Abstract
Bacillus tusciae Bonjour & Aragno 1994 is a hydrogen-oxidizing, thermoacidophilic spore former that lives as a facultative chemolithoautotroph in solfataras. Although 16S rRNA gene sequencing was well established at the time of the initial description of the organism, 16S sequence data were not available and the strain was placed into the genus Bacillus based on limited chemotaxonomic information. Despite the now obvious misplacement of strain T2 as a member of the genus Bacillus in 16S rRNA-based phylogenetic trees, the misclassification remained uncorrected for many years, which was likely due to the extremely difficult, analysis-hampering cultivation conditions and poor growth rate of the strain. Here we provide a taxonomic re-evaluation of strain T2T (= DSM 2912 = NBRC 15312) and propose its reclassification as the type strain of a new species, Kyrpidia tusciae, and the type species of the new genus Kyrpidia, which is a sister-group of Alicyclobacillus. The family Alicyclobacillaceae da Costa and Rainey, 2010 is emended. The 3,384,766 bp genome with its 3,323 protein-coding and 78 RNA genes is part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: hydrogen-oxidizing, aerobe, facultative chemolithoautotroph, thermoacidophile, free-living, solfatara, spore-forming, Bacillaceae, GEBA
Introduction
Strain T2T (= DSM 2912 = NBRC 15312) is the type strain of the Bacillus tusciae [1]. Both strain T2T and strain T201 were isolated from ponds in the solfatara of San Federigo, a geothermal area near Lago, Tuscany, Italy [1,Table 1], as indicated in the Latin species epithet tusciae, meaning from Tuscia [1]. In the original study of Bonjour and Aragno [1], DNA:DNA reassociation studies of strains T2T, Bacillus schlegelii (also autotrophic) and the thermoacidophilic Bacillus acidocaldarius (later on reclassified as Alicyclobacillus acidocaldarius) from another hot spring were carried out. Because of the low levels of hybridization observed, the authors proposed that strains T2T and T201 formed a distinct taxonomic unit [1]. However, only limited chemotaxonomic data was available at the time and 16S rRNA sequence data was not available, which led the authors to place the species into the genus Bacillus [1]. Rather difficult cultivation conditions and the poor growth rate of strain T2T likely delayed an earlier re-classification although Rainey et al. previously noted the link to the genus Alicyclobacillus [19]. This was recently corroborated by the observation that strain T2T as well as A. acidocaldarius lack the sspE gene for acid-soluble spore proteins frequently found in members of the Bacillales [20]. Here we present a summary classification and a set of features for B. tusciae strain T2T, a description of the complete genome sequencing and annotation and a proposal to reclassify B. tusciae as a member of the new genus Kyrpidia as Kyrpidia tuscae comb. nov.
Table 1. Classification and general features of Kyrpidia tusciae strain T2T according to the MIGS recommendations [2] and the NamesforLife database [3].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [4] | |
| Phylum Firmicutes | TAS [5-7] | ||
| Class Bacilli | TAS [8,9] | ||
| Order Bacillales | TAS [10,11] | ||
| Family Bacillaceae | TAS [10,12] | ||
| Genus Bacillus | TAS [10,13,14] | ||
| Species Bacillus tusciae | TAS [1,15] | ||
| Type strain T2 | TAS [1] | ||
| Revised classification | Family Alicyclobacillaceae | TAS [9,16] | |
| Genus Kyrpidia | NAS | ||
| Species Kyrpidia tusciae | NAS | ||
| Gram stain | positive | TAS [1] | |
| Cell shape | straight rods | TAS [1] | |
| Motility | not reported, but lateral flagella visible | TAS [1] | |
| Sporulation | sporulating | TAS [1] | |
| Temperature range | thermophile, grows > 47°, < 65°C | TAS [1] | |
| Optimum temperature | 55°C | TAS [1] | |
| Salinity | not reported | ||
| MIGS-22 | Oxygen requirement | aerobic | TAS [1] |
| Carbon source | short chain fatty acids, amino acids and alcohols | TAS [1] | |
| Energy metabolism | facultatively chemolithoautotroph | TAS [1] | |
| MIGS-6 | Habitat | hot, acidic solfatara fields | TAS [1] |
| MIGS-15 | Biotic relationship | free living | TAS [1] |
| MIGS-14 | Pathogenicity | none | NAS |
| Biosafety level | 1 | TAS [17] | |
| Isolation | ponds in solfatara | TAS [1] | |
| MIGS-4 | Geographic location | San Frederigo, near Lago, Tuscany (Italy) | TAS [1] |
| MIGS-5 | Sample collection time | about or before 1984 | TAS [1] |
| MIGS-4.1 MIGS-4.2 |
Latitude Longitude |
43.33 10.50 |
NAS |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | about 155 m | NAS |
Evidence codes - TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [18].
