Abstract
In an effort to discover anaerobic bacteria capable of lignin degradation, we isolated “Enterobacter lignolyticus” SCF1 on minimal media with alkali lignin as the sole source of carbon. This organism was isolated anaerobically from tropical forest soils collected from the Short Cloud Forest site in the El Yunque National Forest in Puerto Rico, USA, part of the Luquillo Long-Term Ecological Research Station. At this site, the soils experience strong fluctuations in redox potential and are net methane producers. Because of its ability to grow on lignin anaerobically, we sequenced the genome. The genome of “E. lignolyticus” SCF1 is 4.81 Mbp with no detected plasmids, and includes a relatively small arsenal of lignocellulolytic carbohydrate active enzymes. Lignin degradation was observed in culture, and the genome revealed two putative laccases, a putative peroxidase, and a complete 4-hydroxyphenylacetate degradation pathway encoded in a single gene cluster.
Keywords: Anaerobic lignin degradation, tropical forest soil isolate, facultative anaerobe
Introduction
One of the biggest barriers to efficient lignocellulose deconstruction is the problem of lignin, both occluding the action of cellulases and as wasteful lignin by-products. Tropical forest soils are the sites of very high rates of decomposition, accompanied by very low and fluctuating redox potential conditions [1,2]. Because early stage decomposition is typically dominated by fungi and the free-radical generating oxidative enzymes phenol oxidase and peroxidase [3,4], we targeted anaerobic tropical forest soils with the idea that they would be dominated by bacterial rather than fungal decomposers. To discover organisms that were capable of breaking down lignin without the use of oxygen free radicals, we isolated “Enterobacter lignolyticus” SCF1 under anaerobic conditions using lignin as the sole carbon source. In addition to this, it has been observed to withstand high concentrations of ionic liquids [5], and thus was targeted for whole genome sequencing.
Organism information
“E. lignolyticus” SCF1 was isolated from soil collected from the Short Cloud Forest site in the El Yunque experimental forest, part of the Luquillo Long-Term Ecological Research Station in Luquillo, Puerto Rico, USA (Table 1). Soils were diluted in water and inoculated into roll tubes containing MOD-CCMA media with alkali lignin as the source of carbon. MOD-CCMA media consists of 2.8 g L-1 NaCl, 0.1 g L-1 KCl, 27 mM MgCl2, 1 mM CaCl2, 1.25 mM NH4Cl, 9.76 g L-1 MES, 1.1 ml L-1 K2HPO4, 12.5 ml L-1 trace minerals [19,20], and 1 ml L-1 Thauer’s vitamins [21]. Tubes were incubated at room temperature for up to 12 weeks, at which point the colony was picked, grown in 10% tryptic soy broth (TSB), and characterized.
Table 1. Classification and general features of “Enterobacter lignolyticus” SCF1.
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS[6] | |
| Phylum Proteobacteria | TAS[7] | ||
| Class Gammaproteobacteria | TAS[8,9] | ||
| Order Enterobacteriales | TAS[10] | ||
| Family Enterobacteriaceae | TAS[11-13] | ||
| Genus Enterobacter | TAS[11,13-16] | ||
| Species “Enterobacter lignolyticus” | |||
| Strain SCF | |||
| Gram stain | negative | NAS | |
| Cell shape | rod | IDA | |
| Motility | motile via flagella | IDA | |
| Sporulation | non-sporulating | IDA | |
| Temperature range | Mesophile | ||
| Optimum temperature | 30°C | ||
| Carbon source | glucose, xylose, others; see Table 8 | IDA | |
| Energy source | |||
| Terminal electron receptor | |||
| MIGS-6 | Habitat | Soil collected from a subtropical lower montane wet forest | TAS [17] |
| MIGS-6.3 | Salinity | Can tolerate up to 0.75 M NaCl, 1 M KCl, 0.3 M NaOAc, 0.3 M KOAc. Growth in 10% trypticase soy broth is improved with 0.125 M NaCl |
TAS [5] |
| MIGS-22 | Oxygen | facultative aerobe; grows well under completely oxic and anoxic conditions | IDA |
| MIGS-15 | Biotic relationship | free-living | IDA |
| MIGS-14 | Pathogenicity | no | |
| MIGS-4 | Geographic location | Luquillo Experimental Forest, Puerto Rico | IDA |
| MIGS-5 | Sample collection time | July 2009 | IDA |
| MIGS-4.1 | Latitude | 18.268N | IDA |
| MIGS-4.2 | Longitude | 65.760 W | IDA |
| MIGS-4.3 | Depth | 10 cm | IDA |
| MIGS-4.4 | Altitude | 1027 msl | IDA |
a) Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [18].
When grown on 10% TSB agar plates, SCF1 colonies are translucent white, slightly irregular in shape with wavy margins, and have a shiny smooth surface. SCF1 was determined to be a non-sporulating strain based on a Pasteurization test. To do this, a suspension of SCF1 cells was heated at 80°C for 10 minutes. 5μl of heated culture and non-heated control culture were both spotted onto 10% TSB agar and incubated for growth for 3 days at room temperature. The non-heated cells grew while the heated culture did not, indicating the absence of heat-resistant spores.
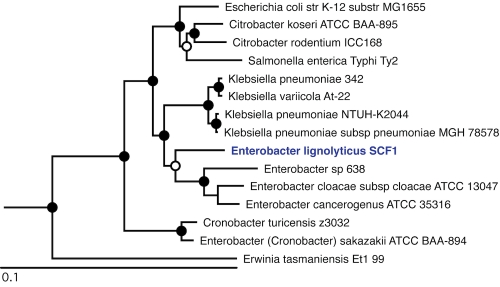
For initial genotyping and for validating the isolation, the small subunit ribosomal RNA gene was sequenced by Sanger sequencing using the universal primers 8F and 1492R [22].The 16S rRNA sequence places “Enterobacter lignolyticus” SCF1 in the family Enterobacteriaceae. However, 16S rRNA sequence is not sufficient to clearly define the evolutionary history of this region of the Gammaproteobacteria, and initially led to the incorrect classification of “E. lignolyticus” SCF1 as a member of the Enterobacter cloacae species. We have rectified its phylogenetic placement using the MicrobesOnline species tree [23], which is generated using 69 single-copy near-universal protein families [24] aligned by MUSCLE [25] with tree construction using FastTree-2 [26] (Figure 1).
