Abstract
Haemophilus parasuis is a member of the family Pasteurellaceae and is the etiologic agent of Glässer’s disease in pigs, a systemic syndrome associated with only a subset of isolates. The genetic basis for virulence and systemic spread of particular H. parasuis isolates is currently unknown. Strain 29755 is an invasive isolate that has long been used in the study of Glässer’s disease. Accordingly, the genome sequence of strain 29755 is of considerable importance to investigators endeavoring to understand the molecular pathogenesis of H. parasuis. Here we describe the features of the 2,224,137 bp draft genome sequence of strain 29755 generated from 454-FLX pyrosequencing. These data comprise the first publicly available genome sequence for this bacterium.
Keywords: Haemophilus parasuis, Glässer’s disease, swine
Introduction
H. parasuis is an obligate pathogen of swine [1]. The bacterium is often carried in the nasal passages [2], but not the lungs [3], of healthy pigs. Through unknown mechanisms some strains can spread systemically and may be isolated from the meninges, lungs, serosa, joints, and blood. H. parasuis strain 29755 (IA84-29755), though not the type strain, has been used extensively in a variety of investigations [4-8] and is the most fully characterized strain of the species. Originally cultured at Iowa State University from a pig exhibiting Glässer’s disease, 29755 is a serovar 5 isolate [9], a class recognized as highly virulent and frequently isolated from respiratory and systemic sites [9,10]. Of the 15 recognized serovars, serovar 5 strains are isolated more frequently worldwide than any other [11]. Strain 29755 has been used as a component of at least one commercially available H. parasuis vaccine (Suvaxyn M. hyo – parasuis, Fort Dodge Animal Health).
Classification and features
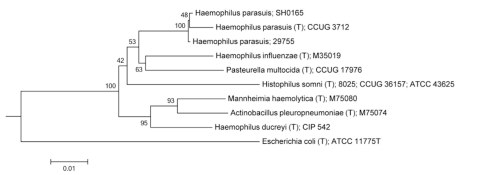
The genus Haemophilus belongs to the Gammaproteobacteria and is classified in the family Pasteurellaceae [12] (Table 1). A phylogenetic tree based on 16S ribosomal RNA sequences is depicted in Figure 1 for H. parasuis and related organisms.
Table 1. MIGS classification and general features of H. parasuis strain 29755.
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [13] | |
| Phylum Proteobacteria | TAS [14] | ||
| Class Gammaproteobacteria | TAS [15,16] | ||
| Order Pasteurellales | TAS [15,17] | ||
| Family Pasteurellaceae | TAS [18,19] | ||
| Genus Haemophilus | TAS [20-22] | ||
| Species Haemophilus parasuis | TAS [20,23] | ||
| Strain 29755 | |||
| Serotype 5 | |||
| Gram stain | negative | TAS [1] | |
| Cell shape | rods (pleomorphic) | TAS [1] | |
| Motility | nonmotile | TAS [1] | |
| Sporulation | non-sporulating | TAS [1] | |
| Temperature range | mesophile (20°C-37°C) | TAS [12] | |
| Optimum temperature | 35°C-37°C | TAS [12] | |
| Carbon source | saccharolytic | TAS [24] | |
| Energy source | chemoorganotroph | TAS [24] | |
| Terminal electron receptor | Oxygen | TAS [25] | |
| MIGS-6 | Habitat | Host, swine upper respiratory tract | TAS [1] |
| MIGS-6.3 | Salinity | 1-1.5% | TAS [12] |
| MIGS-22 | Oxygen requirement | facultative | TAS [12] |
| MIGS-15 | Biotic relationship | obligate pathogen of swine | TAS [1] |
| MIGS-14 | Pathogenicity | mild to severe | TAS [1] |
| MIGS-4 | Geographic location | Iowa | NAS |
| MIGS-5 | Sample collection time | 1970s | NAS |
| MIGS-4.1 | Latitude | not reported | |
| MIGS-4.2 | Longitude | not reported | |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | not reported |
Evidence codes - NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [26]
Figure 1.
