Abstract
Ktedonobacter racemifer corrig. Cavaletti et al. 2007 is the type species of the genus Ktedonobacter, which in turn is the type genus of the family Ktedonobacteraceae, the type family of the order Ktedonobacterales within the class Ktedonobacteria in the phylum ‘Chloroflexi’. Although K. racemifer shares some morphological features with the actinobacteria, it is of special interest because it was the first cultivated representative of a deep branching unclassified lineage of otherwise uncultivated environmental phylotypes tentatively located within the phylum ‘Chloroflexi’. The aerobic, filamentous, non-motile, spore-forming Gram-positive heterotroph was isolated from soil in Italy. The 13,661,586 bp long non-contiguous finished genome consists of ten contigs and is the first reported genome sequence from a member of the class Ktedonobacteria. With its 11,453 protein-coding and 87 RNA genes, it is the largest prokaryotic genome reported so far. It comprises a large number of over-represented COGs, particularly genes associated with transposons, causing the genetic redundancy within the genome being considerably larger than expected by chance. This work is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: aerobic, heterotrophic, filamentous, non-motile, Gram-positive, moderately acidophilic, sporulating, transposon, broken-stick distribution, entropy, Ktedonobacteraceae, Chloroflexi, GEBA
Introduction
Strain SOSP1-21T (= DSM 44963 = NRRL B-41538) is the type strain of the species Ktedonobacter racemifer, which is the type species of the monotypic genus Ktedonobacter, the type genus of the family Ktedonobacteraceae [1]. K. racemifer was first described in 2006 [1,2] as an aerobic, non-motile, filamentous, mesophilic, Gram-positive heterotroph also capable of growing under microaerophilic conditions [1]. The genus name was derived from the Greek word ktedon -onos, fiber, and the Neo-Latin bacter, a rod, meaning a filamentous rod [1]. The species epithet is derived from the Latin adjective racemifer, carrying clusters of grapes [1]. The original spelling, Ktedobacter racemifer was corrected in 2007 on validation according to Rule 61 and Recommendation 6(7) [2]. Strain SOSP1-21T was originally isolated from a soil sample of a black locust wood in Gerenzano, Northern Italy. Ten phylogenetically (class level) related strains were also isolated from soil samples collected at different locations in Northern Italy [1]. Only recently, a nearest cultivated neighbor, Thermosporothrix hazakensis, was isolated from hot compost in Japan [3]. Here we present a summary classification and a set of features for K. racemifer strain SOSP1-21T, together with the description of the complete genomic sequencing and annotation.
Classification and features
Using NCBI BLAST [4], a representative genomic 16S rRNA sequence of K. racemifer SOSP1-21T was compared under default settings (e.g., considering only the high-scoring segment pairs (HSPs) from the best 250 hits) with the most recent release of the Greengenes database [5] and the relative frequencies of taxa and keywords (reduced to their stem [6]) were determined, weighted by BLAST scores. The most frequently occurring genus was 'Ktedobacter' (100.0%) (1 hit in total; this represents the original, incorrect spelling of Ktedonobacter). No hits to sequences with (other) species names were found. (Note that the Greengenes database uses the INSDC (= EMBL/NCBI/DDBJ) annotation, which is not an authoritative source for nomenclature or classification.) The highest-scoring environmental sequence was AM180157 ('New lineage filamentous spore-forming soil isolate SOSP1-30SOSP1-30 str. SOSP1-30'), which showed an identity of 99.0% and an HSP coverage of 95.2%. The most frequently occurring keywords within the labels of environmental samples which yielded hits were 'soil' (11.2%), 'prari, tallgrass' (4.9%), 'miner, weather' (1.9%), 'new' (1.8%) and 'filament, lineag, spore-form' (1.6%) (249 hits in total). These keywords reflect some of the ecological properties reported for strain SOSP1-21T in the original description [1]. Environmental samples which yielded hits of a higher score than the highest scoring species were not found.
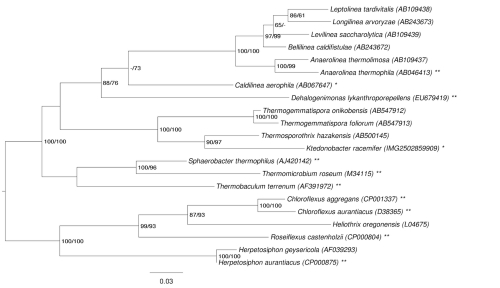
Figure 1 shows the phylogenetic neighborhood of K. racemifer in a 16S rRNA based tree. The sequences of the eight 16S rRNA genes copies in the genome differ by up to nine nucleotides from each other and by up to five nucleotides from the previously published 16S rRNA sequence (AM180156), which contains two ambiguous base calls.
Figure 1.