Classification and features
A representative genomic 16S rRNA sequence of B. tusciae T2T was compared using NCBI BLAST [21] under default settings (e.g., considering only the high-scoring segment pairs (HSPs) from the best 250 hits) with the most recent release of the Greengenes database [22] and the relative frequencies of taxa and keywords (reduced to their stems [23]) were determined, weighted by BLAST scores. The most frequently occurring genera were Alicyclobacillus (67.8%), Bacillus (18.2%), Thermoactinomyces (6.0%), Paenibacillus (5.6%) and Exiguobacterium (1.6%) (99 hits in total). Regarding the seven hits to sequences from members of the species, the average identity within HSPs was 99.6%, whereas the average coverage by HSPs was 99.5%. Among all other species, the one yielding the highest score was A. acidiphilus NR_028637, which corresponds to an identity of 92.4% and an HSP coverage of 57.7%. (Note that the Greengenes database uses the INSDC (= EMBL/NCBI/DDBJ) annotation, which is not an authoritative source for nomenclature or classification.) The highest-scoring environmental sequence was EU638396 ('Ecological Role Firmicutes Identified Thermophilic Microbial Fuel Cells thermophilic microbial fuel cell acetate-fed experiment clone SHBZ1905')), which showed an identity of 99.4% and an HSP coverage of 90.0%. The most frequently occurring keywords within the labels of all environmental samples that were hits were 'microbi' (13.3%), 'thermophil' (12.5%), 'cell, fuel' (12.4%), 'ecolog, firmicut, identifi, role' (6.2%) and 'experi' (6.1%) (151 hits in total). The most frequently occurring keywords within the labels of those environmental samples which yielded hits of a higher score than the highest scoring species were 'microbi' (13.9%), 'cell, fuel, thermophil' (13.1%), 'ecolog, firmicut, identifi, role' (6.5%), 'experi' (6.4%) and 'acetate-f' (4.7%) (124 hits in total). These keywords corroborate the features of the environment from which strain T2T was isolated.
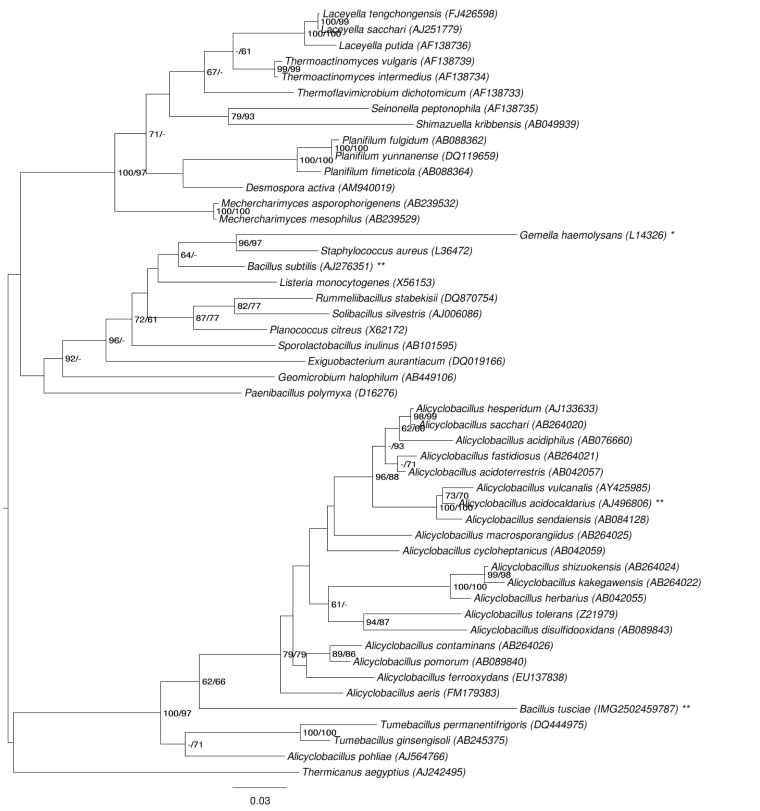
Figure 1 shows the phylogenetic neighborhood of B. tusciae in a 16S rRNA tree. The sequences of the five 16S rRNA gene copies in the genome differ from each other by up to eight nucleotides, and differ by up to six nucleotides from the previously published 16S rRNA sequence AB042062.
Figure 1.
Phylogenetic tree highlighting the position of B. tusciae relative to the type strains within the families Alicyclobacillaceae and Thermoactinomycetaceae, which gave the best hits when conducting a BLAST search [22] against the sequences contained in the last release from the All-Species-Living-Tree Project [24], and the type strains of all other families within the order Bacillales. The tree was inferred from 1,403 aligned characters [25,26] of the 16S rRNA gene sequence under the maximum likelihood (ML) criterion [27]. Rooting was done initially using the midpoint method [28] and then checked for its agreement with the current classification (Table 1). The branches are scaled in terms of the expected number of substitutions per site. Numbers adjacent to the branches are support values from 450 ML bootstrap replicates [29] (left) and from 1,000 maximum parsimony (MP) bootstrap replicates [30] (right) if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [31] are labeled with one asterisk, those also listed as 'Complete and Published' with two asterisks [32,33].