Phylogenetic tree based on 16S rRNA of H. parasuis 29755 and type strains of some closely related species and other genera within the Pasteurellaceae. Also included is the only additional H. parasuis strain for which a genome sequence has been reported, SH0165. The tree was generated with the tree-builder available from the Ribosomal Database Project[27] using the Weighbor (weighted neighbor-joining) algorithm [28] with Jukes-Cantor distance correction [29]. Numbers to the left of branches indicate the percentage of trees in which each branch was represented in 100 replicates. An E. coli type strain was used as an outgroup.
H. parasuis is a small, non-motile, rod-shaped bacterium [1] (Figure 2). The presence of a capsule is variable and may affect colony and cellular morphology [30]. Growth of the bacterium in vitro is dependent on the coenzyme nicotinamide adenine dinucleotide (NAD, or V factor) [31] but, in contrast to some other members of the genus, does not require porphyrins like hemin (X factor) [32]. Plating on Casman Agar Base (BBL) supplemented with 1% (w/v) NAD (Sigma) and 5% GIBCO filtered horse serum (Invitrogen) or on chocolate agar produces small, translucent colonies that appear within 24 hours and reach full size in approximately two days. Colonies are nonhemolytic when grown on blood agar [1]. H. parasuis grows under normal atmosphere at 37°C, although added humidity and 5% CO2 may improve growth.
Figure 2.
Scanning electron micrograph of H. parasuis 29755
Genome sequencing and annotation
Genome project history
H. parasuis strain 29755 was selected for sequencing because it has long been used in the study of Glässer’s disease. Pyrosequencing (454 Life Sciences) was performed at the State University of New York, University at Buffalo Center of Excellence in Bioinformatics and Life Sciences. The draft genome sequence is deposited in GenBank (NZ_ABKM00000000). Summary project information is shown in Table 2 according to the Minimum Information about a Genomic Sequence (MIGS) recommendations [34] and the genome content is summarized in Table 3.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-28 | Libraries used | one 454 pyrosequence standard library |
| MIGS-29 | Sequencing platforms | 454 (FLX) |
| MIGS-30 | Assemblers | Newbler |
| MIGS-31 | Finishing quality | draft |
| MIGS-31.2 | Fold coverage | 28× |
| MIGS-32 | Gene calling method | Glimmer, GeneMark [33] |
| Genome Database release | February 14, 2008 | |
| Genbank ID | NZ_ABKM00000000 | |
| Genbank Date of Release | February 14, 2008 | |
| GOLD ID | - | |
| Project relevance | food animal pathogenesis |
Table 3. Genome statistics.
| Attribute | Value | % of totala |
|---|---|---|
| Size (bp) | 2,224,137 | 100.0% |
| G+C content (bp) | 867,413 | 39.0% |
| Coding region (bp) | 1,890,516 | 85.0% |
| Total genes | 2,309 | 100.0% |
| RNA genes | 58 | 2.5% |
| Protein-coding genes | 2,244 | 97.2% |
| Pseudogenes | noneb | 0.0% |
| Genes in paralog clusters | ndc | - |
| Genes assigned to COGs | 1,926 | 83.4% |
| PSORT cytoplasmic | 1,181 | 50.4% |
| PSORT extracellular | 5 | 0.2% |
| PSORT outer membrane | 51 | 2.2% |
| PSORT periplasmic | 52 | 2.2% |
| PSORT unknown | 1,055 | 45.0% |
a Based either on the size of the genome in base pairs or the total number of protein coding genes in the annotated genome
b Based on preliminary analysis the of draft genome
c nd = not determined
Growth conditions and DNA isolation
H. parasuis 29755 was grown from a frozen seed stock for two days under 5% CO2 at 37°C on Casman Agar Base (BBL) supplemented with 1% (w/v) NAD (Sigma) and 5% GIBCO filtered horse serum (Invitrogen). Following growth, a single colony was used to inoculate 5 ml of brain-heart infusion medium supplemented with 10 μg/ml NAD and 10 μg/ml hemin (sBHI) and the culture was incubated overnight at 37°C and 185 rpm. The next day, 2 ml of the culture were added to 100 ml of sBHI and the bacterium was again allowed to grow overnight to stationary phase at 37°C and 185 rpm. Bacterial cells were pelleted by centrifugation at 4000 × g for 10 minutes. The pellet was resuspended and used as the source of genomic DNA purified with the QIAGEN Blood & Cell Culture DNA Kit, as recommended by the manufacturer. The final preparation contained 1.12 μg/ul genomic DNA as determined by UV absorption spectrometry.