Figure 1.
Phylogenetic tree highlighting the position of “Enterobacter lignolyticus” SCF1 relative to other type and non-type strains within the Enterobacteriaceae. Strains shown are those within the Enterobacteriaceae having corresponding NCBI genome project ids listed within [27]. The tree is based on a concatenated MUSCLE alignment [25] of 69 near-universal single-copy COGs (COGs 12, 13, 16, 18, 30, 41, 46, 48, 49, 52, 60, 72, 80, 81, 86, 87, 88, 89, 90, 91, 92, 93, 94, 96, 97, 98, 99, 100, 102, 103, 104, 105, 124, 126, 127, 130, 143, 149, 150, 162, 164, 172, 184, 185, 186, 197, 198, 200, 201, 202, 215, 237, 244, 256, 284, 441, 442, 452, 461, 504, 519, 522, 525, 528, 532, 533, 540, 541, 552). The tree was constructed using FastTree-2 [26] using the JTT model of amino acid evolution [28]. FastTree-2 infers approximate maximum-likelihood phylogenetic placements and provides local support values based on the Shimodaira-Hasegawa test [29]. Solid circles represent local support values over 90% and open circles over 80%. Erwinia tasmaniensis was used as an outgroup.
Genome sequencing information
Genome project history
The genome was selected based on the ability of “E. lignolyticus” SCF1 to grow on and degrade lignin anaerobically. The genome sequence was completed on August 9, 2010, and presented for public access on 15 October 2010 by Genbank. Finishing was completed at Los Alamos National Laboratory. A summary of the project information is shown in Table 2, which also presents the project information and its association with MIGS version 2.0 compliance [30].
Table 2. Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Illumina GAii shotgun, 454 Titanium Standard, and two 454 paired-end |
| MIGS-29 | Sequencing platforms | Illumina, 454 |
| MIGS-31.2 | Fold coverage | 40× for 454 and 469× for Illumina |
| MIGS-30 | Assemblers | Newbler, Velvet, Phrap |
| MIGS-32 | Gene calling method | Prodigal 1.4, GenePRIMP |
| Genbank ID | CP002272 | |
| Genbank Date of Release | October 15, 2010 | |
| GOLD ID | Gc01746 | |
| Project relevance | Anaerobic lignin, switchgrass decomposition |
Growth conditions and DNA isolation
“E. lignolyticus” SCF1 grows well aerobically and anaerobically, and was routinely cultivated aerobically in 10% tryptic soy broth (TSB) with shaking at 200 rpm at 30°C. DNA for sequencing was obtained using the Qiagen Genomic-tip kit and following the manufacturer’s instructions for the 500/g size extraction. Three column preparations were necessary to obtain 50 μg of high molecular weight DNA. The quantity and quality of the extraction were checked by gel electrophoresis using JGI standards.
Genome sequencing and assembly
The draft genome of “Enterobacter lignolyticus” SCF1 was generated at the DOE Joint Genome Institute (JGI) using a combination of Illumina [31] and 454 technologies [32]. For this genome we constructed and sequenced an Illumina GAii shotgun library which generated 50,578,565 reads totaling 3,844 Mb, a 454 Titanium standard library which generated 643,713 reads and two paired end 454 libraries with average insert sizes of 12517 +/- 3129 bp kb and 10286 +/- 2571 bp which generated 346,353 reads totaling 339.3 Mb of 454 data. All general aspects of library construction and sequencing performed at the JGI can be found at the JGI website [33]. The initial draft assembly contained 28 contigs in 1 scaffold. The 454 Titanium standard data and the 454 paired end data were assembled together with Newbler, version 2.3. The Newbler consensus sequences were computationally shredded into 2 kb overlapping fake reads (shreds). Illumina sequencing data was assembled with VELVET, version 0.7.63 [34], and the consensus sequences were computationally shredded into 1.5 kb overlapping fake reads (shreds). We integrated the 454 Newbler consensus shreds, the Illumina VELVET consensus shreds and the read pairs in the 454 paired end library using parallel phrap, version SPS - 4.24 (High Performance Software, LLC). The software Consed [35-37] was used in the following finishing process. Illumina data was used to correct potential base errors and increase consensus quality using the software Polisher developed at JGI (Alla Lapidus, unpublished). Possible mis-assemblies were corrected using gapResolution (Cliff Han, unpublished), Dupfinisher [38], or sequencing cloned bridging PCR fragments with subcloning. Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR (J-F Cheng, unpublished) primer walks. A total of 198 additional reactions were necessary to close gaps and to raise the quality of the finished sequence. The total size of the genome is 4,814,049 bp and the final assembly is based on 191.3 Mb of 454 draft data, which provided an average 40× coverage of the genome, and 2249.8 Mb of Illumina draft data, which provided an average 469× coverage of the genome; the coverage from different technologies is reported separately because they have different error patterns.
Genome annotation
Protein coding genes were identified using Prodigal [39] and tRNA, rRNA and other RNA genes using tRNAscan-SE [40], RNAmmer [41] and Rfam [42] as part of the ORNL genome annotation pipeline followed by a round of manual curation using the JGI GenePRIMP pipeline [43]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGR-Fam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation were performed within the Integrated Microbial Genomes - Expert Review (IMG-ER) platform [44] using the JGI standard annotation pipeline [45,46].