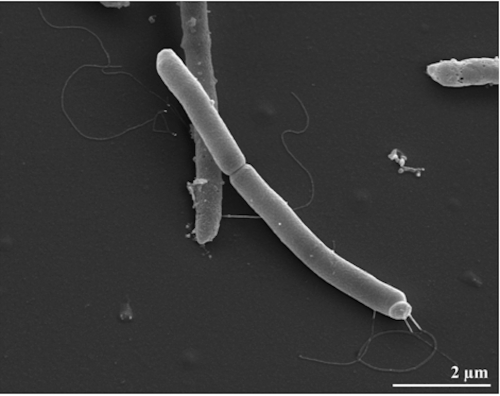
B. tusciae cells are straight rods measuring 0.8 x 4-5 μm length (Figure 2) with single laterally inserted flagella [1]. In young cultures, cells stain Gram-positive and exhibit oval, subterminal spores that swell the sporangium [1]. Cultures grow at 55°C at a pH ranging from 4.2 to 4.8 [1]. Although cells grow best under autotrophic conditions via hydrogen oxidation, they can also grow heterotrophic on alcohols, amino acids and short chain fatty acids, but not with sugars [1]. Substrate usage was described in detail by Bonjour and Aragno [1]. Cells contain a soluble malate-dehydrogenase activity, which cannot reduce pyridine (NAD+/NAD(P)+) [1], nor NADH oxidation was observed [1]. Hydrogenase activity was reported as being inducible [1]. An operational Calvin cycle was reported based on the presence of ribulose-1,5-bisphosphate carboxylase activity in autotrophically-grown cells [1]. Autotrophically grown cells show inclusions of poly-β-hydroxybutyric acid [1].
Figure 2.
Scanning electron micrograph of B. tusciae strain T2T
Chemotaxonomy
The structure of the cell wall of strain T2T was reported as typically Gram-positive, without a protein coat [1]. ω-alicyclic fatty acids are lacking and menaquinone MK-7 was identified as the main quinone (personal communication Brian J. Tindall).
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position [34], and is part of the Genomic Encyclopedia of Bacteria and Archaea project [35]. The genome project is deposited in the Genomes OnLine Database [31] and the complete genome sequence in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Two genomic libraries: Sanger 8 kb pMCL200 and 454 standard library |
| MIGS-29 | Sequencing platforms | ABI3730, 454 GS FLX |
| MIGS-31.2 | Sequencing coverage | 5.5 × Sanger; 25.2 × pyrosequence |
| MIGS-30 | Assemblers | Newbler version 2.0.0-PreRelease-07/15/2008, phrap |
| MIGS-32 | Gene calling method | Prodigal 1.4, GenePRIMP |
| Genbank ID | CP002017 | |
| Genbank Date of Release | April 5, 2010 | |
| GOLD ID | Gc01268 | |
| NCBI project ID | 31345 | |
| Database: IMG-GEBA | 646564511 | |
| MIGS-13 | Source material identifier | DSM 2912 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
B. tusciae strain T2T, DSM 2912, was grown in DSMZ medium 369 (Bacillus tusciae medium) [36] at 50°C. DNA was isolated from 1-1.5 g of cell paste using Qiagen Genomic 500 DNA Kit (Qiagen, Hilden, Germany) with lysis modification st/LALMP according to Wu et al. [35]. DNA is available through the DNA Bank Network [37].
Genome sequencing and assembly
The genome was sequenced using a combination of Sanger and 454 sequencing platforms. All general aspects of library construction and sequencing can be found at the JGI website [38]. Pyrosequencing reads were assembled using the Newbler assembler (Roche). Large Newbler contigs were broken into 3,650 overlapping fragments of 1,000 bp and entered into assembly as pseudo-reads. The sequences were assigned quality scores based on Newbler consensus q-scores with modifications to account for overlap redundancy and adjust inflated q-scores. A hybrid 454/Sanger assembly was made using the phrap assembler [39]. Possible mis-assemblies were corrected with Dupfinisher and gaps between contigs were closed by editing in Consed, by custom primer walks from sub-clones or PCR products [40]. A total of 549 Sanger finishing reads were needed to close gaps, to resolve repetitive regions, and to raise the quality of the finished sequence. The error rate of the completed genome sequence is less than 1 in 100,000. Together, the combination of the Sanger and 454 sequencing platforms provided 30.7 × coverage of the genome. The final assembly contains 18,870 Sanger reads and 413,112 pyrosequencing reads.
Genome annotation
Genes were identified using Prodigal [41] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [42]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) non-redundant database, UniProt, TIGR-Fam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review platform [43].