Phylogenetic tree highlighting the position of K. racemifer relative to the other type strains within the phylum ‘Chloroflexi’. The tree was inferred from 1,359 aligned characters [7,8] of the 16S rRNA gene sequence under the maximum likelihood (ML) criterion [9]. Rooting was done initially using the midpoint method [10] and then checked for its agreement with the current classification (Table 1). The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 750 ML bootstrap replicates [11] (left) and from 1,000 maximum parsimony bootstrap replicates [12] (right) if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [13] are labeled with one asterisk, those also listed as 'Complete and Published' with two asterisks [14-17] as well as CP001337, CP000804, CP000909, CP002084, and AP012029.
Table 1. Classification and general features of K. racemifer SOSP1-21T according to the MIGS recommendations [18] and the NamesforLife database [19].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [20] | |
| Phylum Chloroflexi | TAS [21,22] | ||
| Class Ktedonobacteria | TAS [1-3] | ||
| Order Ktedonobacterales | TAS [1,2] | ||
| Family Ktedonobacteraceae | TAS [1,2] | ||
| Genus Ktedonobacter | TAS [1,2] | ||
| Species Ktedonobacter racemifer | TAS [1] | ||
| Type strain SOSP1-21 | TAS [1] | ||
| Gram stain | positive | TAS [1] | |
| Cell shape | filamentous | TAS [1] | |
| Motility | non-motile | TAS [1] | |
| Sporulation | spherical spore-forming | TAS [1] | |
| Temperature range | mesophile | TAS [1] | |
| Optimum temperature | 28-33°C | TAS [1] | |
| Salinity | NaCl up to 10 g/l growth w/o problem, inhibited at 30 g/l | TAS [1] | |
| MIGS-22 | Oxygen requirement | aerobic and microaerophilic | TAS [1] |
| Carbon source | sugars and peptides | TAS [1] | |
| Energy metabolism | heterotrophic | TAS [1] | |
| MIGS-6 | Habitat | soil | TAS [1] |
| MIGS-15 | Biotic relationship | free-living | NAS |
| MIGS-14 | Pathogenicity | none | NAS |
| Biosafety level | 1 | TAS [23] | |
| Isolation | soil from a black locust wood | TAS [1] | |
| MIGS-4 | Geographic location | Gerenzano, Northern Italy | TAS [1] |
| MIGS-5 | Sample collection time | November 2001 | NAS |
| MIGS-4.1 | Latitude | 45.64 | NAS |
| MIGS-4.2 | Longitude | 9.00 | NAS |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | about 210 m | NAS |
Evidence codes - TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [24].
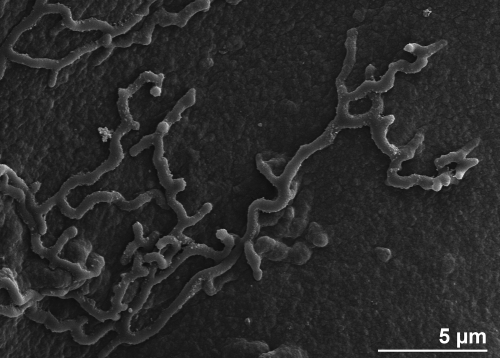
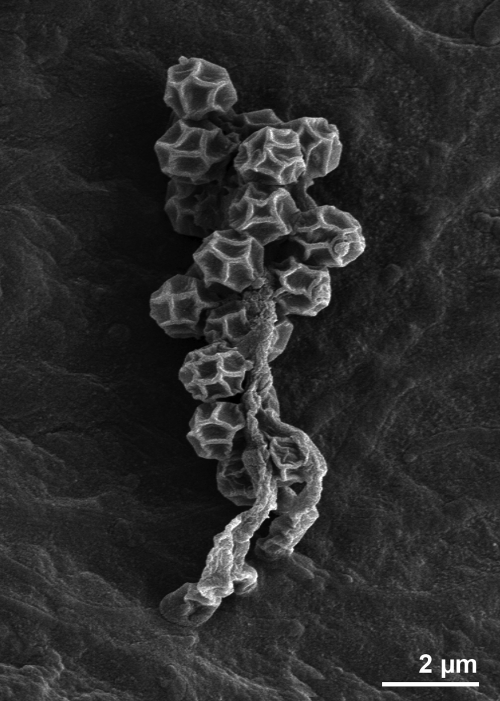
K. racemifer strain SOSP1-21T cells are rod-shaped, filamentous and grow both vegetative and aerial mycelia on solid medium (Figure 2a). The large aerial hyphae produce spherical spores that cluster together with a grape-like appearance (Figure 2b). All other K. racemifer strains produced rounded spores, although they were arranged differently on the aerial hyphae [1]. Filamentous growth of strain SOSP1-21T also occurred in submerged cultures, which contained the branched mycelia known from actinomycetes [1]. SOSP1-21T stains Gram-positive and is not acid fast [1]. It produces pigments ranging from cream to pinkish orange on all media [1]. Although essentially aerobic, SOSP1-21T is capable of growing under microaerophilic conditions [1]. The optimal growth temperature is 28-33°C [1]. It grows well at pH values between 4.8 and 6.8 with an optimum at pH 6 [1]. Salinity up to 10 g per liter does not inhibit the growth of the strain [1].