Genome sequencing and assembly
Library preparation yielded 9.65 × 108 molecules/μl of DNA with a mean size of approximately 600 nucleotides, as determined with a RNA6000 Pico chip on an Agilent 2100 Bioanalyzer. Emulsion PCR was performed at a concentration of 2 molecules per bead. Following sequencing, contigs were assembled using the 454 Newbler assembler.
Genome annotation
Genes were identified manually using GeneMark and automatically using Glimmer as part of the NCBI draft genome submission pipeline. Translated protein sequences were analyzed using PSORTb v.2.0 [35] to predict final location within the cell and assigned to COG functional categories (Table 4).
Table 4. Number of genes associated with the general COG functional categories.
| Code | Value | %agea | Description |
|---|---|---|---|
| J | 168 | 6.55 | Translation |
| A | 1 | 0.03 | RNA processing and modification |
| K | 127 | 4.96 | Transcription |
| L | 166 | 6.48 | Replication, recombination and repair |
| B | 0 | 0.00 | Chromatin structure and dynamics |
| D | 33 | 1.29 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.00 | Nuclear structure |
| V | 32 | 1.25 | Defense mechanisms |
| T | 48 | 1.87 | Signal transduction mechanisms |
| M | 134 | 5.23 | Cell wall/membrane biogenesis |
| N | 16 | 0.62 | Cell motility |
| Z | 0 | 0.00 | Cytoskeleton |
| W | 24 | 0.94 | Extracellular structures |
| U | 75 | 2.93 | Intracellular trafficking and secretion |
| O | 101 | 3.94 | Posttranslational modification, protein turnover, chaperones |
| C | 115 | 4.49 | Energy production and conversion |
| G | 139 | 5.42 | Carbohydrate transport and metabolism |
| E | 175 | 6.83 | Amino acid transport and metabolism |
| F | 57 | 2.22 | Nucleotide transport and metabolism |
| H | 97 | 3.78 | Coenzyme transport and metabolism |
| I | 43 | 1.68 | Lipid transport and metabolism |
| P | 116 | 4.53 | Inorganic ion transport and metabolism |
| Q | 25 | 0.96 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 234 | 9.13 | General function prediction only |
| S | 197 | 7.69 | Function unknown |
| - | 440 | 17.16 | Not in COGs |
a Based on the total number of protein coding genes in the annotated genome
Genome properties
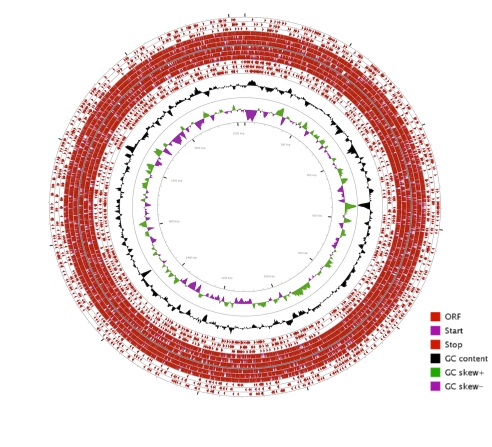
The draft genome is 2,224,137 bp and is likely comprised of one circular chromosome with a G+C content of approximately 39% (Figure 3). For display, contigs were assembled end-to-end with twenty “N” bases between contigs. Orientation and order of contigs will change when the genome sequence is closed.