Genome properties
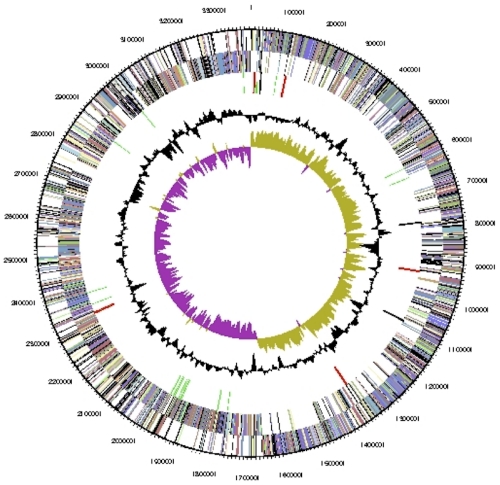
The genome consists of a 3,384,766 bp long circular chromosome with a G+C content of 59.1% (Table 3 and Figure 3). Of the 3,401 genes predicted, 3,323 were protein-coding genes, and 78 RNAs; 173 pseudogenes were also identified. The majority of the protein-coding genes (70.7%) were assigned a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 3,384,766 | 100.00% |
| DNA coding region (bp) | 2,896,588 | 85.58% |
| DNA G+C content (bp) | 2,000,875 | 59.11% |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 3,401 | 100.00% |
| RNA genes | 78 | 2.29% |
| rRNA operons | 5 | |
| Protein-coding genes | 3,323 | 97.71% |
| Pseudo genes | 173 | 5.09% |
| Genes with function prediction | 2,404 | 70.69% |
| Genes in paralog clusters | 718 | 21.11% |
| Genes assigned to COGs | 2,456 | 74.21% |
| Genes assigned Pfam domains | 2,657 | 78.12% |
| Genes with signal peptides | 530 | 15.58% |
| Genes with transmembrane helices | 728 | 21.41% |
| CRISPR repeats | 4 |
Figure 3.
Graphical circular map of the chromosome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 153 | 5.6 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 198 | 7.2 | Transcription |
| L | 192 | 7.0 | Replication, recombination and repair |
| B | 1 | 0.0 | Chromatin structure and dynamics |
| D | 36 | 1.3 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.0 | Nuclear structure |
| V | 44 | 1.6 | Defense mechanisms |
| T | 121 | 4.4 | Signal transduction mechanisms |
| M | 116 | 4.2 | Cell wall/membrane/envelope biogenesis |
| N | 62 | 2.3 | Cell motility |
| Z | 1 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 41 | 1.5 | Intracellular trafficking, secretion and vesicular transport |
| O | 95 | 3.5 | Posttranslational modification, protein turnover, chaperones |
| C | 217 | 7.9 | Energy production and conversion |
| G | 111 | 4.1 | Carbohydrate transport and metabolism |
| E | 255 | 9.3 | Amino acid transport and metabolism |
| F | 68 | 2.5 | Nucleotide transport and metabolism |
| H | 133 | 4.9 | Coenzyme transport and metabolism |
| I | 145 | 5.3 | Lipid transport and metabolism |
| P | 133 | 4.9 | Inorganic ion transport and metabolism |
| Q | 97 | 3.5 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 298 | 10.9 | General function prediction only |
| S | 224 | 8.2 | Function unknown |
| - | 945 | 27.8 | Not in COGs |
Insights into the genome
At the time when the species name B. tusciae was validly published [9], a 16S rRNA gene sequence of the strain was not generated, but DNA-DNA hybridizations (DDHs) were performed with two reference strains, B. schlegelii [44] and B. acidocaldarius [45], to show that strain T2T represents a novel species within the genus Bacillus [1]. While B. schlegelii was named by the same senior author as B. tusciae and most probably is as misplaced in the genus Bacillus as B. tusciae (see below), B. acidocaldarius was subsequently reclassified as Alicyclobacillus acidocaldarius [46]. In retrospect, considering 16S rRNA gene sequence identities of 88.8% (for A. acidocaldarius) and 85.7% (for B. schlegelii) it is not surprising that Bonjour and Aragon observed only 17% and 0% DDH similarity, respectively, for their novel isolate [1]. While the members of the genus Alicyclobacillus do in fact represent the closest relatives of strain T2T (Figure 1) with 87.5% to 89.0% 16S rRNA sequence identity in EzTaxon [47], the appropriate taxonomic placement of B. schlegelii seems to be an unresolved question, with no greater than 89.3% rRNA sequence identity found with other type strains, none of which are members of the genus Bacillus [47]. Figure 1 shows B. tusciae as a sister group of one of the two clades that comprise Alicyclobacillus, with A. pohliae more closely related to Tumebacillus than to the other members of the genus and likely being misclassified based on the current data. However, while the clade comprising B. tusciae, Alicyclobacillus and Tumebacillus has significant statistical support (100% under ML and 97% under MP), the claim that the clade comprises only Tumebacillus and A. pohliae is unsupported.