Genome properties
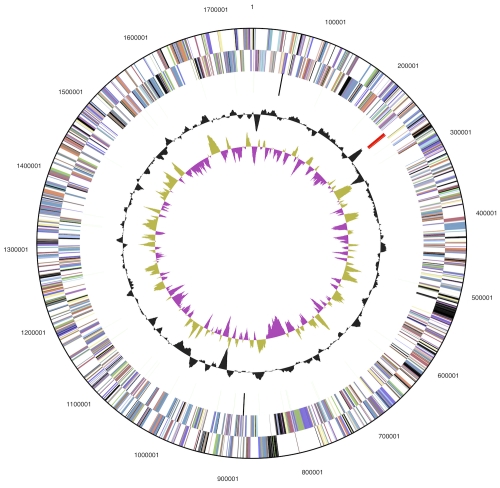
The genome consists of a 4,814,049 bp circular chromosome with a GC content of 57.02% (Table 3 and Figure 2). Of the 4,556 genes predicted, 4,449 were protein-coding genes, and 107 RNAs; 50 pseudogenes were also identified. The majority of the protein-coding genes (85.8%) were assigned with a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4, Table5 and Table 6.
Table 3. Nucleotide content and gene count levels of the genome.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 4,814,049 | 100.00% |
| DNA coding region (bp) | 4,312,328 | 89.58% |
| DNA G+C content (bp) | 2,744,879 | 57.02% |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 4,556 | 100.00% |
| RNA genes | 107 | 2.35% |
| rRNA operons | 7 | |
| Protein-coding genes | 4,449 | 97.65% |
| Pseudo genes | 50 | 1.10% |
| Genes with function prediction | 3,909 | 85.80% |
| Genes in paralog clusters | 823 | 18.06% |
| Genes assigned to COGs | 3,743 | 82.16% |
| Genes assigned Pfam domains | 3,995 | 87.69% |
| Genes with signal peptides | 1,009 | 22.15% |
| Genes with transmembrane helices | 1,108 | 24.32% |
| CRISPR-associated genes (CAS) | 0 | % of Total |
Figure 2.
Graphical circular map of the genome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the 25 general COG functional categories.
| Code | Value | %agea | Description |
|---|---|---|---|
| J | 184 | 4.37 | Translation |
| A | 1 | 0.02 | RNA processing and modification |
| K | 360 | 8.54 | Transcription |
| L | 155 | 3.68 | Replication, recombination and repair |
| B | 0 | 0 | Chromatin structure and dynamics |
| D | 33 | 0.78 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 48 | 1.14 | Defense mechanisms |
| T | 219 | 5.20 | Signal transduction mechanisms |
| M | 239 | 5.67 | Cell wall/membrane biogenesis |
| N | 138 | 3.27 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 1 | 0.02 | Extracellular structures |
| U | 150 | 3.56 | Intracellular trafficking and secretion |
| O | 140 | 3.32 | Posttranslational modification, protein turnover, chaperones |
| C | 275 | 6.52 | Energy production and conversion |
| G | 432 | 10.25 | Carbohydrate transport and metabolism |
| E | 415 | 9.85 | Amino acid transport and metabolism |
| F | 98 | 2.33 | Nucleotide transport and metabolism |
| H | 176 | 4.18 | Coenzyme transport and metabolism |
| I | 108 | 2.56 | Lipid transport and metabolism |
| P | 235 | 5.58 | Inorganic ion transport and metabolism |
| Q | 85 | 2.02 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 409 | 9.70 | General function prediction only |
| S | 314 | 7.45 | Function unknown |
| - | 813 | 17.84 | Not in COGs |
a) The total is based on the total number of protein coding genes in the annotated genome.
Table 5. Number of non-orthologous protein-coding genes found in “Enterobacter lignolyticus” SCF1 with respect to related genomes.
| Species |
Number of distinct genes in “E. lignolyticus” SCF1 |
|---|---|
| Enterobacter sp. 638 | 1,580 |
| Enterobacter cancerogenus ATCC 35316 | 1,551* |
| Enterobacter cloacae ATCC 13047 | 2,891* |
| Klebsiella pneumoniae 342 | 1,389 |
| Klebsiella pneumoniae MGH 78578 | 1,451 |
| Klebsiella pneumoniae NTUH-K2044 | 1,424 |
| Klebsiella variicola At-22 | 1,394 |
| Citrobacter koseri ATCC BAA-895 | 1,507 |
| Citrobacter rodentium ICC168 | 1,682 |
| Escherichia coli K-12 MG1655 | 1,654 |
| Salmonella enterica Typhi Ty2 | 1,811 |
| Cronobacter turicensis z3032 | 1,875 |
| Cronobactersakazakii ATCC BAA-894 | 1,918 |
| Erwinia tasmaniensis Et1/99 | 2,392 |
| Protein-coding genes distinct in “E. lignolyticus” SCF1 compared with all orthologous genes found in above genomes | 643 |
* Based on incompletely annotated genome.
Table 6. Number of genes not found in near-relatives associated with the 25 general COG functional categories*.
| Code | Value | Description |
|---|---|---|
| - | 151 | Hypothetical (no conserved gene family) |
| - | 17 | Transposase / Integrase (annotation-based) |
| - | 80 | Transport (annotation-based) |
| - | 66 | Signaling and Regulation |
| J | 6 | Translation |
| A | 0 | RNA processing and modification |
| K | 51 | Transcription |
| L | 18 | Replication, recombination and repair |
| B | 0 | Chromatin structure and dynamics |
| D | 2 | Cell cycle control, mitosis and meiosis |
| Y | 0 | Nuclear structure |
| V | 7 | Defense mechanisms |
| T | 30 | Signal transduction mechanisms |
| M | 41 | Cell wall/membrane biogenesis |
| N | 20 | Cell motility |
| Z | 0 | Cytoskeleton |
| W | 1 | Extracellular structures |
| U | 22 | Intracellular trafficking and secretion |
| O | 9 | Posttranslational modification, protein turnover, chaperones |
| C | 20 | Energy production and conversion |
| G | 68 | Carbohydrate transport and metabolism |
| E | 28 | Amino acid transport and metabolism |
| F | 5 | Nucleotide transport and metabolism |
| H | 5 | Coenzyme transport and metabolism |
| I | 14 | Lipid transport and metabolism |
| P | 23 | Inorganic ion transport and metabolism |
| Q | 8 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 43 | General function prediction only |
| S | 23 | Function unknown |
| - | 255 | Not in COGs |
* Number of genes from set of 643 genes not found in near-relatives associated with the 25 general COG functional categories and several annotation-based classifications. Note that counts do not sum to 643 genes as a given gene is sometimes classified in more than one COG functional category.