Figure 3.
Graphical circular map of the H. parasuis 29755 draft pseudogenome. From the outside to the center: open reading frames (ORFs) on the forward strand (one ring for each reading frame), start and stop codons for forward and reverse strands, ORFs on the reverse strand, GC content, and GC skew. The map was generated using CGView Server [36,37].
Acknowledgements
The authors wish to thank David Alt, USDA/ARS/National Animal Disease Center for technical advice and the State University of New York, University at Buffalo Center of Excellence in Bioinformatics and Life Sciences for performing pyrosequencing. This work was supported, in part, by grants from the NIH/NCRR (D.W. Dyer, Grant #P2PRR016478), National Pork Board (G.J. Phillips and D.W. Dyer) and Iowa Healthy Livestock Initiative (G.J. Phillips and K.B. Register).
References
- 1.Rapp-Gabrielson VJ, Oliveira SR, Pijoan C. Haemophilus parasuis In: Straw BE, Zimmerman JJ, D’Allaire S, Taylor DJ (eds), Diseases of Swine, Ninth Edition, Wiley-Blackwell, Ames, Iowa, 2006, p. 681-690. [Google Scholar]
- 2.Harris DL, Ross RF, Switzer WP. Incidence of certain microorganisms in nasal cavities of swine in Iowa. Am J Vet Res 1969; 30:1621-1624 [PubMed] [Google Scholar]
- 3.Little TW. Haemophilus infection in pigs. Vet Rec 1970; 87:399-402 10.1136/vr.87.14.399 [DOI] [PubMed] [Google Scholar]
- 4.Blanco I, Galina-Pantoja L, Oliveira S, Pijoan C, Sánchez C, Canals A. Comparison between Haemophilus parasuis infection in colostrums-deprived and sow-reared piglets. Vet Microbiol 2004; 103:21-27 10.1016/j.vetmic.2004.06.011 [DOI] [PubMed] [Google Scholar]
- 5.Sgales J, Domingo M, Solano GI, Pijoan C. Immunohistochemical detection of Haemophilus parasuis serovar 5 in formalin-fixed, paraffin-embedded tissues of experimentally infected swine. J Vet Diagn Invest 1997; 9:237-243 10.1177/104063879700900303 [DOI] [PubMed] [Google Scholar]
- 6.Solano GI, Segalés J, Collins JE, Molitor TW, Pijoan C. Porcine reproductive and respiratory syndrome virus (PRRSv) interaction with Haemophilus parasuis. Vet Microbiol 1997; 55:247-257 10.1016/S0378-1135(96)01325-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveira S, Galina L, Blanco I, Canals A, Pijoan C. Naturally-farrowed, artificially-reared pigs as an alternative model for experimental infection by Haemophilus parasuis. Can J Vet Res 2003; 67:146-150 [PMC free article] [PubMed] [Google Scholar]
- 8.Solano-Aguilar GI, Pijoan C, Rapp-Gabrielson V, Collins J, Carvalho LF, Winkelman N. Protective role of maternal antibodies against Haemophilus parasuis infection. Am J Vet Res 1999; 60:81-87 [PubMed] [Google Scholar]
- 9.Rapp-Gabrielson VJ, Gabrielson DA. Prevalence of Haemophilus parasuis serovars among isolates from swine. Am J Vet Res 1992; 53:659-664 [PubMed] [Google Scholar]
- 10.Blackall PJ, Rapp-Gabrielson VJ, Hampson DJ. Serological characterisation of Haemophilus parasuis isolates from Australian pigs. Aust Vet J 1996; 73:93-95 10.1111/j.1751-0813.1996.tb09984.x [DOI] [PubMed] [Google Scholar]
- 11.Oliveira S, Pijoan C. Haemophilus parasuis: new trends on diagnosis, epidemiology and control. Vet Microbiol 2004; 99:1-12 10.1016/j.vetmic.2003.12.001 [DOI] [PubMed] [Google Scholar]
- 12.Kilian M. Genus III. Haemophilus In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume Two, The Proteobacteria, Part B, Springer, New York, 2005, p. 883-904. [Google Scholar]