As two of the genera selected for Figure 1, Alicyclobacillus and Bacillus, appeared as polyphyletic in the tree, we conducted both unconstrained heuristic searches for the best tree under the ML [27] and MP criterion [30] as well as searches constrained for the monophyly of these two genera, respectively (for details of the data matrix see the figure caption). The best-known ML tree had a log likelihood of -13,289.73, whereas the best trees found under the constraint of Alicyclobacillus monophyly had a log likelihood of -13,297.23 and was not significantly worse in the Shimodaira-Hasegawa test as implemented in RAxML [27] (α = 0.05). However, when enforcing Bacillus monophyly, the resulting log likelihood was -13,412.24, significantly worse than the best tree (α = 0.01). The best-known MP trees had a score of 2,362, whereas the best trees found under the constraint of Alicyclobacillus monophyly had a score of 2,374 and were not significantly worse in the Kishino-Hasegawa test as implemented in PAUP* [30] (α = 0.05). When enforcing Bacillus (Bacillus subtilis and B. tusciae) monophyly, the resulting score was 2,439, significantly worse than the best tree (p < 0.0001). (See, e.g., chapter 21 in [48] for an in-depth description of such paired-site tests.). Accordingly, the current classification of B. tusciae in Bacillus is at odds with the 16S rRNA data and does not reflect the natural relationships based on that gene. In contrast, the placement of A. pohliae in Alicyclobacillus is not significantly disputed by the data.
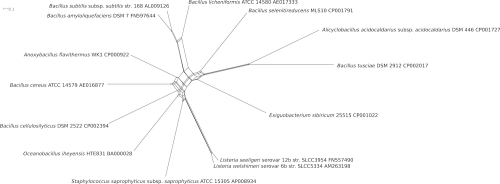
Table 5 shows the whole-genome distances between B. tusciae, A. acidocaldarius [32] and B. subtilis [33] as calculated using the genome-to-genome distance calculator [49-51]. The lower left triangle shows those distances derived by dividing the total sequence length not covered by HSPs through total genome length (left) and by dividing the total number of non-identical base pairs within HSPs by total HSP length (right); the upper right triangle shows the distance derived by dividing total genome length minus total number of identical base pairs within HSPs by total genome length. As expected, those distances relating HSP coverage and number of identical base pairs within HSPs to total genome length are higher between B. tusciae and B. subtilis than between B. tusciae and A. acidocaldarius. That the distances relating the number of identical base pairs to total HSP length behave differently indicates that the genomic similarities between B. tusciae and B. subtilis are more strongly restricted to more conserved sequences, a kind of saturation phenomenon [50]. Figure 4 shows an unrooted phylogenetic network inferred using the Neighbor-Net algorithm from whole-genome distances calculated with GGDC [49-51]. The grouping of B. tusciae and A. acidocaldarius, as well as the very tree-like appearance of this part of the network indicate that genomic data are also in conflict with the placement of B. tusciae within Bacillus.
Table 5. Genome-to-genome distances as calculated using GGDC [49-51].
Figure 4.
Unrooted phylogenetic network inferred with the Neighbor-Net algorithm as implemented in SplitsTree version 4.10 [52] from genome-to-genome distances calculated using GGDC [49-51]. The analysis includes all completely sequenced Bacillales type strain genomes as registered in GOLD at the time of publication [31]. Here, the logarithmic version of the distance calculated as the total genome length minus total number of identical base pairs within HSPs, divided by total genome length (see Table 5), was used as GGDC distance.
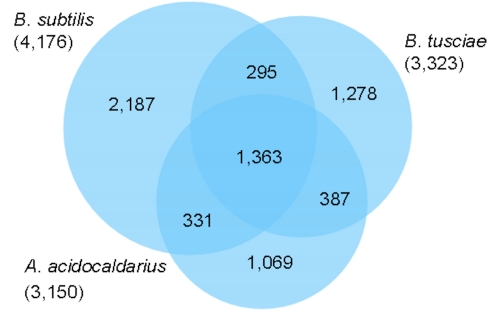
The fraction of shared genes in the genomes of B. tusciae T2T, A. acidocaldarius [32] and B. subtilis [33] is shown in a Venn diagram (Figure 5). The numbers of pairwise shared genes were calculated with the phylogenetic profiler function of the IMG-ER platform [43]. The homologous genes within the genomes were detected with a maximum e-value of 10-5 and a minimum identity of 30%.
Figure 5.
Venn diagram depicting the intersections of protein sets between the target genome and representative genomes of Bacillus and Alicyclobacillus.
A total of 1,363 of the genes are shared by all three genomes, with about equal numbers of genes (295-387) shared between pairs of genomes to the exclusion of the third genome. Within the 1,278 unique genes of B. tusciae that have no detectable homologues in the genomes of A. acidocaldarius and B. subtilis (under the sequence similarity thresholds used for the comparison) are the genes encoding the key enzymes for the Calvin cycle: ribulose-1,5-bisphosphate carboxylase (Btus_2871 large subunit, Btus_2872 small subunit) and the phosphoribulokinase (Btus_2868). A closer look at the genome of B. tusciae revealed additional genes coding for enzymes of the Calvin cycle located in the immediate neighborhood of the ribulose-1,5-bisphosphate carboxylase genes: phosphoglyceratekinase (Btus_2865), fructose-bisphosphate aldolase (Btus_2867), phosphoribulokinase (Btus_2868), glyceraldehyde-3-phosphate dehydrogenase (Btus_2869), fructose-1,6-bisphosphatase (Btus_2870) ribulose-1,5-bisphosphate carboxylase (Btus_2871 large subunit, Btus_2872 small subunit) which are probably organized as an operon. In autotrophically-grown cells of B. tusciae, an active ribulose-1,5-bisphosphate carboxylase in an operating Calvin cycle was reported [1].