Figure 2a.
Scanning electron micrographs of K. racemifer SOSP1-21T mycelium.
Figure 2b.
Scanning electron micrographs of K. racemifer SOSP1-21T spores.
Strain SOSP1-21T was capable of hydrolyzing starch, casein, gelatin, and (to a lesser extent) keratin but not cellulose, xylan, or chitin [1]. Strain SOSP1-21T was catalase positive and produced H2S but could not reduce nitrates [1]. It is sensitive to 5 ug/ml novobiocin or ramoplanin and to 20 mg/ml apramycin and the glycopeptide A40926.
Chemotaxonomy
The peptidoglycan of strain SOSP1-21T contains ornithine, alanine, glutamic acid, serine, and glycine at a molar ratio of approximately 0.7:1.8:1.0:0.8:1.9 [1]. Serine was identified at the N-terminus of the interpeptide bridge [1]. When originally described, a detailed peptidoglycan structure had not been determined but A-type cross-linkage was suggested [1]. The cellular fatty acid pattern of strain SOSP1-21T was reported to be characterized by an unusual high abundance of C16:1 2-OH (30%) with other dominant lipids being branched-chain saturated fatty acids iso-C17:0 (25%), iso-C16:0 (11.5%) and anteiso-C17:0 (9.6%), as well as C16:0 10-Me (7.8%) and C16:0 (6.7%) [1]. Our own data (DSMZ) did not confirm this fatty acid spectrum, but revealed iso-C16:0 (20.1%) as the most frequent fatty acid, followed by anteiso-C17:0 (18.5%), iso-C17:0 (15.0%), only 13.1% C16:1 2-OH and 11.6% C16:0 10-Me. Polar lipids consisted of phosphatidylinisitol, phosphatitylglycerol, diphosphatidylglycerol and an unknown glycolipid [1]. MK-9(H2) was the only menaquinone reported for strain SOSP1-21T [1].
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position [25], and is part of the Genomic Encyclopedia of Bacteria and Archaea project [26]. The genome project is deposited in the Genomes OnLine Database [13] and the complete genome sequence is deposited in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Non-contiguous finished |
| MIGS-28 | Libraries used | Two Sanger 8 kb pMCL200 and fosmid libraries; one 454 pyrosequence standard library |
| MIGS-29 | Sequencing platforms | ABI3730, 454 GS FLX |
| MIGS-31.2 | Sequencing coverage | 10.1 × Sanger; 24.6 × pyrosequence |
| MIGS-30 | Assemblers | Newbler version 1.1.02.15, phrap |
| MIGS-32 | Gene calling method | Prodigal 1.4, Genemark 4.6b, tRNAScan-SE-1.23, infernal 0.81 |
| INSDC ID | ADVG00000000 | |
| Genbank Date of Release | June 14, 2010 | |
| GOLD ID | Gi02261 | |
| NCBI project ID | 27943 | |
| Database: IMG-GEBA | 648276680 | |
| MIGS-13 | Source material identifier | DSM 44963 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
K. racemifer SOSP1-21T, DSM 44963, was grown in DSMZ medium 65 (GYM Streptomyces medium) [27] adjusted to pH 6.0, at 28°C. DNA was isolated from 0.5-1 g of cell paste using Qiagen Genomic 500 DNA Kit (Qiagen 10262) following the manufacturer’s protocol, with cell lysis protocol st/LALMP as described in Wu et al. [26]. DNA is available through the DNA Bank Network [28].
Genome sequencing and assembly
The genome was sequenced using a combination of Sanger and 454 sequencing platforms. All general aspects of library construction and sequencing can be found at the JGI website [29]. Pyrosequencing reads were assembled using the Newbler assembler (Roche). The initial Newbler contigs were broken into 14,080 overlapping fragments of 1,000 bp and entered as pseudo-reads into the subsequence assembly. The sequences were assigned quality scores based on Newbler consensus q-scores with modifications to account for overlap redundancy and to adjust inflated q-scores. A hybrid 454/Sanger assembly was produced using parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with Dupfinisher [30], or transposon bombing of bridging clones (Epicentre Biotechnologies, Madison, WI) [31]. Some gaps between contigs were closed by editing in Consed [32], custom primer walking or PCR amplification. A total of 3,354 Sanger finishing reads and five shatter libraries were produced to close gaps, to resolve some repetitive regions, and to raise the quality of the finished sequence. Illumina reads were also used to correct potential base errors and increase consensus quality using a software Polisher developed at JGI [33]. The error rate of the completed genome sequence is less than 1 in 100,000. Together, the combination of the Sanger and 454 sequencing platforms provided 34.7 × coverage of the genome. The final assembly contained 165,050 pyrosequence and 2,305,667 Illumina reads.