Lignocellulose degradation pathways
“E. lignolyticus” SCF1 has a relatively small arsenal of lignocellulolytic carbohydrate active enzymes, including a single GH8 endoglucanase, and a GH3 beta-glucosidase, but no xylanase or beta-xylosidase. Table 7 provides a more complete list of lignocellulolytic enzymes. The genome also contains a large number of saccharide and oligosaccharide transporters, including several ribose ABC transporters, a xylose ABC transporter (Entcl_0174-0176), and multiple cellobiose PTS transporters (Entcl_1280, Entcl_2546-2548, Entcl_3764, Entcl_4171-4172).
Table 7. Selection of lignocellulolytic carbohydrate active, lignin oxidative (LO) and lignin degrading auxiliary (LDA) enzymes [47,48]†.
| Locus Tag | Family | Function |
|---|---|---|
| Entcl_0212 | GH8 | endoglucanase (EC 3.2.1.4) |
| Entcl_1570 | GH3 | beta-glucosidase (EC 3.2.1.21) |
| Entcl_0851 | GH1 | 6-phospho-beta-glucosidase (EC 3.2.1.86) |
| Entcl_0991 | GH1 | 6-phospho-beta-glucosidase (EC 3.2.1.86) |
| Entcl_1274 | GH1 | 6-phospho-beta-glucosidase (EC 3.2.1.86) |
| Entcl_3004 | GH1 | 6-phospho-beta-glucosidase (EC 3.2.1.86) |
| Entcl_3339 | GH2 | beta-galactosidase (EC 3.2.1.23) |
| Entcl_0624 | GH2 | beta-galactosidase (EC 3.2.1.23) |
| Entcl_2579 | GH2 | beta-mannosidase (EC 3.2.1.25) |
| Entcl_2687 | GH3 | beta-N-acetylhexosaminidase (EC 3.2.1.52) |
| Entcl_3271 | GH4 | alpha-galactosidase (EC 3.2.1.22) |
| Entcl_0170 | GH13 | alpha-amylase (EC 3.2.1.1) |
| Entcl_3416 | GH13 | alpha-glucosidase (EC 3.2.1.20) |
| Entcl_2926 | GH18 | chitinase (EC 3.2.1.14) |
| Entcl_2924 | GH19 | chitinase (EC 3.2.1.14) |
| Entcl_4037 | GH35 | beta-galactosidase (EC 3.2.1.23) |
| Entcl_3090 | GH38 | alpha-mannosidase (EC 3.2.1.24) |
| Entcl_0250 | CE4 | polysaccharide deacetylase (EC 3.5.-.-) |
| Entcl_3596 | CE4 | polysaccharide deacetylase (EC 3.5.-.-) |
| Entcl_3059 | CE8 | pectinesterase (EC 3.1.1.11) |
| Entcl_2112 | LDA2 | vanillyl-alcohol oxidase (EC 1.1.3.38) |
| Entcl_1569 | LDA2 | D-lactate dehydrogenase (EC 1.1.1.28) |
| Entcl_4187 | LDA2 | UDP-N-acetylmuramate dehydrogenase (EC 1.1.1.158) |
| Entcl_3603 | LO1 | putative laccase (EC 1.10.3.2) |
| Entcl_0735 | LO1 | putative laccase (EC 1.10.3.2) |
| Entcl_4301 | LO2 | catalase/peroxidase (EC 1.11.1.6, 1.11.1.7) |
† Enzyme families are as per the CAZy and FOLy databases
The mechanisms for lignin degradation in bacteria are still poorly understood. Two multi-copper oxidases (putative laccases) and a putative peroxidase (see Table 7) may be involved in oxidative lignin degradation. We also found multiple glutathione S-transferase proteins, and it is possible that one or more of these may be involved in cleavage of beta-aryl ether linkages, as is the case with LigE/LigF in Sphingomonas paucimobilis [49]. However, “E. lignolyticus” SCF1 does not seem to posses the core protocatechuate and 3-O-methylgallate degradation pathways responsible for lignin catabolism in S. paucimobilis. Instead, lignin catabolism may proceed via homoprotocatechuate through the 4-hydroxyphenylacetate degradation pathway, encoded on a gene cluster conserved between other Enterobacter, Klebsiella, and some E. coli strains (Figures 3, 4).
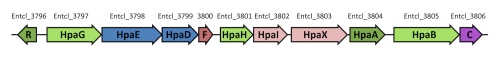
Figure 3.
The entire 4-hydroxyphenylacetate degradation pathway is encoded in a single gene cluster HpaRGEDFHIXABC, including a divergently expressed regulator (HpaR), and a 4-hydroxyphenylacetate permease (HpaX).
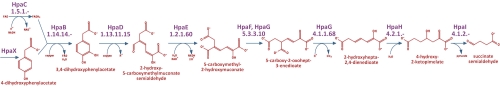
Figure 4.
The 4-hydroxyphenylacetate degradation pathway via homoprotocatechuate (3,4-dihydroxyphenylacetate).