The phylogenetic position of strain T2T as shown in Figures 1 and 4, corroborated by its primarily autotrophic energy metabolism [1] (Tables 1 and 6), and the lack of sspE genes for acid-soluble spore proteins [20] (Table 6), indicated a clear need to reclassify B. tusciae. The joint but distinct phylogenetic position (Figure 1) supports the establishment of a novel genus rather than the inclusion of B. tusciae in Alicyclobacillus. As shown above, 16S rRNA data do not indicate the monophyly of the remaining Alicyclobacillus spp., but are not in significant conflict with it. The branch leading to B. tusciae is rather long, indicating a considerable degree of independent evolution (relative to the other taxa included in the tree, which include the closest relatives of B. tusciae in the current LTP release, see above). Whether the lack of ω-alicyclic fatty acids in A. pohliae and perhaps other Alicyclobacillus spp. can be used in later studies for a revision of Alicyclobacillus remains to be seen.
Table 6. Typical features of reference taxa.
| Bacillus tusciae T2T DSM 2912 [1] | Genus Alicyclobacillus [53,54] | Genus Bacillus [54] |
|
|---|---|---|---|
| cell shape | straight rods, 4-5 μm long | rods, 1.5-6.3 μm long | rods, normally up to 5 µm long |
| Gram stain | positive | positive | positive |
| oxygen requirement | aerobic, facultative anaerobic, but not with nitrate | aerobic, facultative anaerobic | aerobic, facultative anaerobic, a few species are strictly anaerobic |
| endospores | subterminal, oval | terminal up to subterminal | ellipsoidal, central |
| sspE genes for acid-soluble spore proteins | missing | missing | frequent |
| growth temperature | >47°C to <67°C, opt. 55°C | 4-70°C, opt. 35-65°C | 10-60°C |
| pH optimum | 4.2-4.8 | < 4.5, (range 1.5-5) | 5-10 |
| phenotype | thermoacidophile | thermoacidophiles | some obligate or facultative thermophiles |
| habitat | ponds in solfatara of geothermal area | acidic geothermal sites (soil and water), fruit juices, ores | often saprophytes in decaying organic matter |
| flagellation | single lateral flagellum | motility reported for some species | motile or nonmotile; lateral |
| menaquinones | MK-7 (personal communication Brian J. Tindall) |
MK-7 | MK-7 |
| major membranous lipid components | no ω-alicyclic fatty acids (personal communication Brian J. Tindall) | Large amounts (>80%) of ω-alicyclic fatty acids with six- or seven carbon rings, such as ω-cyclohexane undecanoic acid -C17:0 and ω-cyclohexane tridecanoic acid -C19:0. Three species do not possess these fatty acids. Some strains are known to form hopanoids. | cellular fatty acids: ai-C15:0, i-C15:0, ai-C15:0; no ω-alicyclic fatty acids |
| oxidase/catalase | weak/weak | ±/± | ±/± |
| Inclusion bodies | poly-β-hydroxybutyric acid when grown autotrophically under ammonium starvation | not specified | some species |
| energy metabolism | grows best under autotrophic conditions, chemolithoautotrophic with H2 and CO2, but also chemoorganoheterotrophic; does not metabolize sugars | chemoorganotrophic or mixotrophic; carbohydrates, organic acids and amino acids can be utilized. Mixotrophic species utilize Fe2+ and S0 |
chemoorganotrophic |
On the basis of the above-mentioned physiological, chemotaxonomic and phylogenetic characteristics of strain T2T, a novel genus is proposed, Kyrpidia, as the second genus in the parent family Alicyclobacillaceae, and a novel species is proposed, Kyrpidia tusciae sp. nov., comb. nov. Comparative characteristics of strain T2T are given in Table 1.
Furthermore the phylogenetic analysis as shown in Figure 1 clearly supports the assignment of the genus Tumebacillus to the family Alicyclobacillaceae.
Emended description of the family Alicyclobacillaceae da Costa and Rainey 2010.
The description of the family Alicyclobacillaceae is given by da Costa and Rainey 2010 in [54]. Acid may be produced from carbohydrates or not. The family is comprised of the genera Alicyclobacillus, Tumebacillus and Kyrpidia.
Description of Kyrpidia gen. nov.
Kyrpidia (Kyr.pi´di.a N.L. fem. n. Kyrpidia named in honor of Nikolaos C. Kyrpides, a Greek-American genomics scientist, who co-initiated the Genomic Encyclopedia of Archaea and Bacteria).
Cells are straight rods, 1.5 to 5 μm long, facultatively anaerobic, Gram-positive, chemolitooautotrophic or chemoorganoheterotrophic. Thermoacidophilic; growth occurs above 42°C and below 67°C, with an optimum at 55°C, and at pH 4.2-7.5. Endospores are formed, but sspE genes for acid-soluble spore proteins are not found. The predominant menaquinone is MK-7. Major fatty acids are iso-C15:0 and iso-C17:0; ω-alicyclic fatty acids are not present. The mol% G + C content of the type strain of the type species is 59.11 mol%. The type species is Kyrpidia tusciae. Kyrpidia is a member of the Alicyclobacillaceae.