Genome annotation
Genes were identified using Prodigal [34] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [35]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) non-redundant database, UniProt, TIGR-Fam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation were performed within the Integrated Microbial Genomes - Expert Review (IMG-ER) platform [36].
Genome properties
The non-contiguous finished genome consists of ten contigs ranging in size from 1,579 bp to almost four Mbp, with five contigs being longer than one Mb (1,302,518 bp, 2,713,222 bp, 2,766,182 bp, 2,916,502 bp, and 3,837,106 bp) and a G+C content of 53.8% (Table 3 and Figure 3). Of the 11,540 genes predicted, 11,453 were protein-coding genes, and 87 RNAs; No pseudogenes were identified. The majority of the protein-coding genes (61.2%) were assigned a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 13,661,586 | 100.00% |
| DNA coding region (bp) | 10,422,932 | 76.29% |
| DNA G+C content (bp) | 7,348,426 | 53.79% |
| Number of contigs | 10 | |
| Extrachromosomal elements | unknown | |
| Total genes | 11,540 | 100.00% |
| RNA genes | 87 | 0.75% |
| rRNA operons | 8 | |
| Protein-coding genes | 11,453 | 99.25% |
| Pseudo genes | 0 | |
| Genes with function prediction | 7,065 | 61.22% |
| Genes in paralog clusters | 4,919 | 42.63% |
| Genes assigned to COGs | 6,654 | 57.66% |
| Genes assigned Pfam domains | 7,250 | 62.82% |
| Genes with signal peptides | 2,660 | 23.05% |
| Genes with transmembrane helices | 2,581 | 22.27% |
| CRISPR repeats | 7 |
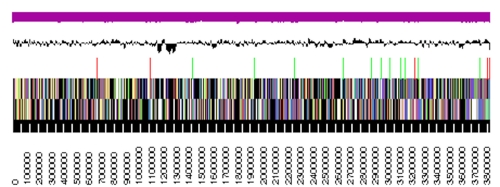
Figure 3.
Graphical linear map of the largest, 3,837,106 bp long contig. From bottom to the top: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | %age | Description |
|---|---|---|---|
| J | 224 | 2.9 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 893 | 11.6 | Transcription |
| L | 975 | 12.6 | Replication, recombination and repair |
| B | 3 | 0.0 | Chromatin structure and dynamics |
| D | 34 | 0.4 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.0 | Nuclear structure |
| V | 215 | 2.8 | Defense mechanisms |
| T | 617 | 8.0 | Signal transduction mechanisms |
| M | 257 | 3.3 | Cell wall/membrane/envelope biogenesis |
| N | 20 | 0.3 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 54 | 0.7 | Intracellular trafficking, secretion, and vesicular transport |
| O | 195 | 2.5 | Posttranslational modification, protein turnover, chaperones |
| C | 416 | 5.4 | Energy production and conversion |
| G | 612 | 7.9 | Carbohydrate transport and metabolism |
| E | 474 | 6.2 | Amino acid transport and metabolism |
| tF | 135 | 1.8 | Nucleotide transport and metabolism |
| H | 264 | 3.4 | Coenzyme transport and metabolism |
| I | 236 | 3.1 | Lipid transport and metabolism |
| P | 255 | 3.3 | Inorganic ion transport and metabolism |
| Q | 217 | 2.8 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 1,098 | 14.4 | General function prediction only |
| S | 519 | 6.7 | Function unknown |
| - | 4,886 | 42.3 | Not in COGs |
Insights from the genome sequence
Genome structure
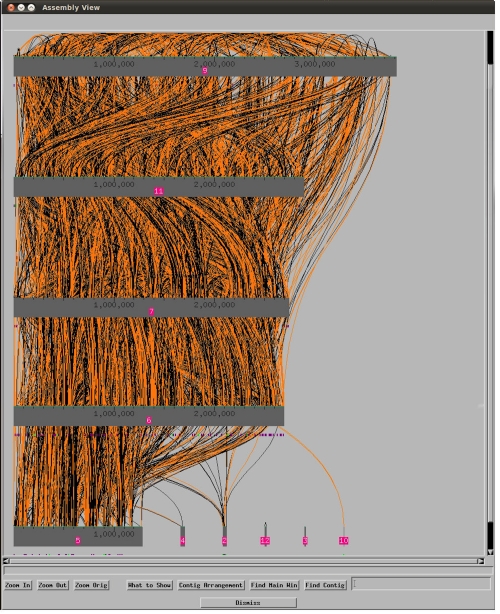
With a length of 13,661,586 bp for the ten contigs (Table 3) K. racemifer SOSP1-21T has the largest of all completely sequenced 1,760 archaeal and bacterial genomes [37] thus far, followed by Sorangium cellulosum, 13.0 Mbp [38], Steptomyces bingchenggensis, 11.9 Mbp [39], Catenulispora acidiphila, 10.5 Mbp [40], and Streptosporangium roseum, 10.4 Mbp [41]. However, this genome was also one of the most difficult to assemble. Figure 4 shows the unusually high number of identical sequence fragments across the genome, which caused the termination of the project as non-contiguous finished genome without closure of the last ten sequence gaps.