Lignin degradation
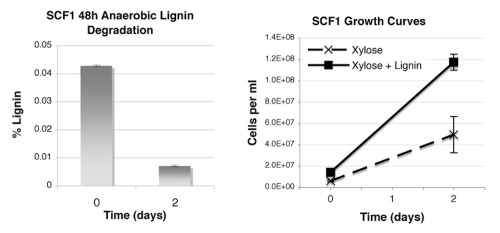
We have grown SCF1 in xylose minimal media with and without lignin, and measured both cell counts (by acridine orange direct counts) and lignin degradation (by change in absorbance at 280 nm) over time. Lignin degradation was substantial after two days (left), and significantly enhanced growth of cells in culture (right); data are expressed as mean with standard deviation (n=3, Figure 5). Further studies will explore the moieties of lignin used in anaerobic growth as well as explore growth on and utilization of other types of lignin.
Figure 5.
Anaerobic lignin degradation by “E. lignolyticus” SCF1 after 48 hours in culture, grown with xylose minimal media.
Phenotypic Microarray
We used the Biolog phenotypic microarray to test the range of growth conditions. For each of the eight plates in the array, “E. lignolyticus” SCF1 cells were grown up on 10% TSB agar plates, scraped off and resuspended in 20mM D-Glucose MOD-CCMA, adjusted to 0.187 OD, 1× concentrate of Biolog Dye Mix G added, and then inoculated. PM plates include two plates with different carbon sources (PM 1 and 2a), one plate of different simple nitrogen sources (PM 3b), one plates of phosphorous and sulfur sources (PM4A), one plate of nutritional supplements (PM5), and three plates of amino acid dipeptides as nitrogen sources (PM6, PM7, PM8). Carbon source, D-Glucose, was omitted from MOD-CCMA when used to inoculate PM1 and 2a. Similarly, NH4Cl, KH2PO4 and vitamins were omitted from 20mM D-Glucose MOD CCMA when inoculating plates containing nitrogen sources, phosphorus/sulfur sources, and nutrient supplements, respectively. On plates 6-8, the positive control is L-Glutamine. The phenotypic microarray revealed a number of carbon and nitrogen sources that resulted in four times the growth or more compared to the negative control based on duplicate runs (Table 8 and 9), as well as sulfur and phosphorous sources that improved growth by 10% or more (Tables 10 and 11). None of the dipeptides resulted in an increase in growth more than twice the background, and so are not reported here. Of the nutritional supplements tested in PM5, 2'-deoxyuridine and 2'-deoxyadenosine resulted in 10% growth improvement, while (5) 4-amino-imidazole-4(5)-carboxamide, Tween 20, Tween 40, Tween 60, and Tween 80 resulted in 20% growth improvement.
Table 8. Carbon source by phenotypic array (PM 1 and 2a).
| Chemical Name | KEGG | CAS | Ratio to background |
|---|---|---|---|
| D-Fructose | C00095 | 57-48-7 | 8.48 |
| D-Sorbitol | C00794 | 50-70-4 | 8.36 |
| N-Acetyl-D-Glucosamine | C03000 | 7512-17-6 | 8.30 |
| D-Gluconic Acid | C00257 | 527-07-1 | 8.28 |
| D-Trehalose | C01083 | 99-20-7 | 8.18 |
| D-Mannose | C00159 | 3458-28-4 | 8.10 |
| D-Xylose | C00181 | 58-86-6 | 8.09 |
| a-D-Glucose | C00031 | 50-99-7 | 8.07 |
| N-Acetyl-D-Mannosamine | C00645 | 7772-94-3 | 7.92 |
| D-Mannitol | C00392 | 69-65-8 | 7.92 |
| D-Galactose | C00124 | 59-23-4 | 7.92 |
| D-Glucosaminic Acid | C03752 | 3646-68-2 | 7.85 |
| D-Ribose | C00121 | 50-69-1 | 7.76 |
| b-Methyl-D-Glucoside | 709-50-2 | 7.70 | |
| D-Glucuronic Acid | C00191 | 14984-34-0 | 7.69 |
| D-Glucosamine | C00329 | 66-84-2 | 7.68 |
| D-Galactonic Acid-g-Lactone | C03383 | 2782-07-2 | 7.67 |
| Maltose | C00208 | 69-79-4 | 7.62 |
| 2-Deoxy-D-Ribose | C01801 | 533-67-5 | 7.57 |
| Glycerol | C00116 | 56-81-5 | 7.52 |
| m-Hydroxyphenyl Acetic Acid | C05593 | 621-37-4 | 7.42 |
| L-Arabinose | C00259 | 87-72-9 | 7.40 |
| m-Inositol | C00137 | 87-89-8 | 7.39 |
| L-Serine | C00065 | 56-45-1 | 7.38 |
| 3-Methylglucose | 13224-94-7 | 7.36 | |
| Maltotriose | C01835 | 1109-28-0 | 7.30 |
| D-Melibiose | C05402 | 585-99-9 | 7.25 |
| L-Fucose | C01019 | 2438-80-4 | 7.25 |
| D-Arabinose | C00216 | 10323-20-3 | 7.10 |
| Hydroxy-L-Proline | C01015 | 51-35-4 | 7.08 |
| 2'-Deoxyadenosine | C00558 | 16373-93-6 | 7.02 |
| L-Alanine | C00041 | 56-41-7 | 6.94 |
| Tyramine | C00483 | 60-19-5 | 6.93 |
| Gly-Pro | 704-15-4 | 6.93 | |
| D-Galacturonic Acid | C00333 | 91510-62-2 | 6.91 |
