Abstract
The incorporation of radiotherapy into multimodality treatment plans has led to significant improvements in glioma patient survival. However, local recurrence from glioma resistance to ionizing radiation remains a therapeutic challenge. The tumoricidal effect of radiation therapy is largely attributed to the induction of dsDNA breaks (DSBs). In the past decade, there have been tremendous strides in understanding the molecular mechanisms underlying DSB repair. The identification of gene products required for DSB repair has provided novel therapeutic targets. Recent studies revealed that many US FDA-approved cancer agents inhibit DSB repair by interacting with repair proteins. This article will aim to provide discussion of DSB repair mechanisms to provide molecular targets for radiation sensitization of gliomas and a discussion of FDA-approved cancer therapies that modulate DSB repair to highlight opportunities for combination therapy with radiotherapy for glioma therapy.
Keywords: chemotherapy, DNA damage response, DNA double strand break repair, glioblastoma, radiosensitization, radiotherapy
Glioblastomas & radiation therapy
Glioblastoma is the most common form of primary brain tumor [1]. Approximately 10,000 new patients each year in the USA are diagnosed with glioblastoma multiformes (GBMs). Despite its comparatively low incidence of approximately two to three new cases per 100,000 people per year (for comparison, the incidence of colon cancer in the USA is approximately one new case per 1800 people per year), the total number of deaths per year attributable to glioblastoma rivals that of other major cancers. This disparity reflects the lethal nature of the disease. If untreated, patients with glioblastoma generally die within 3–4 months of their diagnosis [2,3]. Diffuse invasion into the surrounding normal cerebral parenchyma is a cardinal feature of glioblastoma, preventing surgical cure. In this context, it is not surprising that surgical resection alone does not significantly prolong patient survival [2]. The current standard of care is based on the landmark trial by the EORTC–NCIC, in which maximal safe surgical resection is combined with radiation and the chemotherapeutic agent, temozolomide. This treatment strategy has resulted in an improvement of median survival to 14.6 months [4]. While this regimen achieves an approximate 10% 5-year progression-free survival [5], the majority of the afflicted patients succumb to the disease. Effective therapeutic strategies are desperately needed.
Despite the abundance of therapeutic strategies and agents that have been tested over the last half century for the treatment of GBM, the single most efficacious modality and significant advance has been the use of postresection radiation therapy. The efficacy of radiation therapy in glioblastoma was established in three randomized control trials. Brain Tumor Study Group (BTSG 66-01), randomized patients with newly diagnosed malignant gliomas to whole-brain radiotherapy (WBRT) with or without mithramycin [6]. While mithramycin did not significantly impact survival, the mean survival of patients who received adjuvant WBRT was extended to approximately 5 months. BTSG 69-01 randomized patients, who had undergone surgical resection, to supportive care or chemotherapy (1,3-bis[2-chloroethyl]-1-nitrosourea [BCNU]), with or without WBRT. Overall, patient survival was improved with radiation therapy (with or without BCNU) when compared with supportive care alone. The median survival for the surgery-only patients was 14 weeks. The median survival for patients who underwent surgery followed by radiation therapy alone increased to 36 weeks (p < 0.05). Similar results were seen in the third randomized trial, BTSG72-01 [7]. A meta-analysis of BTSG66-01, BTSG69-01 and BTSG72-01 [8] demonstrated that patients who did not receive radiation or were treated with less than 4500 cGy exhibited poor survival (median survival of 4.2 and 3.1 months, respectively). By contrast, patients treated with 5000, 5500 and 6000 cGy were associated with improved survival (median survival of 6.5, 8.4 and 9.8 months, respectively). In a Medical Research Council (MRC) randomized trial, comparing 4500 cGy with 6000 cGy postoperatively, the median survival improved from 9 to 12 months with the higher dose [9]. Subsequent dose escalation beyond 6000 cGy revealed increased neuro-toxicity without significant therapeutic gain [10]. These studies collectively established the role of radiotherapy for modern glioblastoma therapy.
It is important to note that, as a single agent, the efficacy of radiation therapy as a glioblastoma therapeutic has yet to be matched by any chemotherapy [10]. For this reason, radiation therapy remains a universal component in clinical trials with curative intent for treatment of newly diagnosed glioblastomas. Identification of radio-sensitizing agents that augment the efficacy of radiation therapy remains a promising strategy for glioblastoma therapy. In this context, we will review the molecular mechanism underlying cellular response to radiation.
DNA repair & damage response following ionizing radiation
While ionizing radiation (IR) has been shown to induce a plethora of cellular effects, it is widely held that its tumoricidal activity is largely related to its effect on DNA [11]. IR induces multiple types of DNA lesions, including damage to the nucleobase, sugar and phosphate back bone [11]. Ultimately, these lesions, if unrepaired, are converted into dsDNA breaks (DSBs). DSBs are highly cytotoxic as the presence of a single DSB in a cell is sufficient to induce clonogenic death [12–17]. Our understanding of the DNA damage response (DDR) and DSB repair has expanded exponentially in the past decade. To make this vast information more accessible to the reader, we will organize the information into the following sections:
General mechanism of DSB repair
Mechanism by which the DDR activates DSB repair
Effect of DNA damage on cell cycle progression
General mechanism of dsDNA break repair
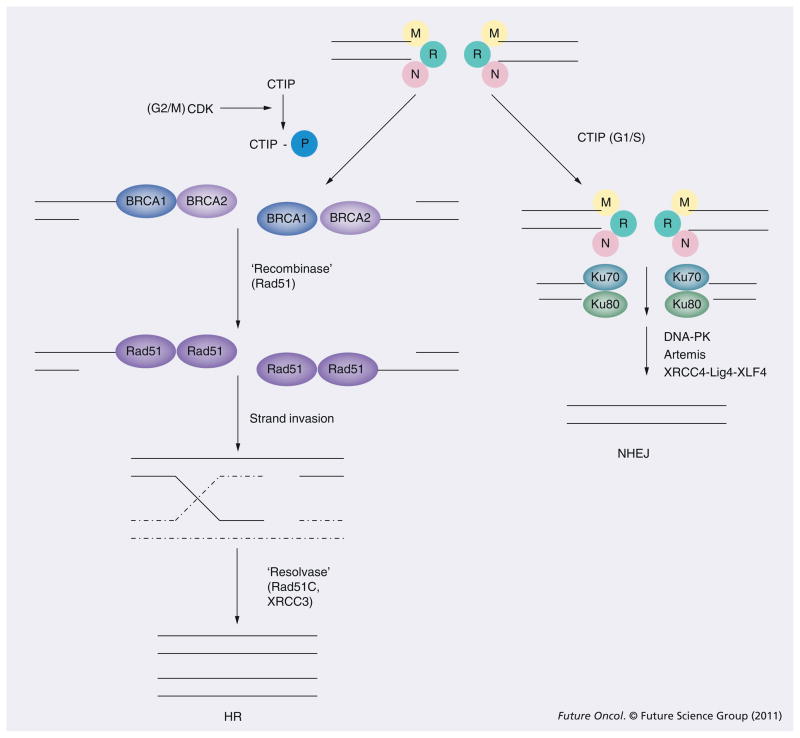
dsDNA breaks are repaired by two major mechanisms: nonhomologous end-joining (NHEJ) or homologous recombination (HR) [18,19]. The simplest way of repairing DSB is by re-ligation. This process is termed NHEJ since no significant DNA sequence homology at the broken ends is required (Figure 1). Instead, the NHEJ enzymatic machinery relies on limited processing of the DNA ends followed by annealing short stretches of complementary DNA sequences. Because short stretches of complementary sequences occur at some regularity in the human genome, NHEJ is prone to the generation of mutations and deletions, as well as chromosomal rearrangements [20].