Description of Kyrpidia tusciae (Bonjour & Aragno 1984) comb. nov.
Kyrpidia tusciae (tus´ci.ae L. gen.n. tusciae from Tuscia; named after Tuscia, a region in central Italy where the Etruscians (Tuscii) lived and where the organism was found.)
Basonym: Bacillus tusciae Bonjour and Aragno 1984.
The genus Kyrpidia is comprised of one species Kyrpidia tusciae. The characteristics of the species are given in the genus description and the description given by Bonjour and Aragno [1].
The type strain is T2T (= DSM 2912 = NBRC 15312).
Acknowledgements
We would like to gratefully acknowledge the help of Claudia Wahrenburg for growing the B. tusciae cells, and Susanne Schneider for DNA extraction and quality analysis (both at DSMZ). This work was performed under the auspices of the US Department of Energy Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, as well as German Research Foundation (DFG) INST 599/1-1.
References
- 1.Bonjour F, Aragno M. Bacillus tusciae, a new species of thermoacidophilic, facultatively chemoautotrophic, hydrogen oxidizing sporeformer from a geothermal area. Arch Microbiol 1984; 139:397-401 10.1007/BF00408386 [DOI] [Google Scholar]
- 2.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thompson N, Allen MJ, Anguiuoli SV, et al. Towards a richer description of our complete collection of genomes and metagenomes: the “Minimum Information about a Genome Sequence” (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrity G. NamesforLife. BrowserTool takes expertise out of the database and puts it right in the browser. Microbiol Today 2010; 37:9 [Google Scholar]
- 4.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibbons NE, Murray RGE. Proposals concerning the higher taxa of bacteria. Int J Syst Bacteriol 1978; 28:1-6 10.1099/00207713-28-1-1 [DOI] [Google Scholar]
- 6.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 7.Murray RGE. The Higher Taxa, or, a Place for Everything...? In: Holt JG (ed), Bergey's Manual of Systematic Bacteriology, First Edition, Volume 1, The Williams and Wilkins Co., Baltimore, 1984, p. 31-34. [Google Scholar]
- 8.Ludwig W, Schleifer KH, Whitman WB. Class I. Bacilli class nov. In: De Vos P, Garrity GM, Jones D, N.R. Krieg W. Ludwig W, Rainey EA, Schleifer KH, Withman WB (eds) Bergey's Manual of Systematic Bacteriology, second edition, vol. 3 (The Firmicutes), Springer, Dordrecht, Heidelberg, London, New York, 2009, pp. 19-20. [Google Scholar]
- 9.List Editor List of new names and new combinations previously effectively, but not validly, published. List no. 132. Int J Syst Evol Microbiol 2010; 60:469-472 10.1099/ijs.0.022855-0 [DOI] [PubMed] [Google Scholar]
- 10.Skerman VBD, McGowan V, Sneath PHA, eds. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [Google Scholar]
- 11.Prévot AR. In: Hauduroy P, Ehringer G, Guillot G, Magrou J, Prévot AR, Rosset, Urbain A (eds) Dictionnaire des Bactéries Pathogènes, 2nd ed., Masson, Paris, 1953, pp. 1-692. [Google Scholar]
- 12.Fischer A. Untersuchungen über bakterien. Jahrbücher für Wissenschaftliche Botanik 1895; 27:1-163 [Google Scholar]
- 13.Cohn F. Untersuchungen über Bakterien. Beitr Biol Pflanz 1872; 1:127-224 [Google Scholar]
- 14.Gibson T, Gordon RE. Genus I. Bacillus Cohn 1872, 174; Nom. gen. cons. Nomencl. Comm. Intern. Soc. Microbiol. 1937, 28; Opin. A. Jud. Comm. 1955, 39. In: Buchanan RE, Gibbons NE (eds), Bergey's Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 529-550. [Google Scholar]
- 15.List Editor Validation List no. 17. Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1985; 35:223-225 10.1099/00207713-35-2-223 [DOI] [Google Scholar]
- 16.da Costa MS, Rainey FA. Family II. Alicyclobacillaceae fam. nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 3, Springer-Verlag, New York, 2009, p. 229. [Google Scholar]
- 17.BAuA Classification of Bacteria and Archaea in risk groups. http://www.baua.de. TRBA 2010; 466:37 [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rainey FA, Fritze D, Stackebrandt E. The phylogenetic diversity of thermophilic members of the genus Bacillus as revealed by 16S rDNA analysis. FEMS Microbiol Lett 1994; 115:205-211 10.1111/j.1574-6968.1994.tb06639.x [DOI] [PubMed] [Google Scholar]
- 20.Vyas J, Cox J, Setlow B, Coleman WH, Setlow P. Extremely variable conservation of γ-type small, acid-soluble proteins from spores of some species in the bacterial order Bacillales. J Bacteriol 2011; 193:1884-1892 10.1128/JB.00018-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-410 [DOI] [PubMed] [Google Scholar]
- 22.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-5072 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porter MF. An algorithm for suffix stripping. Program: electronic library and information systems 1980; 14:130-137.