| L-Rhamnose | C00507 | 3615-41-6 | 6.86 |
| p-Hydroxyphenyl Acetic Acid | C00642 | 156-38-7 | 6.83 |
| Acetic Acid | C00033 | 127-09-3 | 6.81 |
| L-Proline | C00148 | 147-85-3 | 6.80 |
| Fumaric Acid | C00122 | 17013-01-3 | 6.80 |
| D,L-Malic Acid | C00497 | 6915-15-7 | 6.75 |
| D,L-Lactic acid | C01432 | 312-85-6 | 6.71 |
| Dihydroxyacetone | C00184 | 96-26-4 | 6.69 |
| Tween 20 | C11624 | 9005-64-5 | 6.57 |
| N-Acetyl-D-Galactosamine | 14215-68-0 | 6.45 | |
| Inosine | C00294 | 58-63-9 | 6.45 |
| Ala-Gly | 687-69-4 | 6.43 | |
| L-Histidine | C00135 | 5934-29-2 | 6.37 |
| D-Alanine | C00133 | 338-69-2 | 6.29 |
| D-Fructose-6-Phosphate | C00085 | 26177-86-637250-85-4 | 6.25 |
| L-Glutamine | C00064 | 56-85-9 | 6.08 |
| Gly-Glu | 7412-78-4 | 6.00 | |
| D-Cellobiose | C00185 | 528-50-7 | 5.98 |
| D-Glucose-1-Phosphate | C00103 | 56401-20-8 | 5.95 |
| D-Psicose | C06468 | 551-68-8 | 5.92 |
| Citric Acid | C00158 | 6132-04-3 | 5.91 |
| L-Glutamic Acid | C00025 | 6106-04-3 | 5.84 |
| b-Methyl-D-Galactoside | C03619 | 1824-94-8 | 5.70 |
| L-Aspartic Acid | C00049 | 3792-50-5 | 5.65 |
| D-Serine | C00740 | 312-84-5 | 5.63 |
| Methylpyruvate | 600-22-6 | 5.62 | |
| Pyruvic Acid | C00022 | 113-24-6 | 5.56 |
| Propionic Acid | C00163 | 137-40-6 | 5.48 |
| Melibionic Acid | 70803-54-2 | 5.43 | |
| D-Malic Acid | C00497 | 636-61-3 | 5.38 |
| D-Aspartic Acid | C00402 | 1783-96-6 | 5.38 |
| 5-Keto-D-Gluconic Acid | C01062 | 91446-96-7 | 5.37 |
| Succinic Acid | C00042 | 6106-21-4 | 5.35 |
| Gly-Asp | C02871 | 5.28 | |
| D,L-a-Glycerol Phosphate | C00093 | 3325-00-6 | 5.26 |
| Putrescine | C00134 | 333-93-7 | 5.14 |
| Gentiobiose | C08240 | 554-91-6 | 5.00 |
| D-Glucose-6-Phosphate | C00092 | 3671-99-6 | 4.90 |
| a-Methyl-D-Galactoside | C03619 | 3396-99-4 | 4.84 |
| Uridine | C00299 | 58-96-8 | 4.68 |
| Bromosuccinic Acid | 923-06-8 | 4.68 | |
| Thymidine | C00214 | 50-89-5 | 4.63 |
| L-Asparagine | C00152 | 70-47-3 | 4.55 |
| a-Hydroxybutyric Acid | C05984 | 19054-57-0 | 4.38 |
| L-Malic Acid | C00149 | 138-09-0 | 4.34 |
| L-Ornithine | C00077 | 3184-13-2 | 4.28 |
| N-Acetyl-D-glucosaminitol | 4271-28-7 | 4.23 | |
| L-Lyxose | C01508 | 1949-78-6 | 4.23 |
| L-Threonine | C00188 | 72-19-5 | 4.21 |
| g-Amino-N-Butyric Acid | C00334 | 56-12-2 | 4.19 |
| Arbutin | C06186 | 497-76-7 | 4.17 |
Table 9. Nitrogen sources by phenotypic array (PM 3b).
Table 10. Phosphorous source by phenotypic array (PM 4a).
| Chemical Name | KEGG | CAS | Ratio to background |
|---|---|---|---|
| O-Phospho-D-Serine | 73913-63-0 | 1.42 | |
| Phospho-Glycolic Acid | C00988 | 1.28 | |
| Carbamyl Phosphate | C00416 | 72461-86-0 | 1.26 |
| O-Phospho-L-Threonine | 1114-81-4 | 1.25 | |
| Tripolyphosphate | C02466 | 1.24 | |
| O-Phospho-L-Serine | 407-41-0 | 1.23 | |
| Cysteamine-S-Phosphate | 3724-89-8 | 1.22 | |
| Cytidine 2'-Monophosphate | C03104 | 85-94-9 | 1.21 |
| Guanosine 5'-Monophosphate | C00144 | 5550-12-9 | 1.21 |
| Guanosine 3'-Monophosphate | C06193 | 1.20 | |
| Phosphoenol Pyruvate | C00074 | 5541-93-5 | 1.20 |
| Cytidine 3'-Monophosphate | C05822 | 84-52-6 | 1.20 |
| Cytidine 5'-Monophosphate | C00055 | 6757-06-8 | 1.20 |
| Adenosine 2',3'-Cyclic Monophosphate | 37063-35-7 | 1.20 | |
| Phospho-L-Arginine | 108321-86-4 | 1.20 | |
| Adenosine 3'-Monophosphate | C01367 | 84-21-9 | 1.20 |
| Guanosine 2',3'-Cyclic Monophosphate | 15718-49-7 | 1.19 | |
| D-3-Phospho-Glyceric Acid | C00631 | 80731-10-8 | 1.19 |
| Phosphate | C00009 | 10049-21-5 | 1.19 |
| Guanosine 2'-Monophosphate | 6027-83-4 | 1.19 | |
| Thiophosphate | 10489-48-2 | 1.18 | |
| Thymidine 3'-Monophosphate | 108320-91-8 | 1.18 | |
| Thymidine 5'-Monophosphate | C00364 | 33430-62-5 | 1.16 |
| 6-Phospho-Gluconic Acid | 53411-70-4 | 1.16 | |
| Dithiophosphate | 1.16 | ||
| 2-Aminoethyl Phosphonic Acid | C03557 | 2041-14-7 | 1.15 |
| Phosphoryl Choline | C00588 | 4826-71-5 | 1.14 |
| D,L-a-Glycerol Phosphate | C00093 | 3325-00-6 | 1.13 |
| Trimetaphosphate | C02466 | 7785-84-4 | 1.13 |
Table 11. Sulfur source by phenotypic array (PM 4a).