Figure 1. Molecular mechanism of dsDNA repair.
CTIP is a key determinant of whether DSBs are repaired by HR and NHEJ. CTIP is activated by CDK phosphorylation during the G2 phase of the cell cycle. Phosphorylated CTIP binds to the MRN complex to facilitate processing of DSBs into ssDNA tails. This processing enables the initiation of HR. CTIP is not phosphorylated during the G1 phase of the cell cycle. In this context, the MRN complex recruits the Ku proteins to initiate NHEJ.
CDK: Cyclin-dependent kinase; CTIP: C-terminal binding protein interacting protein; DSB: dsDNA break; HR: Homologous recombination; NHEJ: Nonhomologous end-joining; MRN complex: Mre11–Rad50–Nbs1; P: Phosphorylation.
The second major DSB repair process is termed HR (Figure 1). HR initiates with extensive 5′ to 3′ end processing of the broken ends into large regions of single-stranded tails. The resultant 3′ single-stranded tail invades a homologous donor sequence. This strand invasion is mediated by enzyme(s) termed ‘recombinase’, which coats the single-stranded tail. Subsequent strand extension and Holliday junction resolution result in restoration of DNA continuity. The resolution is mediated by specific enzymes(s) termed ‘resolvase’ [19]. Since the process cannot proceed without extensive sequence homology, HR tends to be less prone to mutagenesis relative to NHEJ. Owing to the requirement for a homologous donor sequence, HR occurs only in the S/G2/M phases of the cell cycle while NHEJ occurs throughout the cell cycle. Both HR and NHEJ contribute to the repair of radiation therapy-induced DSBs, suggesting that these pathways may be functionally complementary [21–23]. Given the critical role of these two processes, it is not surprising that they are subject to complex regulation.
In mammalian cells, both processes are carried out by multistep processes facilitated by a large number of proteins. The mechanistic details of these processes remain an active area of investigation. Working models are described as follows.
For NHEJ, upon DDR activation (see below), the Ku70–Ku80 heterodimer is recruited to the break site. This protein complex forms a ring-shaped structure to protect the broken DNA ends [24]. In addition, the heterodimer serves as a platform for binding of the critical kinase, DNA-PKcs [25] and the XRCC4–Ligase IV-XLF4 complex to the site of damage [26]. DNA-PK performs two important functions:
Phosphorylates the Ligase IV complex to facilitate the joining of DNA ends;
In cases where DNA end processing is required before rejoining, DNA-PK binds to and recruits the Artemis endonuclease to perform this function.
While other proteins also participate in NHEJ [19], in vitro reconstitution of NHEJ with these seven proteins (Ku70, Ku80, DNA-PK, XRCC4–Ligase IV-XLF4 complex and Artemis) suggest that they are essential for this process.
By comparison, the genetics of HR is more complex and far less well understood. Upon DDR activation, it is thought that the BRCA1 and BRCA2 protein are recruited to the site of DNA damage. These two proteins were cloned by virtue of their inactivation in familial breast cancer cohorts (breast cancer genes 1 and 2) [27–29]. Both proteins encode large molecular weight proteins that mediate multiple cellular processes to suppress tumor formation. One of these critical functions involves HR. It is proposed that BRCA1 and 2 bind to aberrant DNA structures related to DSB ends [30,31]. Through BRCA2, the mammalian recombinase, RAD51, is recruited to the site of damage [32]. RAD51 coats the DNA and facilitates the strand exchange reaction in homologous recombination [33–35]. After strand invasion, resolution of the Holliday intermediate is mediated by a protein complex consisting of XRCC3 and RAD51C (two homologues of RAD51) [19,36].
The mechanism by which HR and NHEJ is activated in response to DNA damage is discussed below.
DNA damage response
The DDR refers to the signal transduction cascades that are triggered by DNA damage. These cascades coordinate DNA repair, cell cycle progression and cell death mechanisms to facilitate the faithful transmission of genetic material after DNA damage. The process initiates with the recognition of DNA damage by specialized ‘sensor’ proteins. In turn, these sensor proteins recruit and/or activate ‘transducer’ proteins required for subsequent signaling to ‘effector’ responses such as cell cycle arrest, apoptosis, transcription and DNA repair [37]. Defects in the DDR have been associated with genomic instability, sensitivity to genotoxic agents and cancer predisposition [38].
