13C′) ≥ 3.35 ppm for a central alanine site in a helix.7 The prior estimate of the CSD for 100% helix is clearly too small.
As a result, we have adopted a 13C′ CSD of 3.50 ppm as the 100% folded reference value for both protein and peptide helices. With this value, and the current w(Ala) value (1.54) in Helix 1.54b (the same value has been reported recently for helices templated by a La3+ binding capping/nucleation unit19), the propagation values from the data reported herein are: w(Arg) = 0.98 (for a central helical position), w(Lys) = 0.80 ± 0.05 (for a central helical position), and w(Lys) = 0.87 ± 0.02 (for locations near the helix C-terminus). While the precise w values may be disputed, there should no longer be any question about the effects of either Lys or Arg inserted in long (Ala)n sequences. Neither of these polar residues increases helicity; quite the contrary, helicity decreases and the decrease is in proportion to the propagation constants derived in earlier reports4a,b based on CD measurements for Ala-rich helical peptides.
IR studies of unlabeled and 13C=O-labeled (AAAKA)- repeat helices17b,20 support extensive solvation of the backbone amides of such helices: the amide I′ bands appear at 1627–1636 and 1587–1592 cm−1(or 1598 cm−1)20d (12C=O and 13C=O, respectively). These are the same values as observed for exposed Ala sites in coiled coil species (which display amide I′ bands at 1648–1652 and 1605–1608 cm−1, 12C=O and 13C=O, respectively, for the buried sites)21 and the IR evidence of differential solvation associated with the relative locations of Ala and Lys residues remains ambiguous.22 A desolvated 13C=O amide (1617 cm−1) has, however, been observed upstream from a valine in an Ala-rich helix.17b We will leave the incorporation of the experimental observations in this account into detailed theorizing regarding the effects of helix backbone desolvation to others, but do note that the present results are consistent with the view3b that alanine is uniquely helix-favoring since the small methyl side- chain has no entropic penalty for structuring and does not hinder backbone solvation at its site or upstream in the helix.
Supplementary Material
Acknowledgments
This work was supported by the Organic and Macromolecular Chemistry Program of the US National Science Foundation through grant # CHE-0650318. National Institutes of Health support (GM-059658) for Trp-cage studies is also acknowledged.
Footnotes
Electronic supplementary information (ESI) available: Fig. S1 and procedure for deriving propagation values. See DOI: 10.1039/b807101b
These peptides were synthesized employing standard Fmoc (9-fluorenylmethoxycarbonyl) solid-phase peptide synthesis methods, cleaved with TFA in the presence of protecting group scavengers and purified using RP-HPLC (using C18 and/or C8 stationary phases). The sequences of all peptides were confirmed by the molecular ions observed using ion-trap mass spectrometry and a complete TOCSY/NOESY NMR assignment.
CSDs are defined as (δobs − δcoil), where δcoil is the statistical coil expectation δ value. In the present study, the statistical coil values were obtained from very short peptides with the same local sequence. All CSDs reported are positive values indicating a downfield structuring shift. The quite small effect of temperature on δcoil (see for example ref. 17b and Fig. 1) was included in the CSD calculations; potential errors in this term do not change the propagation values significantly (< ±0.04).