| Chemical Name | KEGG | CAS | Ratio to background |
|---|---|---|---|
| L-Cysteine Sulfinic Acid | C00607 | 1115-65-7 | 1.24 |
| Gly-Met | 554-94-9 | 1.23 | |
| Tetramethylene Sulfone | 126-33-0 | 1.21 | |
| L-Methionine | C00073 | 63-68-3 | 1.21 |
| N-Acetyl-D,L-Methionine | C02712 | 71463-44-0 | 1.20 |
| L-Methionine Sulfoxide | C02989 | 3226-65-1 | 1.19 |
| Tetrathionate | C02084 | 13721-29-4 | 1.18 |
| L-Cysteine | C00097 | 7048-04-6 | 1.17 |
| Sulfate | C00059 | 7727-73-3 | 1.14 |
| L-Djenkolic Acid | C08275 | 28052-93-9 | 1.14 |
| Cys-Gly | 19246-18-5 | 1.13 |
Conclusion
Close relatives of “Enterobacter lignolyticus” SCF1 were isolated seven independent times from Puerto Rico tropical forest soils, growing anaerobically with lignin or switchgrass as the sole carbon source, suggesting that it is relatively abundant in tropical forest soils and has broad capability for deconstruction of complex heteropolymers such as biofuel feedstocks. In a previous study, Enterobacter was one of four isolates from the poplar rhizosphere chosen for genomic sequencing because of its ability to improve the carbon sequestration ability of poplar trees when grown in poor soils [50].
Isolates from the Enterobacteriaceae are extremely genetically diverse despite the near identity of genotypic markers such as small subunit ribosomal (16S) RNA genes. Multi-locus sequence typing and comparative genomic hybridization show that the isolates seem to fall into two distinct clades: the first being more homogeneous and containing isolates found in hospitals, and the second being more diverse and found in a broader array of environments [51].
This organism was determined to grow aerobically and anaerobically, and when screening for enzyme activity, the enzymes isolated showed accelerated phenol oxidase and peroxidase enzyme activity under aerobic conditions. In addition, this organism is capable of growth in 8% ethyl-methylimidazolium chloride ([C2mim]Cl), an ionic liquid being studied for pre-treatment of feedstocks. This extremely high tolerance to ionic liquids is potentially quite useful for industrial biofuels production from feedstocks and the mechanism is currently under investigation.
Acknowledgements
The work conducted in part by the U.S. Department of Energy Joint Genome Institute and in part by the Joint BioEnergy Institute (http://www.jbei.org) supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research, under Contract No. DE-AC02-05CH11231.
Abbreviations:
- EMBL- European Molecular Biology Laboratory
NCBI- National Center for Biotechnology Information (Bethesda, MD, USA), RDP- Ribosomal Database Project (East Lansing, MI, USA)
References
- 1.Parton W, Silver WL, Burke IC, Grassens L, Harmon ME, Currie WS, King JY, Adair EC, Brandt LA, Hart SC, et al. Global-scale similarities in nitrogen release patterns during long-term decomposition. Science 2007; 315:361-364 10.1126/science.1134853 [DOI] [PubMed] [Google Scholar]
- 2.Cusack DF, Chou WW, Yang WH, Harmon ME, Silver WL. Controls on long-term root and leaf litter decomposition in neotropical forests. Glob Change Biol 2009; 15:1339-1355 10.1111/j.1365-2486.2008.01781.x [DOI] [Google Scholar]
- 3.Paul EA, Clark FE. Soil microbiology and biochemistry, Second edition. In: Paul EA, Clark FE, editors. Soil microbiology and biochemistry, Second edition. 1250 Sixth Ave., San Diego, California 92101; 14 Belgrave Square, 24-28 Oval Road, London NW1 70X, England, UK: Academic Press, Inc.; Academic Press Ltd.; 1996. p xiii+340. [Google Scholar]
- 4.Freeman C, Ostle N, Kang H. An enzymic 'latch' on a global carbon store - A shortage of oxygen locks up carbon in peatlands by restraining a single enzyme. Nature 2001; 409:149 10.1038/35051650 [DOI] [PubMed] [Google Scholar]
- 5.Khudyakov J, D’Haeseleer P, Borglin SE, DeAngelis KM, Woo HH, Lindquist, EA, Hazen TC, Simmons BA, Thelen MP. Global transcriptome response to ionic liquid by a tropical rain forest soil bacterium, “Enterobacter lignolyticus” SCF1PNAS plus 2011. In press. [DOI] [PMC free article] [PubMed]
- 6.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 8.List Editor Validation of publication of new names and new combinations previously effectively published outside the IJSEM. List no. 106. Int J Syst Evol Microbiol 2005; 55:2235-2238 10.1099/ijs.0.64108-0 [DOI] [PubMed] [Google Scholar]
- 9.Garrity GM, Bell JA, Lilburn T. Class III. Gammaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 10.Garrity GM, Holt JG. Taxonomic Outline of the Archaea and Bacteria In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 155-166. [Google Scholar]
- 11.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]
- 12.Rahn O. New principles for the classification of bacteria. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg 1937; 96:273-286 [Google Scholar]
- 13.Judicial Commission Conservation of the family name Enterobacteriaceae, of the name of the type genus, and designation of the type species OPINION NO. 15. Int Bull Bacteriol Nomencl Taxon 1958; 8:73-74 10.1099/0096266X-8-1-73 [DOI] [Google Scholar]
- 14.Hormaeche E, Edwards PR. A proposed genus Enterobacter. Int Bull Bacteriol Nomencl Taxon 1960; 10:71-74 10.1099/0096266X-10-2-71 [DOI] [Google Scholar]
- 15.Sakazaki R. Genus VII. Enterobacter Hormaeche and Edwards 1960, 72; Nom. cons. Opin. 28, Jud. Comm. 1963, 38. In: Buchanan RE, Gibbons NE (eds), Bergey's Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 324-325. [Google Scholar]