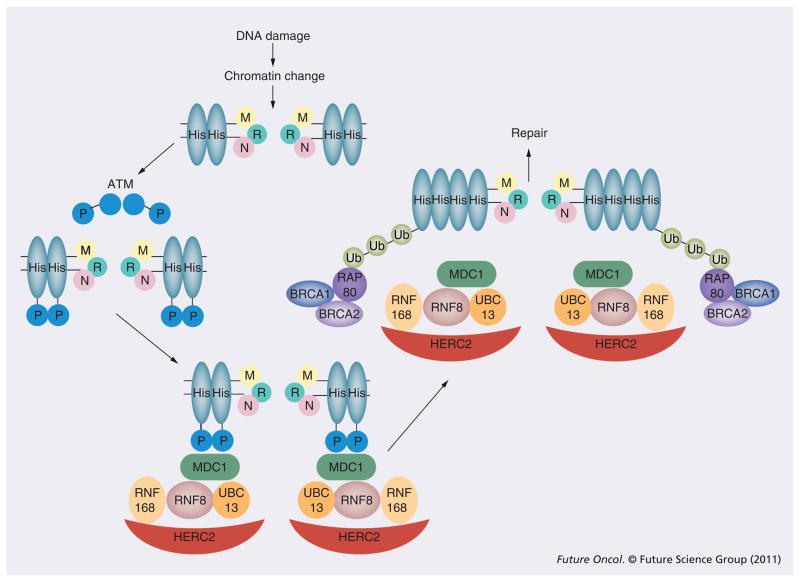
Upon DNA damage, the strand discontinuities trigger complex changes in DNA topology secondary to histone acetylation and phosphorylation of chromatin proteins [39]. The unveiled strand break is recognized by the Mre11–Rad50–Nbs1 (MRN) complex (Figure 2). In addition to serving as an exonuclease/endonuclease to process the DSBs into ssDNA tails [40], the MRN complex also recruits the ATM protein kinase to the site of the DSB [41,42]. When recruited to DSBs, ATM – normally existing in an inactive dimeric form – dissociates and autophosphorylates on multiple residues that are thought to be important for activation of ATM’s kinase activity [43]. The activated ATM phosphorylates the histone protein, H2AX, over a region of megabases surrounding a DSB [44,45]. In turn, the phosphorylated H2AX (also known as γ-H2AX) recruits the MDC1 protein [46,47]. The MDC1 protein serves as a scaffold protein for the docking of the E3 ubiquitin ligase complex, UBC13–RNF8 [48], which serves to polyubiquitinate H2AX. Completion of this polyubiquitination reaction requires a second ubiquitin ligase, RNF168 [49]. RNF168 is recruited to the site of DNA damage through its interaction with HERC2 and RNF8 [50]. The polyubiquitination reaction alters local chromatin structure as well as providing a docking site for the ubiquitin-binding protein, RAP80. RAP80 then recruits the BRCA1–BRCA2–RAD51 repair complex by direct physical interaction [51]. This complex initiates DSB repair by HR as well as arresting cell cycle progression in a process known as DNA damage checkpoint activation (see ensuing section).
Figure 2. Molecular mechanism of the DNA damage response.
DNA damage triggers chromatin remodeling and recruitment of the MRN complex. In turn, the MRN complex recruits and activates the ATM homodimer to phosphorylate His proteins. The phosphorylated His bind MDC1, which serve as a platform for proteins required for His Ub (UBC13, RNF8, RNF168 and HERC2). Finally, the ubiquitinated His recruits the proteins required for DNA repair (BRCA1 and BRCA2) through interaction with RAP80.
His: Histone; MRN complex: Mre11–Rad50–Nbs1; P: Phosphorylation; Ub: Ubiquitination.
It is important to note that, while the above damage response is described in a linear manner, parallel interactions occur at each step. For instance, in addition to recruiting UBC13–RNF8, MDC1 also interacts with ATM [46] and MRN [52] to stabilize the repair complex. The aggregate effect of these other complex interactions induce chromatin state changes surrounding the DSB and the localization of numerous proteins required for coordinating DNA repair and checkpoint regulation.
Similar to HR, the NHEJ process can be initiated by the MRN complex upon DDR activation. The Mre11 protein in the complex can directly interact with the Ku70 subunit [53]. Moreover, the RAD50 protein in the MRN complex encodes a high-affinity DNA binding domain and a second domain that facilitates homodimeric interactions that holds DNA ends in close proximity [54] to facilitate subsequent NHEJ.
Since the MRN complex may initiate either HR or NHEJ, a central question in the field of DNA repair involves the mechanism of this regulation. Inappropriate activation of HR in the G1 phase of the cell cycle could lead to cell death. Similarly, activation of NHEJ during the S/G2 phases of the cell cycle could increase the rate of mutagenesis. One of the key mediators of this regulatory process involves the protein CTIP. In a landmark study, CTIP was found to interact with the MRN complex to promote its exonuclease/endonuclease activity and process DSBs into ssDNA ends [55]. Importantly, this activity is regulated by cell cycle-dependent phosphorylation events mediated by cyclin-dependent kinases (CDKs). In the S/G2 phase of the cell cycle, CTIP is phosphorylated and the MRN complex processes DSBs into single-stranded tails required for the initiation of HR. In the G1 phase of the cell cycle, CTIP remains unphosphorylated and the MRN complex remains inactive as an exonuclease/endonuclease. Without this processing, HR cannot be initiated and NHEJ becomes the predominant repair process.
Effect of DNA damage on cell cycle progression
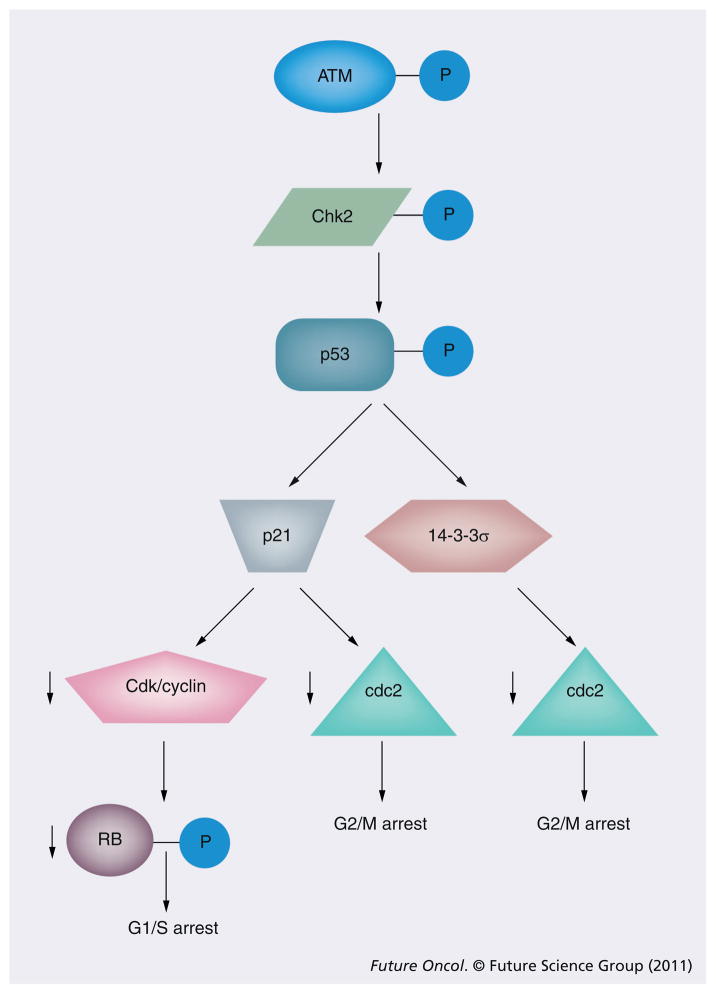
In addition to stimulating the assembly of repair complexes as previously described above, the DNA damage signal also interacts with proteins required for cell cycle progression, such as the CDK–cyclin complex. Generally, DNA damage checkpoint regulation occurs at three distinct phases of the cell cycle (Figure 3):
Figure 3. DNA damage-induced checkpoint activation.