References
- 1.(a) Lin JC, Barua B, Andersen NH. J Am Chem Soc. 2004;126:13679–13684. doi: 10.1021/ja047265o. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Spek EJ, Olson CA, Shi Z, Kallenbach NR. J Am Chem Soc. 1999;121:5571–5572. [Google Scholar]
- 2.(a) Kemp DS, Boyd JG, Muendel CC. Nature. 1991;352:451–454. doi: 10.1038/352451a0. [DOI] [PubMed] [Google Scholar]; (b) Vila JA, Williams RL, Grant JA, Wojcik J, Scheraga HA. Proc Natl Acad Sci U S A. 1992;89:7821–7825. doi: 10.1073/pnas.89.16.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Rohl CA, Fiori W, Baldwin RL. Proc Natl Acad Sci U S A. 1999;96:3682–3687. doi: 10.1073/pnas.96.7.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Luo P, Baldwin RL. Proc Natl Acad Sci U S A. 1999;96:4930–4935. doi: 10.1073/pnas.96.9.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Baldwin RL. J Biol Chem. 2003;278:17581–17588. doi: 10.1074/jbc.X200009200. [DOI] [PubMed] [Google Scholar]
- 4.(a) Chakrabartty A, Kortemme T, Baldwin RL. Protein Sci. 1994;3:843–852. doi: 10.1002/pro.5560030514. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Andersen NH, Tong H. Protein Sci. 1997;6:1920–1936. doi: 10.1002/pro.5560060913. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sun JE, Penel S, Doig AJ. Protein Sci. 2000;9:750–764. doi: 10.1110/ps.9.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Cochran DAE, Doig AJ. Protein Sci. 2001;10:1305–1311. doi: 10.1110/ps.50701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Groebke K, Renold P, Tsang KY, Allen TJ, McClure KF, Kemp DS. Proc Natl Acad Sci U S A. 1996;93:4025–4029. doi: 10.1073/pnas.93.9.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Vila JA, Ripoll DR, Scheraga HA. Proc Natl Acad Sci U S A. 2000;97:13075–13079. doi: 10.1073/pnas.240455797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Williams L, Kather K, Kemp DS. J Am Chem Soc. 1998;120:11033–11043. [Google Scholar]; (b) Miller JS, Kennedy RJ, Kemp DS. Biochemistry. 2001;40:305–309. doi: 10.1021/bi0019500. [DOI] [PubMed] [Google Scholar]; (c) Miller JS, Kennedy RJ, Kemp DS. J Am Chem Soc. 2002;124:945–962. doi: 10.1021/ja011726d. [DOI] [PubMed] [Google Scholar]
- 7.Job GE, Kennedy RJ, Heitmann B, Miller JS, Walker SM, Kemp DS. J Am Chem Soc. 2006;128:8227–8233. doi: 10.1021/ja060094y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Lifson S, Roig A. J Chem Phys. 1961;34:1963–1974. [Google Scholar]; (b) Doig AJ. Biophys Chem. 2002;101–102:281–293. doi: 10.1016/s0301-4622(02)00170-9. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Zhao K, Gong Y, Vologodskii A, Kallenbach NR. J Am Chem Soc. 1998;120:10646–10652. [Google Scholar]
- 10.Couch VA, Cheng N, Nambiar K, Fink W. J Phys Chem B. 2006;110:3410–3419. doi: 10.1021/jp055209j. and references cited therein. [DOI] [PubMed] [Google Scholar]
- 11.(a) Schneider JP, DeGrado WF. J Am Chem Soc. 1998;120:2764–2767. [Google Scholar]; (b) Song K, Stewart JM, Fesinmeyer RM, Andersen NH, Simmerling C. Biopolymers. 2008;89:747–760. doi: 10.1002/bip.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Scheraga HA, Vila JA, Ripoll DR. Biophys Chem. 2002;101–102:255–265. doi: 10.1016/s0301-4622(02)00175-8. [DOI] [PubMed] [Google Scholar]; (b) Garcia AE, Sanbonmatsu KY. Proc Natl Acad Sci U S A. 2002;99:2782–2787. doi: 10.1073/pnas.042496899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury S, Zhang W, Wu C, Xiong G, Duan Y. Biopolymers. 2003;68:63–75. doi: 10.1002/bip.10216. [DOI] [PubMed] [Google Scholar]
- 14.(a) Nymeyer H, Garcia AE. Proc Natl Acad Sci U S A. 100:13934–13939. doi: 10.1073/pnas.2232868100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang W, Lei HX, Chowdhury S, Duan Y. J Phys Chem B. 2004;108:7479–7489. [Google Scholar]
- 15.The classical values for [θ]222 for an infinite 100% helical segment were in the −38000 to −40000 deg-cm2/residue-dmol range4a,b with corrections for the non-H-bonded end residues bringing this value down to the −27000 to −34000° range observed for 15-residue helices (Chakrabartty A, Kortemme T, Padmanabhan S, Baldwin RL. Biochemistry. 1993;32:5560–5565. doi: 10.1021/bi00072a010.Andersen NH, Cort JR, Liu Z, Sjoberg SJ, Tong H. J Am Chem Soc. 1996;118:10309–10310.These values have been used for the majority of the CD studies of residue helix propensities (relative w values). CD studies of fluoroalcohol titrations (Luo P, Baldwin RL. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133.) and of short helices of fixed geometry (Chin DH, Woody RW, Rohl CA, Baldwin RL. Proc Natl Acad Sci U S A. 2002;99:15416–15421. doi: 10.1073/pnas.232591399.) have increased the 100% helical value to −44000° and decreased the fraying correction for shorter sequences. Studies of block co-polymers have yielded [θ]222(100%) values as large as −61000°7.