- 13.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 15.List Editor Validation of publication of new names and new combinations previously effectively published outside the IJSEM. List no. 106. Int J Syst Evol Microbiol 2005; 55:2235-2238 10.1099/ijs.0.64108-0 [DOI] [PubMed] [Google Scholar]
- 16.Garrity GM, Bell JA, Lilburn T. Class III. Gammaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 17.Garrity GM, Bell JA, Lilburn T. Order XIV. Pasteurellales ord. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 850. [Google Scholar]
- 18.List Editor Validation List no. 7. Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1981; 31:382-383 10.1099/00207713-31-3-382 [DOI] [Google Scholar]
- 19.Pohl SPD. Dissertation, Phillips-Universität Marburg, 1979. [Google Scholar]
- 20.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]
- 21.Winslow CEA, Broadhurst J, Buchanan RE, Krumwiede C, Rogers LA, Smith GH. The Families and Genera of the Bacteria: Preliminary Report of the Committee of the Society of American Bacteriologists on Characterization and Classification of Bacterial Types. J Bacteriol 1917; 2:505-566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zinnemann KS, Biberstein EL. Genus Haemophilus Winslow, Broadhurst, Buchanan, Krumwiede, Rogers and Smith 1917, 561. In: Buchanan RE, Gibbons NE (eds), Bergey's Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 364-370. [Google Scholar]
- 23.Biberstein EL, White DC. A proposal for the establishment of two new Haemophilus species. J Med Microbiol 1969; 2:75-78 10.1099/00222615-2-1-75 [DOI] [PubMed] [Google Scholar]
- 24.Fink DK, St. Geme JW, III. The Genus Haemophilus In: Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E. (eds), The Prokaryotes, A Handbook on the Biology of Bacteria: Proteobacteria: Gamma Subclass, Third Edition, Volume 6, Springer, New York, 2006, p. 1034-1061. [Google Scholar]
- 25.Kroppenstedt RM, Mannheim W. Lipoquinones in members of the family Pasteurellaceae. Int J Syst Evol Microbiol 1989; 39:304-308 [Google Scholar]
- 26.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribosomal Database Project http://rdp.cme.msu.edu/treebuilderpub/treeHelp.jsp
- 28.Bruno WJ, Socci ND, Halpern AL. Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol Biol Evol 2000; 17:189-197 [DOI] [PubMed] [Google Scholar]
- 29.Som A. Theoretical foundation to estimate the relative efficiencies of the Jukes-Cantor+gamma model and the Jukes-Cantor model in obtaining the correct phylogenetic tree. Gene 2006; 385:103-110 10.1016/j.gene.2006.03.027 [DOI] [PubMed] [Google Scholar]
- 30.Morozumi T, Nicolet J. Morphological variations of Haemophilus parasuis strains. J Clin Microbiol 1986; 23:138-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biberstein EL, Gunnarsson A, Hurvell B. Cultural and biochemical criteria for the identification of Haemophilus spp from swine. Am J Vet Res 1977; 38:7-11 [PubMed] [Google Scholar]
- 32.Biberstein EL, Mini PD, Gills MG. Action of Haemophilus cultures on δ-aminolevulinic acid. J Bacteriol 1963; 86:814-819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 1998; 26:1107-1115 10.1093/nar/26.4.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 2005; 21:617-623 10.1093/bioinformatics/bti057 [DOI] [PubMed] [Google Scholar]
- 36.Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 2008; 36:W181-W184 10.1093/nar/gkn179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.CGView Server http://stothard.afns.ualberta.ca/cgview_server/index.html