In addition to modulating DNA repair, ATM activation also triggers stabilization of p53 and G1/S arrest (through transcriptional induction of p21) and G2/M arrest (through transcriptional induction of p21 and 14-3-3σ induction).
P: Phosphorylation.
The G1–S transition
The intra-S phase
The G2–M transition
Most of what we understand of this transduction process involves protein phosphorylation cascades, although the importance of other types of reversible modifications, such as ubiquitination and sumoylation, are becoming increasingly apparent [56]. Here, we will review an illustrative example of signal transduction between DNA damage sensors and cell cycle regulation. Upon recognition of DSB, the MRN complex recruits and activates the critical ATM kinase. In turn, the ATM kinase phosphorylates the tumor suppressor, p53, and another kinase, Chk2 [57]. ATM phosphorylation of Chk2 activates its kinase activity which, in turn, phosphorylates both p53 and MDM2. These phosphorylation events stabilize p53 by disrupting its association with MDM2, a protein that normally targets p53 for degradation [58]. Activated p53 then induces the transcription of its target genes, which include the critical regulator of the G1–S transition p21 [59]. The binding of p21 to the G1–S CDK–cyclin complexes prevents phosphorylation of the retinoblastoma protein (pRb). When in a hypophosphorylated state, pRb blocks cellular proliferation by sequestering E2F transcription factors and preventing their transcription of genes required for progression through the G1 and the S phase of the cell cycle, which control transcription of genes required for progression from G1 into S phase. Disruption of the pRb pathway – as occurs with mutant p53 or p21 – liberates E2Fs and allows cell proliferation. This renders cells insensitive to DNA damage-induced antigrowth signals that normally operate to inhibit passage through G1 phase of the cell cycle [60].
With regards to the G2–M checkpoint, ATM activation causes p53-induced transcription of p21 as well as 14-3-3σ. The 14-3-3σ protein sequesters the cyclinB–cdc2 kinase complex in the cytoplasm and prevents nuclear phosphorylation events required for G2/M progression [61]. In addition, p21 binds to any residual cdc2 that enters the nucleus to prevent its activation. These and other ATM-mediated events prevent progression through the G2/M transition and allow time for DSB repair [62].
Strategies for sensitization
A prediction of the model presented in the previous section is that inhibition of any of the proteins required for DDR or DSB repair should lead to radiation sensitization. In general, this prediction has been confirmed [63]. However, therapeutic agents that directly inhibit these critical proteins are still in pre-clinical development. Encouragingly, several US FDA-approved agents have recently been shown to modulate DDR and could be evaluated in combination with radiotherapy as radiation sensitizers.
Molecular rationale for therapeutic window
Before considering the strategy of radiation sensitization, one must first consider the molecular rationale for therapeutic window. After all, if normal and tumor cells were equally sensitized by the agent, then no therapeutic efficacy is gained.
A large body of data suggest that oncogene activation creates a tumor state with increased DNA damage accumulation [64–67]. These damages, if unrepaired, can be converted into DSBs that eventually lead to cell death. To compensate for this increased DNA damage, the tumor cells require increased utilization of DNA repair processes [67]. In this context, the administration of IR introduces additional DNA damage that can further tax the already overutilized repair process. This situation may increase the likelihood of an IR-induced DSB, causing cell death. The same effect can be achieved by inhibition of DSB repair. The following sections will review FDA-approved agents with such properties. It is important to note that these agents induce pleiotropic effects beyond DSB repair inhibition.
DNA-damaging agents
Conventional chemotherapy includes DNA-damaging agents that are often used in conjunction with radiation. In this context, these FDA-approved agents often sensitize the tumoricidal effects of radiation. The mechanism of this sensitization is thought to be related to the generation of DNA damage that sequesters critical DNA repair proteins. For instance, lesions generated by cisplatin bind to and sequester the Ku70–Ku80 hetero-dimer and thereby compromise the efficiency of NHEJ [68]. Furthermore, most DNA damage induced by conventional chemotherapy are ultimately converted to DSBs [69]. These DSBs will titrate away the repair proteins available for repair of DSBs induced by subsequent radiation. These types of mechanisms likely account for the increased glioblastoma patient survival observed in the context of concurrent radiation/temozolomide treatment relative to radiation treatment alone [4,5]. Cell culture experiments with established and primary human glioblastoma cell lines have demonstrated that temozolomide treatment resulted in an increase in IR-mediated DSBs [70,71]. The mechanism of sensitization remains unclear, although it is likely to be similar to those observed with other chemotherapeutic agents such as cisplatin.
Poly(ADP-ribose) polymerase
Poly(ADP-ribose) polymerase (PARP)1 is a nuclear protein that is actively involved in ssDNA break repair, particularly through the base excision repair processes [72]. Recently, several potent and specific chemical inhibitors of PARP activity have been developed [73]. These agents enhance replication-dependent DSB formation [74]. While the mechanism underlying this sensitization remains unclear, it is thought that PARP inhibition disrupts cellular repair of radiation-induced ssDNAs. Subsequent DNA replication at these sites results in the formation of DSBs [74]. Importantly, oncogenic activation in glioblastomas has been shown to cause increased reactive oxidative stress and ssDNA-break accumulation [67], rendering tumor cells more dependent on PARP1-mediated DNA repair than normal cells [67]. Accordingly, PARP1 inhibition effectively sensitizes glioblastoma cells to IR in both in vitro and in vivo models [74,75]. Based on these preclinical observations, two radiation-based PARP1 inhibitor trials were recently initiated. The first examines the safety of incorporating PARP1 inhibitor in the context of WBRT. The second explores the effect of combining PARP1 inhibition with WBRT and temozolomide [73]. These studies should provide the safety profiles of PARP1 inhibition in the context of brain radiation and the foundation for future Phase I/II efficacy trials.
Proteasome inhibitors
As a result of extreme aneuploidy, copy-number variation and transcriptional alteration that are present in many cancer cells, there is increased need for the chaperone pathways (such as heat shock proteins) to maintain folding of over-expressed proteins. When the capacity of these chaperone proteins becomes saturated, the unfolded proteins require degradation by the proteasome complex [76]. Thus, tumor cells exhibit increased dependency on proteasome function. Indeed, proteasome inhibition has been demonstrated to selectively ablate cancer cells both in vitro and in vivo [77]. The proteasome inhibitor, bortezomib, has attained FDA approval as a treatment for multiple myeloma and mantle cell lymphoma.