- 16.Barua B, Lin JC, Williams DV, Neidigh JW, Kummler P, Andersen NH. Protein Eng, Design Selection. 2008;21:171–185. doi: 10.1093/protein/gzm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Park SH, Shalongo W, Stellwagen E. Proteins: Struct, Funct, Genet. 1998;33:167–176. doi: 10.1002/(sici)1097-0134(19981101)33:2<167::aid-prot3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]; (b) Fesinmeyer RM, Peterson ES, Dyer RB, Andersen NH. Protein Sci. 2005;14:2324–2332. doi: 10.1110/ps.051510705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jas GS, Kuczera K. Biophys J. 2004;87:3786–3798. doi: 10.1529/biophysj.104.045419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goch G, Maciejczyk M, Oleszczuk M, Stachowiak D, Malicka J, Bierzyński A. Biochemistry. 2003;42:6840–6847. doi: 10.1021/bi027339d. [DOI] [PubMed] [Google Scholar]
- 20.(a) Andersen NH, Dyer RB, Fesinmeyer RM, Gai F, Liu Z, Neidigh JW, Tong H. J Am Chem Soc. 1999;121:9879–9880. [Google Scholar]; (b) Werner JH, Dyer RB, Fesinmeyer RM, Andersen NH. J Phys Chem B. 2002;106:487–494. [Google Scholar]; (c) Huang CY, Klemke JW, Getahun Z, DeGrado WF, Gai F. J Am Chem Soc. 2001;123:9235–9238. doi: 10.1021/ja0158814. [DOI] [PubMed] [Google Scholar]; (d) Starzyk A, Barber-Armstrong W, Sridharan M, Decatur SM. Biochemistry. 2005;44:369–376. doi: 10.1021/bi0481444. [DOI] [PubMed] [Google Scholar]
- 21.(a) Manas ES, Getahun Z, Wright WW, DeGrado WF. J Am Chem Soc. 2000;122:9883–9890. [Google Scholar]; (b) Walsh STR, Cheng RP, Wright WW, Alonso DOV, Daggett V, Vanderkooi JM, DeGrado WF. Protein Sci. 2003;12:520–531. doi: 10.1110/ps.0223003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calculated IR spectra based on MD ensembles for an (AAAKA)3 peptide suggest that a shift to higher frequency is expected at the alanine C=O four residues upstream from the lysine (Gnanakaran S, Hochstrasser RM, Garcia AE. Proc Natl Acad Sci U S A. 2004;101:9229–9234. doi: 10.1073/pnas.0402933101.). Some experimental evidence for this has been reported by Decatur,20d but that study also indicated that these positions are those that are most notably desolvated by the addition of trifluoroethanol. We have not observed17b,20a,b a resolved 13C=O peak for desolvated sites in (AAAKA)-repeat 13C′-isotopomers.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


