Abstract
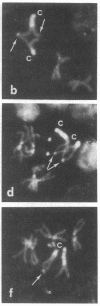
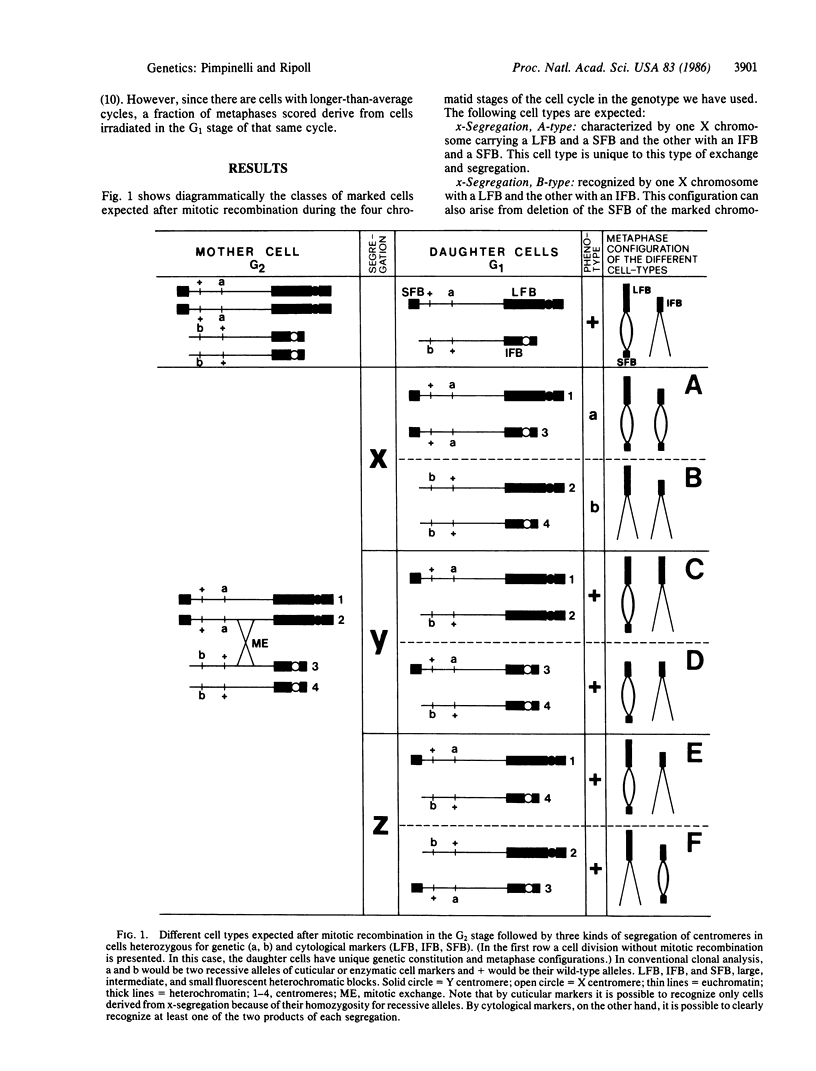
Mitotic recombination is widely used in Drosophila as a technique to study genetic and developmental problems. It has been generally assumed that, following mitotic exchange between homologous chromatids during the G2 stage, the centromeres attached to the chromatids involved in the exchange segregate randomly. As a result, two equally frequent types of segregation, yielding genetically different products, are produced. However, when epidermal or enzymatic cell-marker mutants are used, only one type of segregation gives rise to marked cells. In the present work we test this assumption of random segregation using cytological markers. With cytological markers, larval neuroblast cells resulting from mitotic recombination exhibit recognizably all possible products from mitotic recombination. We find that one type of segregation is favored, in that, after mitotic recombination, the centromeres attached to the chromatids involved in the mitotic exchange preferentially migrate to opposite poles during anaphase. This favored segregation could be the result of exchange between previously oriented chromatids or could be due to the effect of the exchange upon subsequent orientation of homologous chromosomes. In either case, frequencies of mitotic recombination have been overestimated in the past.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker B. S., Carpenter A. T., Ripoll P. The Utilization during Mitotic Cell Division of Loci Controlling Meiotic Recombination and Disjunction in DROSOPHILA MELANOGASTER. Genetics. 1978 Nov;90(3):531–578. doi: 10.1093/genetics/90.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., Pimpinelli S., Santini G. Characterization of Drosophila heterochromatin. I. Staining and decondensation with Hoechst 33258 and quinacrine. Chromosoma. 1976 Sep 24;57(4):351–375. doi: 10.1007/BF00332160. [DOI] [PubMed] [Google Scholar]
- Kennison J. A. The Genetic and Cytological Organization of the Y Chromosome of DROSOPHILA MELANOGASTER. Genetics. 1981 Jul;98(3):529–548. doi: 10.1093/genetics/98.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli S., Pignone D., Gatti M., Olivieri G. X-ray induction of chromatid interchanges is somatic cells of Drosophila melanogaster: variations through the cell cycle of the pattern of rejoining. Mutat Res. 1976 Apr;35(1):101–110. doi: 10.1016/0027-5107(76)90172-x. [DOI] [PubMed] [Google Scholar]
- Pimpinelli S., Santini G., Gatti M. Characterization of Drosophila heterochromatin. II. C- and N-banding. Chromosoma. 1976 Sep 24;57(4):377–386. doi: 10.1007/BF00332161. [DOI] [PubMed] [Google Scholar]
- Stern C. Somatic Crossing over and Segregation in Drosophila Melanogaster. Genetics. 1936 Nov;21(6):625–730. doi: 10.1093/genetics/21.6.625. [DOI] [PMC free article] [PubMed] [Google Scholar]