Recent studies implicate proteasome function in DSB repair. The yeast Sem1 protein is a subunit of the 19S proteasome that is required for efficient HR [78]. The human Sem1 homologue, DSS1, physically interacts with the HR protein, BRCA2 and is required for its stability and function [79–81]. Using the direct repeat–green fluorescent protein assay to directly assess HR efficiency, Murakawa et al. demonstrated that HR efficiency is significantly reduced by proteasome inhibition [82]. As a whole, these studies suggest proteasome inhibition as a means to target HR in cancer therapy.
The mechanism by which proteasome inhibition modulates HR remains an area of investigation. One hypothesis frequently put forward is that the proteins destined for proteasome degradation are typically modified by attachment of multiple ubiquitin moieties [78]. Processing of the tagged protein releases the tagged ubiquitin to replete the intracellular pool. Proteasome inhibition, thus, leads to accumulation of ubiquitinated proteins. In turn, this accumulation depletes the intracellular ubiquitin pool. Since free ubiquitins are required to activate HR, the repair process is compromised by proteasome inhibition.
There have been several clinical series laying the foundation for translation of proteasome inhibition as a radiosensitizing strategy. Kubicek et al. reported a Phase I trial combining bortezomib, temozolomide and radiation therapy for glioblastoma patients [83]. Of the 23 patients studied, no dose-limiting toxicity was reported when bortezomib was escalated to a dose of 1.3 mg/m when combined with temozolomide and radiation. Importantly, the therapeutic schedule of the radiation significantly altered the sensitizing effect of bortezomib in murine glioma models [84,85]; a factor that must be taken into consideration for future clinical trial design.
EGF receptor inhibitors
EGF receptor is frequently amplified or mutated in several cancer types, including non-small-cell lung cancer and glioblastomas [86–88]. As aberrant EGFR signaling is required to sustain tumor survival and proliferation in some cancers, targeted inhibition has led to selective tumor ablation [86]. Clinical trial success has led to FDA approval for treatment of non-small-cell lung cancer.
Several studies have demonstrated that EGFR inhibition sensitized tumor cells to radiation [63]. Insights into the mechanism underlying this sensitization have been provided by several recent studies. One series of studies demonstrate that a subset of EGFR travels to the nucleus where it binds to and enhances DNA-PK activity to enhance NHEJ [89,90]. Indeed, glioblastomas overexpressing an overactive form of EGFR (termed EGFRvIII) exhibit radiation resistance that can be abridged by treatment with DNA-PK inhibitors [91]. Another series of studies reveal that EGFR inhibition leads to retention of BRCA1 in the cytoplasm, thereby causing defective HR [92]. Finally, other downstream effectors of EGFR, including ERK1/2, also modulate HR efficiency [93]. It is likely that the radiation sensitization effect of EGFR inhibition represents a culmination of these individual effects.
Preliminary clinical experience of EGFR inhibition as a radiation-sensitizing strategy in glioblastoma patients yielded mixed results. In one study, Prados et al. treated 65 patients with newly diagnosed glioblastoma or gliosarcoma with erlotinib, and fractionated radiotherapy with concomitant and adjuvant temozolomide [94]. The treatment resulted in a marked improvement of median progression-free and overall survival (8.2 and 19.3 months, respectively) when compared with a combined historical control [94]. However, in another study, Brown et al. treated 89 patients with newly diagnosed glioblastoma with erlotinib in combination with temozolomide and radiotherapy [95]. Median overall survival was reported to be 15.7 months and comparison to the radiotherapy plus temozolomide arm from the European Organisation for Research and Treatment of Cancer 26981/22981 – National Cancer Institute of Canada trial revealed no significant difference. In a third study by Peereboom et al., 27 patients with newly diagnosed glioblastoma were treated witih 150 mg/day erlotinib and radiotherapy, and temozolomide [96]. This trial was terminated preterm because of unacceptable toxicity and lack of efficacy. Median progression-free and overall survival were 2.8 months for the erlotinib group and 8.6 months for the temozolomide/radiation group [96]. Clearly, resolution of these conflicting results will require thoughtful clinical trial design in the future. Importantly, such trials must factor in the ability of the inhibitor to penetrate the blood–brain barrier [97] as well as molecular determinants of sensitivity to EGFR inhibitors [98].
Other late-stage clinical trial agents
There are several other agents that are in mid-to-late clinical trial testing that have also been shown to inhibit DDR. For instance, histone deacetylase inhibitors have been shown to downregulate the transcript level of BRCA1 [99]. These agents also disrupt the chromatin reorganization required for ATM activation [39]. As another example, HSP90 (the prototypical chaperone protein) inhibitor treatment inhibits ATM autophosphorylation upon DNA damage [100] and destabilizes the MRN complex [101,102], thereby inhibiting HR. Finally, CDK1 inhibition causes the loss of a critical phosphorylation event on BRCA1 required for its HR function [103].
There are additional modulators of DDR and DNA repair not described here [63]. Indeed, the number of pharmacologic inhibitors that either directly or indirectly inhibit DSB repair is being uncovered at a rapid pace. Careful consideration should be given for combination with radiation therapy in future clinical trial design.
Conclusion
The infiltrative nature of glioblastoma has precluded surgical cure, necessitating multimodality therapy. Radiotherapy has proven to be the most effective postsurgical single-agent treatment modality in the management of glioblastoma. Radiation therapy alone more than doubled median survival in multiple randomized trials. In regards to radiotherapy dose, maximal benefit of radiotherapy is observed when a total dose of 60 Gy is given in 2-Gy fractions. Attempts at further treatment intensification with radiotherapy dose escalation have been disappointing, with no demonstrable benefit in patient survival or tumor control. This has led to an intense focus to find novel chemotherapy agents that can act as radiosensitizers in addition to their single-agent antitumor activity. Unfortunately, incremental gains with medical therapies have proven elusive, with most agents demonstrating moderate activity in vitro or with encouraging early clinical experience failing to materialize into a clinical benefit in larger trials. The major exception has been the incorporation of temozolomide with radiotherapy. Randomized clinical trials have demonstrated that the addition of temozolomide to radiotherapy has led to improved survival. However, the benefit is modest, with median survival improved to 14.6 months.
The critical lethal event of conventionally fractionated IR is thought to be DSBs. Insight into the mechanisms of DDR has resulted in the identification of ‘druggable’ target proteins. Modulation of the DDR has shown to affect tumor cell survival to IR. These targets are particularly attractive in the context that oncogenes induce increased DNA damage accumulation and cause tumors to become hyper-dependent on DDR. Interestingly, FDA-approved anticancer agents have been shown to act as radiosensitizers by altering the DDR. Some of these agents have begun to enter the clinic in combination with radiation therapy and temozolomide.