- 24.Munoz R, Yarza P, Ludwig W, Euzéby J, Amann R, Schleifer KH, Glöckner FO, Rosselló-Móra R. Release LTPs104 of the All-Species Living Tree. Syst Appl Microbiol 2011; 34:169-170 10.1016/j.syapm.2011.03.001 [DOI] [PubMed] [Google Scholar]
- 25.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 26.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 27.Stamatakis A, Hoover P, Rougemont J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 28.Hess PN, De Moraes Russo CA. An empirical test of the midpoint rooting method. Biol J Linn Soc Lond 2007; 92:669-674 10.1111/j.1095-8312.2007.00864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How Many Bootstrap Replicates Are Necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [Google Scholar]
- 30.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0 b10. Sinauer Associates, Sunderland, 2002. [Google Scholar]
- 31.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346-D354 10.1093/nar/gkp848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mavromatis K, Sikorski J, Lapidus A, Glavina Del Rio T, Copeland A, Tice H, Cheng JF, Lucas S, Chen F, Nolan M, et al. Complete genome sequence of Alicyclobacillus acidocaldarius type strain (104-IAT). Stand Genomic Sci 2010; 2:9-18 10.4056/sigs.591104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotini A, Borchert S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 1997; 390:249-256 10.1038/36786 [DOI] [PubMed] [Google Scholar]
- 34.Klenk HP, Göker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 2010; 33:175-182 10.1016/j.syapm.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 35.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova N, Kunin V, Goodwin L, Wu M, Tindall BJ. A phylogeny-driven genomic encyclopedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.List of growth media used at DSMZ: http://www.dsmz.de/microorganisms/ media_list.php
- 37.Gemeinholzer B, Dröge G, Zetzsche H, Haszprunar G, Klenk HP, Güntsch A, Berendsohn WG, Wägele JW. The DNA Bank Network: the start from a German initiative. Biopreservation and Biobanking 2011; 9:51-55 10.1089/bio.2010.0029 [DOI] [PubMed] [Google Scholar]
- 38.The DOE Joint Genome Institute http://www.jgi.doe.gov
- 39.Phrap and Phred for Windows. MacOS, Linux, and Unix. http://www.phrap.com
- 40.Sims D, Brettin T, Detter JC, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome sequence of Kytococcus sedentarius type strain (541T). Stand Genomic Sci 2009; 1:12-20 10.4056/sigs.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal Prokaryotic Dynamic Programming Genefinding Algorithm. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A Gene Prediction Improvement Pipeline for microbial genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 43.Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]
- 44.Schenk A, Aragano M. Bacillus schlegelii a new species of thermophilic, facultatively chemolithoautotrophic bacterium oxidizing molecular hydrogen. J Gen Microbiol 1979; 115:333-341 [Google Scholar]
- 45.Darland G, Brock TD. Bacillus acidocaldarius sp. nov., an acidophilic thermophilic spore-forming bacterium. J Gen Microbiol 1971; 67:9-15 [Google Scholar]
- 46.Wisotzkey JD, Jurtshuk P, Jr, Fox GE, Deinhard G, Poralla K. Comparative sequence analyses on the 16S rRNA (rDNA) of Bacillus acidocaldarius, Bacillus acidoterrestris, and Bacillus cycloheptanicus and proposal for creation of a new genus, Alicyclobacillus gen. nov. Int J Syst Bacteriol 1992; 42:263-269 10.1099/00207713-42-2-263 [DOI] [PubMed] [Google Scholar]
- 47.Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 2007; 57:2259-2261 10.1099/ijs.0.64915-0 [DOI] [PubMed] [Google Scholar]
- 48.Felsenstein J. Inferring phylogenies. Sinauer Associates Inc., Sunderland, Massachusetts 2004. [Google Scholar]
- 49.Genome-to-genome distance calculator. http://ggdc.gbdp.org
- 50.Auch AF, Klenk HP, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2010; 2:142-148 10.4056/sigs.541628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Auch AF, von Jan M, Klenk HP, Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2010; 2:117-134 10.4056/sigs.531120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 2005; 23:254-267 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 53.Wisotzkey JD, Jurtshuk P, Jr, Fox GE, Deinhard G, Poralla K. Comparative sequence analyses on the 16S rRNA (rDNA) of Bacillus acidocaldarius, Bacillus acidoterrestris, and Bacillus cycloheptanicus and proposal for creation of a new genus, Alicyclobacillus gen. nov. Int J Syst Bacteriol 1992; 42:263-269 10.1099/00207713-42-2-263 [DOI] [PubMed] [Google Scholar]
- 54.da Costa MS, Rainey FA. Family II. Alicyclobacillaceae fam. nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 3, Springer-Verlag, New York, 2009, p. 229. [Google Scholar]