Figure 4.
Screen shot from CROSSMATCH [32] indicating the matches between sequences within and across the contigs. CROSSMATCH options were – minmatch 30 – minscore 60.
Comparative genomics
Lacking an available genome sequence of the closest relative of K. racemifer, Thermosporothrix hazakensis [3] (Figure 1), the following comparative analyses were done with Sphaerobacter thermophilus [42] and Thermomicrobium roseum [43], the closest organisms phylogenetically for which there are publically available genome sequences [15,16].
K. racemifer stands out because of its enormous genome size of more than 13 Mbp. The genomes of S. thermophilus and T. roseum are significantly smaller, 3.9 Mbp and 2.9 Mbp, respectively. Whereas S. thermophilus and T. roseum have similar G+C-contents of 68% and 64%, respectively, the G+C-content of the K. racemifer genome is significantly lower (54%).
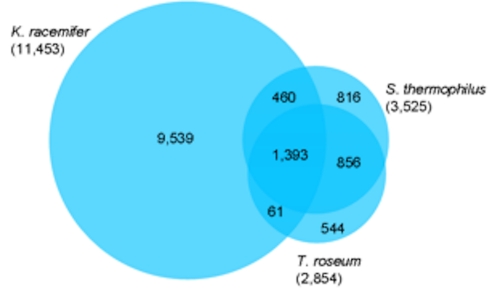
The fraction of shared genes in the three genomes is shown in a Venn diagram (Figure 5). The numbers of pairwise shared genes were calculated with the phylogenetic profiler function of the IMG-ER platform [36]. Homologous genes within the genomes were detected with a maximum E-value of 10-5 and a minimum identity of 30%.
Figure 5.
Venn diagram depicting the intersections of protein sets (total number of derived protein sequences in parentheses) of K. racemifer, S. thermophilus and T. roseum.
A total of 1,393 genes are shared by the three genomes, referring to the whole genome sizes 39% and 48% of the genes in S. thermophilus and T. roseum have homologs in the three genomes, in the case of K. racemifer only 12% of the genes are shared by the other two genomes. The pairwise comparison of S. thermophilus and T. roseum revealed 2,249 genes which are shared by these two organisms, referring to the whole genomes 64% of the S. thermophilus genes and 79% of the T. roseum have homologous genes in the respective other genome.
The genome of K. racemifer encodes an enormously high number of transposon-associated genes; its annotation revealed 601 genes encoding transposases, 151 genes encoding integrases and 107 genes encoding resolvases. The genes coding these enzymes are spread over the whole genome with some regions having a higher density than others. The extremely high number of transposases is due to several gene copies that are to a greater or lesser extent similar in their sequences. The presence of that many mobile elements may explain the unusually high number of identical sequence fragments across the genome and the resulting difficulties occurring during the genome assembly.
Within the 9,539 unique genes of K. racemifer that have no detectable homologs in the genomes of S. thermophilus and T. roseum (under the sequence similarity thresholds used for the comparison) the 29 genes encoding xylose isomerases appear to be especially noteworthy; for 27 of these isomerase genes no homologous genes were detected in the other two genomes; only one gene was identified in T. roseum, and two in S. thermophilus. The high number of xylose isomerase genes suggests a strong utilization of pentoses by K. racemifer. To date K. racemifer was not tested regarding xylose utilization, but the close relative T. hazakensis is able to use xylose as the only carbon source [3]. Furthermore, a high number of genes encoding proteins responsible for resistance against several antibiotics were predicted: 61 bleomycin resistance proteins and 41 aminoglycoside phosphotransferases.
An estimate of the overall similarity between K. racemifer, S. thermophilus and T. roseum, was generated with the GGDC Genome-to-Genome Distance Calculator [44,45]. This system calculates the distances by comparing the genomes to obtain HSPs (high-scoring segment pairs) and interfering distances from a set of formulas (1, HSP length / total length; 2, identities / HSP length; 3, identities / total length). Table 5 shows the results of the pairwise comparison between the three genomes.
Table 5. Pairwise comparison of K. racemifer, S. thermophilus and T. roseum using the GGDC-Calculator.
| HSP length /total length [%] | identities /HSP length [%] | identities /total length [%] | ||
|---|---|---|---|---|
| K. racemifer | S. thermophilus | 0.57 | 86.4 | 0.50 |
| K. racemifer | T. roseum | 0.48 | 87.2 | 0.42 |
| T. roseum | S. thermophilus | 9.41 | 83.1 | 7.82 |
The pairwise comparison (Table 5) of the genomes of K. racemifer with S. thermophilus and T. roseum revealed that only 0.57% and 0.48% of the average of the genome lengths are covered with HSPs. The identity within these HSPs was 86.4% and 87.2%, whereas the identity over the whole genome was only 0.50% and 0.42%, respectively. The comparison of T. roseum with S. thermophilus revealed that 9.41% of the average of both genome lengths are covered with HSPs, with an identity within these HSPs of 83.1%. The identity over the whole genome is 7.82%. These results show how distant the relationship between K. racemifer and S. thermophilus and T. roseum, respectively, is, if genome sizes are taken into consideration.