Future perspective
Given the central role of radiotherapy in glioblastoma therapy, efforts have been focused on finding better radiosensitizers. It is anticipated that novel radiosensitizers that enhance IR-mediated tumor cell killing will be incorporated into the standard treatment regimen for glioblastoma. Such radiosensitizers will target the DDR induced by IR. Encouragingly, several FDA-approved agents modulate critical proteins in DDR/repair, including conventional DNA damaging agents, proteasome inhibitors and EGFR inhibitors. Clinical trials involving such FDA-approved agents will be tested in combination with radiotherapy and temozolomide.
Given the heterogeneity of tumors between patients, it is likely that different portions of the DDR pathway are altered in individual patients. Personalized glioblastoma therapy will begin to enter the clinic. Pretreatment biopsy profiling of patients’ glioblastoma will reveal distinct DDR alterations by mutation, deletion or amplification. Elucidating how an individual patient’s tumor responds to IR-mediated DSBs will result in individualized targeting of the DDR pathway to achieve radiosensitization and improvement in patient outcomes in glioblastoma. Future clinical glioblastroma trials with these FDA-approved drugs and other agents modulating DDR should be designed with these considerations in mind.
Executive summary.
The current standard of care for glioblastoma is surgical resection followed by radiotherapy with temozolomide, which results in a median survival of 14.6 months.
Radiotherapy dose-escalation trials have demonstrated no benefit of radiotherapy below 4500 cGy and an increasing benefit on overall survival as total dose was increased from 5000 to 6000 cGy. Dose escalation beyond 6000 cGy has not resulted in any further improved survival.
Ionizing radiation kills tumor cells by inducing dsDNA breaks. Ionizing radiation (IR)-induced dsDNA breaks are repaired by two major mechanisms: nonhomologous end joining and homologous recombination. dsDNA break repair pathways mediate tumor cell resistance to radiotherapy in part.
DNA breaks activate the DNA damage response (DDR) pathway. This pathway consists of ‘sensors’ of DNA damage that activate ‘transducers’, which mediate ‘effector’ protein responses. The elucidation of the molecular events governing DDR pathway have resulted in new therapeutic targets to radiosensitize glioblastoma cells.
Radiosensitizers are agents that when delivered with IR improve the therapeutic efficacy of radiotherapy by selectively increasing tumor cell kill while sparing normal tissue.
Certain US FDA-approved anticancer drugs are now known to modulate the DDR pathway and act as radiosensitizers. Such agents include direct DNA-damaging agents, EGFR inhibitors and proteasome inhibitors. Rationale incorporation of these drugs with radiotherapy and temozolomide may further improve patient survival in glioblastoma.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
CC Chen is supported by the Doris Duke Charitable Foundation Clinical Scientist Development Award, the Sontag Foundation Distinguished Scientist Award, the Burroughs Wellcome Fund Career Awards for Medical Sciences, the Kimmel Scholar award, a Discovery Grant from the American Brain Tumor Association, and a National Cancer Institute K12 award. This work was supported in part by grants from NIH K08CA124804, ARRA 3P30CA023100-25S8 and a James S McDonnell Foundation award to S Kesari. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 2▪.Walker MD, Alexander E, Jr, Hunt WE, et al. Evaluation of BCNU and/or radiotherapy in the treatment of anaplastic gliomas. A cooperative clinical trial. J Neurosurg. 1978;49(3):333–343. doi: 10.3171/jns.1978.49.3.0333. Initial randomized trial demonstrating the benefit of radiotherapy over supportive care for gliomas. [DOI] [PubMed] [Google Scholar]
- 3.Keime-Guibert F, Chinot O, Taillandier L, et al. Radiotherapy for glioblastoma in the elderly. N Engl J Med. 2007;356(15):1527–1535. doi: 10.1056/NEJMoa065901. [DOI] [PubMed] [Google Scholar]
- 4▪▪.Stupp R, Mason WP, Van Den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. Initial randomized trial that established the benefit of adding temozolomide to radiotherapy for glioblastoma. [DOI] [PubMed] [Google Scholar]
- 5.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 6.Walker MD, Alexander E, Jr, Hunt WE, et al. Evaluation of mithramycin in the treatment of anaplastic gliomas. J Neurosurg. 1976;44(6):655–667. doi: 10.3171/jns.1976.44.6.0655. [DOI] [PubMed] [Google Scholar]
- 7.Walker MD, Green SB, Byar DP, et al. Randomized comparisons of radiotherapy and nitrosoureas for the treatment of malignant glioma after surgery. N Engl J Med. 1980;303(23):1323–1329. doi: 10.1056/NEJM198012043032303. [DOI] [PubMed] [Google Scholar]
- 8.Walker MD, Strike TA, Sheline GE. An analysis of dose–effect relationship in the radiotherapy of malignant gliomas. Int J Radiat Oncol Biol Phys. 1979;5(10):1725–1731. doi: 10.1016/0360-3016(79)90553-4. [DOI] [PubMed] [Google Scholar]
- 9.Bleehen NM, Stenning SP. A Medical Research Council trial of two radiotherapy doses in the treatment of grades 3 and 4 astrocytoma The Medical Research Council Brain Tumour Working Party. Br J Cancer. 1991;64(4):769–774. doi: 10.1038/bjc.1991.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang JE, Khuntia D, Robins HI, Mehta MP. Radiotherapy and radiosensitizers in the treatment of glioblastoma multiforme. Clin Adv Hematol Oncol. 2007;5(11):894–902. 907–815. [PubMed] [Google Scholar]
- 11.Hall EJ, Giaccia AJ. In: Radiobiology for the radiologist. Hall EJ, editor. Lippincott Williams and Wilkins Publishing; PA, USA: 2006. [Google Scholar]
- 12.Bennett CB, Lewis AL, Baldwin KK, Resnick MA. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc Natl Acad Sci USA. 1993;90(12):5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett CB, Westmoreland TJ, Snipe JR, Resnick MA. A double-strand break within a yeast artificial chromosome (YAC) containing human DNA can result in YAC loss, deletion or cell lethality. Mol Cell Biol. 1996;16(8):4414–4425. doi: 10.1128/mcb.16.8.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dugle DL, Gillespie CJ, Chapman JD. DNA strand breaks, repair, and survival in x-irradiated mammalian cells. Proc Natl Acad Sci USA. 1976;73(3):809–812. doi: 10.1073/pnas.73.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Game JC, Mortimer RK. A genetic study of x-ray sensitive mutants in yeast. Mutat Res. 1974;24:281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- 16.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94(3):399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 17.Sandell LL, Zakian VA. Loss of a yeast telomere Arrest, recovery, and chromosome loss. Cell. 1993;75(4):729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 18.Hartlerode AJ, Scully R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J. 2009;423(2):157–168. doi: 10.1042/BJ20090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyman C, Kanaar R. DNA double-strand break repair. All’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 20.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4(9):712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 21.Ristic D, Modesti M, Kanaar R, Wyman C. Rad52 and Ku bind to different DNA structures produced early in double-strand break repair. Nucleic Acids Res. 2003;31(18):5229–5237. doi: 10.1093/nar/gkg729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schar P, Herrmann G, Daly G, Lindahl T. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 1997;11(15):1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takata M, Sasaki MS, Sonoda E, et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17(18):5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412(6847):607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 25.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72(1):131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 26.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4–ligase IV complex to DNA ends. Mol Cell Biol. 2000;20(9):2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Futreal PA, Liu Q, Shattuck-Eidens D, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266(5182):120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 28.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene. BRCA1 Science. 1994;266(5182):66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 29.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene. BRCA2 Nature. 1995;378(6559):789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 30.Paull TT, Cortez D, Bowers B, Elledge SJ, Gellert M. Direct DNA binding by BRCA1. Proc Natl Acad Sci USA. 2001;98(11):6086–6091. doi: 10.1073/pnas.111125998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, Jeffrey PD, Miller J, et al. BRCA2 function in DNA binding and recombination from a BRCA2–DSS1–ssDNA structure. Science. 2002;297(5588):1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 32.Powell SN, Kachnic LA. Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene. 2003;22(37):5784–5791. doi: 10.1038/sj.onc.1206678. [DOI] [PubMed] [Google Scholar]
- 33.Galkin VE, Esashi F, Yu X, Yang S, West SC, Egelman EH. BRCA2 BRC motifs bind RAD51–DNA filaments. Proc Natl Acad Sci USA. 2005;102(24):8537–8542. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin DS, Chahwan C, Huffman JL, Tainer JA. Structure and function of the double-strand break repair machinery. DNA Repair (Amst) 2004;3(8–9):863–873. doi: 10.1016/j.dnarep.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 35.Shivji MK, Venkitaraman AR. DNA recombination, chromosomal stability and carcinogenesis: insights into the role of BRCA2. DNA Repair (Amst) 2004;3(8–9):835–843. doi: 10.1016/j.dnarep.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Masson JY, Shah R, O’Regan P, West SC. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303(5655):243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- 37.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28(5):739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 38▪.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. In-depth review of the DNA damage response pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol. 2005;6(10):757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 40.D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3(5):317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 41.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11–Rad50–Nbs1 complex. Science. 2005;308(5721):551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 42.Paull TT, Lee JH. The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle. 2005;4(6):737–740. doi: 10.4161/cc.4.6.1715. [DOI] [PubMed] [Google Scholar]
- 43.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 44.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10(15):886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 45.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146(5):905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lou Z, Minter-Dykhouse K, Franco S, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21(2):187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 47.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123(7):1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 48.Huen MS, Grant R, Manke I, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131(5):901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doil C, Mailand N, Bekker-Jensen S, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 50.Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol. 2010;12(1):80–86. doi: 10.1038/ncb2008. [DOI] [PubMed] [Google Scholar]
- 51.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316(5828):1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 52.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9(8):795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goedecke W, Eijpe M, Offenberg HH, Van Aalderen M, Heyting C. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet. 1999;23(2):194–198. doi: 10.1038/13821. [DOI] [PubMed] [Google Scholar]
- 54.Dinkelmann M, Spehalski E, Stoneham T, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol. 2009;16(8):808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sartori AA, Lukas C, Coates J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450(7169):509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15(17):2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 57.Peters M, Deluca C, Hirao A, et al. Chk2 regulates irradiation-induced, p53-mediated apoptosis in Drosophila. Proc Natl Acad Sci USA. 2002;99(17):11305–11310. doi: 10.1073/pnas.172382899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng Q, Chen J. Mechanism of p53 stabilization by ATM after DNA damage. Cell Cycle. 2010;9(3):472–478. doi: 10.4161/cc.9.3.10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55(22):5187–5190. [PubMed] [Google Scholar]
- 60.Shapiro GI, Harper JW. Anticancer drug targets: cell cycle and checkpoint control. J Clin Invest. 1999;104(12):1645–1653. doi: 10.1172/JCI9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bunz F, Dutriaux A, Lengauer C, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 62.Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14-3-3σ is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401(6753):616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 63.Ljungman M. Targeting the DNA damage response in cancer. Chem Rev. 2009;109(7):2929–2950. doi: 10.1021/cr900047g. [DOI] [PubMed] [Google Scholar]
- 64.Denko NC, Giaccia AJ, Stringer JR, Stambrook PJ. The human Ha-ras oncogene induces genomic instability in murine fibroblasts within one cell cycle. Proc Natl Acad Sci USA. 1994;91(11):5124–5128. doi: 10.1073/pnas.91.11.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee AC, Fenster BE, Ito H, et al. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J Biol Chem. 1999;274(12):7936–7940. doi: 10.1074/jbc.274.12.7936. [DOI] [PubMed] [Google Scholar]
- 66.Lo HW, Hsu SC, Ali-Seyed M, et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7(6):575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 67.Nitta M, Kozono D, Kennedy R, et al. Targeting EGFR induced oxidative stress by PARP1 inhibition in glioblastoma therapy. PLoS One. 2010;5(5):E10767. doi: 10.1371/journal.pone.0010767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turchi JJ, Henkels KM, Zhou Y. Cisplatin-DNA adducts inhibit translocation of the Ku subunits of DNA-PK. Nucleic Acids Res. 2000;28(23):4634–4641. doi: 10.1093/nar/28.23.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stojic L, Mojas N, Cejka P, et al. Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes Dev. 2004;18(11):1331–1344. doi: 10.1101/gad.294404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chakravarti A, Erkkinen MG, Nestler U, et al. Temozolomide-mediated radiation enhancement in glioblastoma: a report on underlying mechanisms. Clin Cancer Res. 2006;12(15):4738–4746. doi: 10.1158/1078-0432.CCR-06-0596. [DOI] [PubMed] [Google Scholar]