- 16.Editorial Board OPINION 28 Rejection of the Bacterial Generic Name Cloaca Castellani and Chalmers and Acceptance of Enterobacter Hormaeche and Edwards as a Bacterial Generic Name with Type Species Enterobacter cloacae (Jordan) Hormaeche and Edwards. Int Bull Bacteriol Nomencl Taxon 1963; 13:38 10.1099/0096266X-13-1-38 [DOI] [Google Scholar]
- 17.DeAngelis KM, Silver WL, Thompson AW, Firestone MK. Microbial communities acclimate to recurring changes in soil redox potential status. Environ Microbiol 2010; 12:3137-3149 10.1111/j.1462-2920.2010.02286.x [DOI] [PubMed] [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tschech A, Pfennig N. Growth-Yield Increase Linked to Caffeate Reduction in Acetobacterium-Woodii. Arch Microbiol 1984; 137:163-167 10.1007/BF00414460 [DOI] [Google Scholar]
- 20.Widdel F, Kohring GW, Mayer F. Studies on Dissimilatory Sulfate-Reducing Bacteria That Decompose Fatty-Acids. 3. Characterization of the Filamentous Gliding Desulfonema-Limicola gen-nov sp-nov, and Desulfonema-Magnum Sp-Nov. Arch Microbiol 1983; 134:286-294 10.1007/BF00407804 [DOI] [Google Scholar]
- 21.Janssen PH, Schuhmann A, Morschel E, Rainey FA. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl Environ Microbiol 1997; 63:1382-1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson KH, Blitchington RB, Greene RC. Amplification of bacterial-16S ribosomal DNA with polymerase chain-reaction. J Clin Microbiol 1990; 28:1942-1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, et al. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res 2010; 38:D396-D400 10.1093/nar/gkp919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science 2006; 311:1283-1287 10.1126/science.1123061 [DOI] [PubMed] [Google Scholar]
- 25.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792-1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price MN, Dehal PS, Arkin AP. FastTree 2-Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010; 5:e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrity GM, Lilburn TG, Cole JR, Harrison SH, Euzeby J, Tindall BJ. The Taxonomic Outline of Bacteria and Archaea version 7.7. Michigan State University Board of Trustees; 2007. [Google Scholar]
- 28.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutant data matrices from protein sequences. Comput Appl Biosci 1992; 8:275-282 [DOI] [PubMed] [Google Scholar]
- 29.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 1999; 16:1114-1116 [Google Scholar]
- 30.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett S. Solexa Ltd. Pharmacogenomics 2004; 5:433-438 10.1517/14622416.5.4.433 [DOI] [PubMed] [Google Scholar]
- 32.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437:376-380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DOE Joint Genome Institute http://www.jgi.doe.gov/
- 34.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18:821-829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998; 8:175-185 [DOI] [PubMed] [Google Scholar]
- 36.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 1998; 8:186-194 [PubMed] [Google Scholar]
- 37.Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res 1998; 8:195-202 [DOI] [PubMed] [Google Scholar]
- 38.Han C, Chain P. Finishing repeat regions automatically with Dupfinisher. In: Arabnia HR, Valafar J, editors. Proceedings of the 2006 international Conference on Bioinformatics and Computational Biology: CSREA Press; 2006. p 141-146. [Google Scholar]
- 39.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translatin initiation site identification. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955-964 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. Rfam: Wikipedia, clans and the "decimal" release. Nucleic Acids Res 2011; 39:D141-D145 10.1093/nar/gkq1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 44.Markowitz VM, Mavromatis K, Ivanova NN, Chen IMA, Chu K, Krypides NC. IMG ER: a system tool for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]
- 45.Mavromatis K, Ivanova NN, Chen IMA, Szeto E, Markowitz VM, Kyrpides NC. The DOE-JGI Standard Operating Procedure for the Annotations of Microbial Genomes. Stand Genomic Sci 2009; 1:63-67 10.4056/sigs.632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.User Guide IMG.https://img.jgi.doe.gov/er/doc/userGuide.pdf
- 47.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 2009; 37:D233-D238 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levasseur A, Plumi F, Coutinho PM, Rancurel C, Asther M, Delattre M, Henrissat B, Pontarotti P, Record E. FOLy: An integrated database for the classification and functional annotation of fungal oxidoreductases potentially involved in the degradation of lignin and related aromatic compounds. Fungal Genet Biol 2008; 45:638-645 10.1016/j.fgb.2008.01.004 [DOI] [PubMed] [Google Scholar]
- 49.Masai E, Katayama Y, Fukuda M. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem 2007; 71:1-15 10.1271/bbb.60437 [DOI] [PubMed] [Google Scholar]
- 50.Taghavi S, Garafola C, Monchy S, Newman L, Hoffman A, Weyens N, Barac T, Vangronsveld J, van der Lelie D. Genome Survey and Characterization of Endophytic Bacteria Exhibiting a Beneficial Effect on Growth and Development of Poplar Trees. Appl Environ Microbiol 2009; 75:748-757 10.1128/AEM.02239-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paauw A, Caspers MPM, Schuren FHJ, Leverstein-van Hall MA, Deletoile A, Montijn RC, Verhoef J, Fluit AC. Genomic Diversity within the Enterobacter cloacae Complex. PLoS ONE 2008; 3:e3018 10.1371/journal.pone.0003018 [DOI] [PMC free article] [PubMed] [Google Scholar]