In order to quantify the differences in gene redundancy between the three genomes, as well as to determine over-represented genes, we used approaches based on entropy and the broken-stick distribution, respectively, applied to the set of genes from either genome assigned to COGs. Shannon's entropy (see, e.g., pp. 214, 243 in [46]) H can be used as a measure of disorder for discrete distributions; it is maximum (Hmax) if all categories (COGs in our case) are represented by exactly one item (gene) and then equal to the logarithm of the number of items (genes). Thus, one can measure the evenness (non-redundancy) within such a distribution as H/Hmax and the corresponding redundancy as 1.0 – H/Hmax. The broken-stick distribution reflects the relative abundance of a given number of categories within a random population of items (see, e.g., p. 244 and 410 in [46]). Over-represented items (here: COGs) are those whose real relative frequencies (here: number of genes assigned to this COG relative to the total number of genes assigned to COGs) are larger than the broken-stick value of the corresponding rank within the list of frequencies sorted in decreasing order. Moreover, the entropy Hexp of the broken-stick distribution can be used as an estimate for the expected entropy, yielding 1.0 – H/Hexp as an alternative measure of redundancy (which becomes negative when the evenness is larger than expected by chance).
The 2,022 genes assigned to 1,300 distinct COGs in the genome of T. roseum corresponded to an entropy of 6.912, an expected entropy of 6.748 and, hence, a redundancy of 9.20% if measured using Hmax and of -2.42% using Hexp, whereas S. thermophilus (2,619 genes assigned to 1,383 COGs) yielded an entropy of 6.837 (expected: 6.810) and a redundancy of 13.14% with Hmax and -0.39% with Hexp. In contrast, the 6,654 genes assigned to 1,731 distinct COGs in the genome of K. racemifer yielded an entropy of only 6.455 (expected: 7.034) and a redundancy of 26.67% (using Hmax) and 8.24% (using Hexp). That is, in contrast to the other two genomes the genes within the genome of K. racemifer are distributed less even than expected by chance.
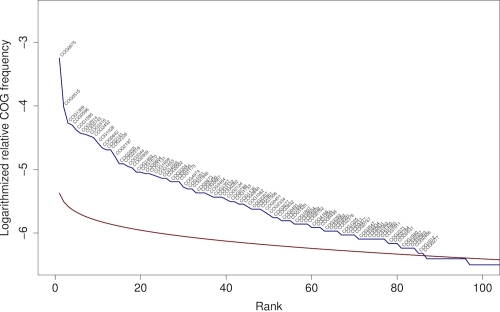
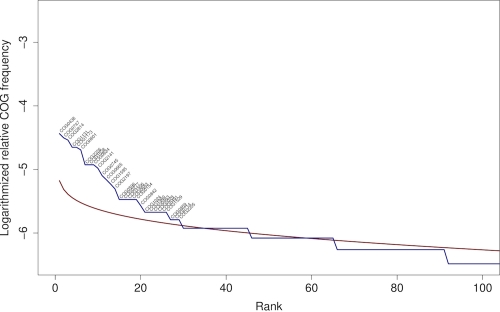
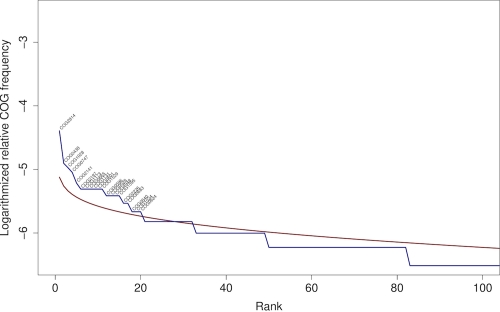
Figure 6 compares the relative frequencies of the COGs in the genome of K. racemifer compared to their expected frequency. More than 80 COGs were judged as over-represented by this comparison, considerably more than in the genomes of S. thermophilus [33; Figure 7] and T. roseum ([15]; Figure 8). A closer look onto the 20 most over-represented COGs in K. racemifer, S. thermophilus and T. roseum revealed differences between the three organisms. Not surprisingly the genes coding transposases (COG0675; by far the most frequent one), integrases (COG3316) and resolvases (COG2452) can be found among the over-represented COGs in K. racemifer (Figure 6).
Figure 6.
Relative frequencies of the 100 most frequent COGs in the genome of K. racemifer (blue line) compared to their expected frequency as estimated using the broken-stick distribution (red line). Over-represented COGs are labeled.
Figure 7.
Relative frequencies of the 100 most frequent COGs in the genome of S. thermophilus (blue line) compared to their expected frequency as estimated using the broken-stick distribution (red line). Over-represented COGs are labeled.