- 71.Kil WJ, Cerna D, Burgan WE, et al. In vitro and in vivo radiosensitization induced by the DNA methylating agent temozolomide. Clin Cancer Res. 2008;14(3):931–938. doi: 10.1158/1078-0432.CCR-07-1856. [DOI] [PubMed] [Google Scholar]
- 72.Chen CC, Kahle KT, Ng K, Nitta M, D’Andrea A. Of Escherichia coli and man: understanding glioma resistance to temozolomide therapy. In: Meir EG, editor. CNS Cancer. Humana; NY, USA: 2009. pp. 679–713. [Google Scholar]
- 73.Chalmers AJ, Lakshman M, Chan N, Bristow RG. Poly(ADP-ribose) polymerase inhibition as a model for synthetic lethality in developing radiation oncology targets. Semin Radiat Oncol. 2010;20(4):274–281. doi: 10.1016/j.semradonc.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 74.Dungey FA, Loser DA, Chalmers AJ. Replication-dependent radiosensitization of human glioma cells by inhibition of poly(ADP-Ribose) polymerase: mechanisms and therapeutic potential. Int J Radiat Oncol Biol Phys. 2008;72(4):1188–1197. doi: 10.1016/j.ijrobp.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 75.Russo AL, Kwon HC, Burgan WE, et al. In vitro and in vivo radiosensitization of glioblastoma cells by the poly (ADP-ribose) polymerase inhibitor E7016. Clin Cancer Res. 2009;15(2):607–612. doi: 10.1158/1078-0432.CCR-08-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136(5):823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rajkumar SV, Richardson PG, Hideshima T, Anderson KC. Proteasome inhibition as a novel therapeutic target in human cancer. J Clin Oncol. 2005;23(3):630–639. doi: 10.1200/JCO.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 78.Krogan NJ, Lam MH, Fillingham J, et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell. 2004;16(6):1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 79.Gudmundsdottir K, Lord CJ, Witt E, Tutt AN, Ashworth A. DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep. 2004;5(10):989–993. doi: 10.1038/sj.embor.7400255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li J, Zou C, Bai Y, Wazer DE, Band V, Gao Q. DSS1 is required for the stability of BRCA2. Oncogene. 2006;25(8):1186–1194. doi: 10.1038/sj.onc.1209153. [DOI] [PubMed] [Google Scholar]
- 81.Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol. 1999;19(7):4633–4642. doi: 10.1128/mcb.19.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Murakawa Y, Sonoda E, Barber LJ, et al. Inhibitors of the proteasome suppress homologous DNA recombination in mammalian cells. Cancer Res. 2007;67(18):8536–8543. doi: 10.1158/0008-5472.CAN-07-1166. [DOI] [PubMed] [Google Scholar]
- 83.Kubicek GJ, Werner-Wasik M, Machtay M, et al. Phase I trial using proteasome inhibitor bortezomib and concurrent temozolomide and radiotherapy for central nervous system malignancies. Int J Radiat Oncol Biol Phys. 2009;74(2):433–439. doi: 10.1016/j.ijrobp.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Labussiere M, Pinel S, Vandamme M, Plenat F, Chastagner P. Radiosensitizing properties of bortezomib depend on therapeutic schedule. Int J Radiat Oncol Biol Phys. 2011;79(3):892–900. doi: 10.1016/j.ijrobp.2010.09.051. [DOI] [PubMed] [Google Scholar]
- 85.Ng K, Nitta M, Hu L, et al. A small interference RNA screen revealed proteasome inhibition as strategy for glioblastoma therapy. Clin Neurosurg. 2009;56:107–118. [PubMed] [Google Scholar]
- 86.Da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- 87.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88▪▪.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. The Cancer Genome Atlas comprehensive genomic evaluation of samples from 206 glioblastoma patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dittmann K, Mayer C, Fehrenbacher B, et al. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280(35):31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 90.Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 2011;71(3):1103–1114. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mukherjee B, Mcellin B, Camacho CV, et al. EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009;69(10):4252–4259. doi: 10.1158/0008-5472.CAN-08-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li L, Wang H, Yang ES, Arteaga CL, Xia F. Erlotinib attenuates homologous recombinational repair of chromosomal breaks in human breast cancer cells. Cancer Res. 2008;68(22):9141–9146. doi: 10.1158/0008-5472.CAN-08-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Golding SE, Rosenberg E, Neill S, Dent P, Povirk LF, Valerie K. Extracellular signal-related kinase positively regulates ataxia telangiectasia mutated, homologous recombination repair, and the DNA damage response. Cancer Res. 2007;67(3):1046–1053. doi: 10.1158/0008-5472.CAN-06-2371. [DOI] [PubMed] [Google Scholar]
- 94.Prados MD, Chang SM, Butowski N, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27(4):579–584. doi: 10.1200/JCO.2008.18.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brown PD, Krishnan S, Sarkaria JN, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North Central Cancer Treatment Group Study N0177. J Clin Oncol. 2008;26(34):5603–5609. doi: 10.1200/JCO.2008.18.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peereboom DM, Shepard DR, Ahluwalia MS, et al. Phase II trial of erlotinib with temozolomide and radiation in patients with newly diagnosed glioblastoma multiforme. J Neurooncol. 2010;98(1):93–99. doi: 10.1007/s11060-009-0067-2. [DOI] [PubMed] [Google Scholar]
- 97.Clarke HA, Tarshis J, Lam-McCulloch J, Kay J. Saddle block analgesia for high-dose-rate brachytherapy: a prospective study. Brachytherapy. 2009;8(4):335–338. doi: 10.1016/j.brachy.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 98▪.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353(19):2012–2024. doi: 10.1056/NEJMoa051918. Study developing biomarkers to predict glioblastoma patient response to EGF receptor kinase inhibitors. [DOI] [PubMed] [Google Scholar]
- 99.Burkitt K, Ljungman M. Phenylbutyrate interferes with the Fanconi anemia and BRCA pathway and sensitizes head and neck cancer cells to cisplatin. Mol Cancer. 2008;7:24. doi: 10.1186/1476-4598-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koll TT, Feis SS, Wright MH, et al. HSP90 inhibitor, DMAG, synergizes with radiation of lung cancer cells by interfering with base excision and ATM-mediated DNA repair. Mol Cancer Ther. 2008;7(7):1985–1992. doi: 10.1158/1535-7163.MCT-07-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Camphausen K, Tofilon PJ. Inhibition of Hsp90: a multitarget approach to radiosensitization. Clin Cancer Res. 2007;13(15 Pt 1):4326–4330. doi: 10.1158/1078-0432.CCR-07-0632. [DOI] [PubMed] [Google Scholar]
- 102.Dote H, Burgan WE, Camphausen K, Tofilon PJ. Inhibition of HSP90 compromises the DNA damage response to radiation. Cancer Res. 2006;66(18):9211–9220. doi: 10.1158/0008-5472.CAN-06-2181. [DOI] [PubMed] [Google Scholar]
- 103.Johnson N, Cai D, Kennedy RD, et al. Cdk1 participates in BRCA1-dependent S phase checkpoint control in response to DNA damage. Mol Cell. 2009;35(3):327–339. doi: 10.1016/j.molcel.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]