Figure 8.
Relative frequencies of the 100 most frequent COGs in the genome of T. roseum (blue line) compared to their expected frequency as estimated using the broken-stick distribution (red line). Over-represented COGs are labeled.
Our analyses also showed that genes belonging to the category COG3344 are over-represented in the genome of K. racemifer. COG3344 represents retron type reverse transcriptases, which are found in group II introns. Group II introns are large catalytic RNA molecules that act as mobile genetic elements [47]. They were first identified in mitochondria and chloroplast genomes, but with the increasing number of bacterial genome sequencing projects, the number of group II intron sequences in the databases also increased. Dai and Zimmerly reported in 2003 that a quarter of the sequenced bacterial genomes contain group II introns [48,49]. By using the IMG-ER platform [36] we calculated that approximately one third of the 2,727 sequenced bacterial genomes contain group II introns. In the genome of K. racemifer, 34 genes coding reverse transcriptases could be identified, all of them having the same domain structure with the reverse transcriptase domain followed by a maturase-specific domain and the C-terminal HNH-endonuclease domain.
Acknowledgements
We would like to gratefully acknowledge the help of Marlen Jando for growing K. racemifer cultures and Susanne Schneider for DNA extraction and quality control (both at DSMZ). This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, UT-Battelle and Oak Ridge National Laboratory under contract DE-AC05-00OR22725, as well as German Research Foundation (DFG) INST 599/1-1.
References
- 1.Cavaletti L, Monciardini P, Bamonte R, Schumann P, Rohde M, Sosio M, Donadio S. New lineage of filamentous, spore-forming, Gram-positive bacteria from Soil. Appl Environ Microbiol 2006; 72:4360-4369 10.1128/AEM.00132-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Validation list No. 114. Int J Syst Evol Microbiol 2007; 57:433-434 10.1099/ijs.0.65052-0 [DOI] [PubMed] [Google Scholar]
- 3.Yabe S, Aiba Y, Sakai Y, Hazaka M, Yokota A. Thermosporothrix hazakensis gen. nov., sp. nov., isolated from compost, description of Thermosporotrichaceae fam. nov. within the class Ktedonobacter Cavaletti et al. 2007 and emended description of the class Ktedonobacteria. Int J Syst Evol Microbiol 2010; 60:1794-1801 10.1099/ijs.0.018069-0 [DOI] [PubMed] [Google Scholar]
- 4.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-410 [DOI] [PubMed] [Google Scholar]
- 5.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl Environ Microbiol 2006; 72:5069-5072 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porter MF. An algorithm for suffix stripping. Program: electronic library and information systems 1980; 14:130-137.
- 7.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 8.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 9.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 10.Hess PN, De Moraes Russo CA. An empirical test of the midpoint rooting method. Biol J Linn Soc Lond 2007; 92:669-674 10.1111/j.1095-8312.2007.00864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [PubMed] [Google Scholar]
- 12.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0 b10. Sinauer Associates, Sunderland, 2002. [Google Scholar]
- 13.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes OnLine Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2010; 38:D346-D354 10.1093/nar/gkp848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiss H, Nett M, Domin N, Martin K, Maresca JA, Copeland A, Lapidus A, Lucas S, Berry KW, Glavina Del Rio T, et al. Complete genome sequence of the filamentous predatory bacterium Herpetosiphon aurantiacus type strain (114-95T). Stand Genomic Sci 2011; (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu D, Raymond J, Wu M, Chatterji S, Ren Q, Graham JE, Bryant DA, Robb F, Colman A, Tallon LJ, et al. Complete genome sequence of the aerobic CO-oxidizing thermophile Thermomicrobium roseum. PLoS ONE 2009; 4:e4207 10.1371/journal.pone.0004207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pati A, LaButti K, Pukall R, Nolan M, Glavina Del Rio T, Tice H, Cheng JF, Lucas S, Chen F, Copeland A, et al. Complete genome sequence of Sphaerobacter thermophilus type strain (S 6033T). Stand Genomic Sci 2010; 2:49-56 10.4056/sigs.601105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiss H, Cleland D, Lapidus A, Lucas S, Glavina Del Rio T, Nolan M, Tice H, Han C, Goodwin L, Pitluck S, et al. Complete genome sequence of 'Thermobaculum terrenum' type strain (YNP1T). Stand Genomic Sci 2010; 3:153-162 10.4056/sigs.1153107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrity G. NamesforLife. BrowserTool takes expertise out of the database and puts it right in the browser. Microbiol Today 2010; 37:9 [Google Scholar]
- 20.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrity GM, Holt JG. Phylum BVI. Chloroflexi phy. nov. In: Boone DR, Castenholz RW, Garrity GM (eds), Bergey's Manual of Systematic Bacteriology, second edition, vol. 1 (The Archaea and the deeply branching and phototrophic Bacteria) Springer, New York, 2001, p. 427-446. [Google Scholar]
- 22.Hugenholtz P, Stackebrandt E. Reclassification of Sphaerobacter thermophilus from the subclass Sphaerobacteridae in the phylum Actinobacteria to the class Thermomicrobia (emended description) in the phylum Chloroflexi (emended description). Int J Syst Evol Microbiol 2004; 54:2049-2051 10.1099/ijs.0.03028-0 [DOI] [PubMed] [Google Scholar]
- 23.BAuA Classification of bacteria and archaea in risk groups. http://www.baua.de. TRBA 2010; 466:112 [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klenk HP, Göker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 2010; 33:175-182 10.1016/j.syapm.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 26.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 2009; 462:1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.List of growth media used at DSMZ: http//www.dsmz.de/microorganisms/media_list.php
- 28.Gemeinholzer B, Dröge G, Zetzsche H, Haszprunar G, Klenk HP, Güntsch A, Berendsohn WG, Wägele JW. The DNA Bank Network: the start from a German initiative. Biopreservation and Biobanking 2011; 9:51-55 10.1089/bio.2010.0029 [DOI] [PubMed] [Google Scholar]
- 29.The DOE Joint Genome Institute www.jgi.doe.gov
- 30.Han C, Chain P. Finishing repeat regions automatically with Dupfinisher. in Proceeding of the 2006 international conference on bioinformatics & computational biology. Edited by Hamid R. Arabnia & Homayoun Valafar, CSREA Press. June 26-29, 2006: 141-146. [Google Scholar]
- 31.Sims D, Brettin T, Detter JC, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome sequence of Kytococcus sedentarius type strain (strain 541T). Stand Genomic Sci 2009; 1:12-20 10.4056/sigs.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phrap and Phred for Windows. MacOS, Linux, and Unix. http://www.phrap.com
- 33.Lapidus A, LaButti K, Foster B, Lowry S, Trong S, Goltsman E. POLISHER: An effective tool for using ultra short reads in microbial genome assembly and finishing. AGBT, Marco Island, FL, 2008. [Google Scholar]
- 34.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 36.Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]
- 37.NCBI Complete Microbial Genomes http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi
- 38.Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, Bartels D, Bekel T, Beyer S, Bode E, et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol 2007; 25:1281-1289 10.1038/nbt1354 [DOI] [PubMed] [Google Scholar]
- 39.Wang XJ, Yan YJ, Zhang B, An J, Wang JJ, Tian J, Jiang L, Chen YH, Huang SX, Yin M, et al. Genome sequence of the Milbemycin-producing bacterium Streptomyces bingchenggensi. J Bacteriol 2010; 192:4526-4527 10.1128/JB.00596-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Copeland A, Lapidus A, Glavina Del Rio T, Nolan M, Lucas S, Chen F, Tice H, Cheng JF, Bruce D, Goodwin L, et al. Complete genome sequence of Catenulispora acidiphila type strain (ID 139908T). Stand Genomic Sci 2009; 1:119-125 10.4056/sigs.17259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nolan M, Sikorski J, Jando M, Lucas S, Lapidus A, Glavina Del Rio T, Chen F, Tice H, Pitluck S, Cheng JF, et al. Complete genome sequence of Streptosporangium roseum type strain (NI 9100T). Stand Genomic Sci 2010; 2:29-37 10.4056/sigs.631049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demharter W, Hensel R, Smida J, Stackebrandt E. Sphaerobacter thermophilus gen. nov., sp. nov. A deeply rooting member of the actinomycetes subdivision isolated from thermophilically treated sewage sludge. Syst Appl Microbiol 1989; 11:261-266 [Google Scholar]
- 43.Skerman VBD, McGowan V, Sneath PHA, eds. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [Google Scholar]
- 44.Auch AF, von Jan M, Klenk HP, Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2010; 2:117-134 10.4056/sigs.531120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auch AF, Klenk HP, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2010; 2:142-148 10.4056/sigs.541628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Legendre P, Legendre L. Numerical Ecology. 2nd edn. Elsevier, Amsterdam, 1998. [Google Scholar]
- 47.Martínez-Abarca F, Toro N. Group II introns in the bacterial world. Mol Microbiol 2000; 38:917-926 10.1046/j.1365-2958.2000.02197.x [DOI] [PubMed] [Google Scholar]
- 48.Dai L, Zimmerly S. ORF-less and reverse-transcriptase-encoding group II introns in archaebacteria, with a pattern of homing into related group II intron ORFs. RNA 2003; 9:14-19 10.1261/rna.2126203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kuever J, Rainey FA, Widdel F. Family I. Desulfurellaceae fam. nov. In: Brenner DJ, Krieg NR, Staley JT Garrity GM (eds), Bergey's Manual of Systematic Bacteriology, second edition, vol. 2 (The Proteobacteria), part C (The Alpha-, Beta-, Delta-, and Epsilonproteobacteria), Springer, New York, 2005, p. 923. [Google Scholar]