Summary
Background
MicroRNAs (miRNAs) are ~22 nt small RNAs that control development, physiology and pathology in animals and plants. Production of miRNAs involves the sequential processing of primary hairpin -containing RNA polymerase II transcripts by the RNase III enzymes Drosha in the nucleus and Dicer in the cytoplasm. miRNA duplexes then assemble into Argonaute proteins to form the RNA-induced silencing complex (RISC). In mature RISC, a single-stranded miRNA directs the Argonaute protein to bind partially complementary sequences, typically in the 32 untranslated regions of messenger RNAs, repressing their expression.
Results
Here, we show that after loading into Ago1 more than a quarter of all Drosophila miRNAs undergo 32 end trimming by the 32-to-5′ exoribonuclease Knabber (CG9247). Depletion of Knabber by RNAi reveals that miRNAs are frequently produced by Dicer-1 as intermediates that are longer than ~22 nucleotides. Trimming of miRNA 32 ends occurs after removal of the miRNA* strand from pre-RISC and may be the final step in RISC assembly, ultim ately enhancing target mRNA repression. In vivo, depletion of Knabber by RNAi causes developmental defects.
Conclusions
We provide a molecular explanation for the previously reported heterogeneity of miRNA 32 ends and propose a model in which Knabber converts miRNAs into isoforms that are compatible with the preferred length of Ago1-bound small RNAs.
Introduction
MicroRNAs regulate mRNA stability and translation in plants, green algae and animals [1, 2]. Originally discovered in Caenorhabditis elegans, these ~22 nucleotide (nt) small RNAs regulate development and physiology, and have been implicated in diseases such as cancer, diabetes, and viral infection [3–5]. Loss of proteins required for the production or function of miRNAs typically results in severe developmental defects or lethality.
miRNA genes are generally transcribed by RNA polymerase II to generate 52 capped and 32 polyadenylated primary miRNAs (pri-miRNAs) that are then sequentially processed into mature miRNA duplexes [6]. Pri-miRNAs contain one or more characteristic stem-loops that are recognized and cleaved by the nuclear RNase III enzyme Drosha [7] to generate ~70 nt long precursor miRNAs (pre-miRNAs) [8]. Pre-miRNAs comprise a single-stranded loop and a partially base-paired stem whose termini bear the hallmarks of RNase III processing: a two-nucleotide 3′ overhang, a 52 phosphate and a 32 hydroxyl group.
Nuclear pre-miRNAs are exported by Exportin 5 to the cytoplasm, where the RNase III enzyme Dicer liberates ~22 nt mature miRNA/ miRNA* duplexes from the pre-miRNA stem [9–12]. Like all Dicer products, miRNA duplexes contain two-nucleotide 32 overhangs, 52 phosphate and 32 hydroxyl groups. In flies, Dicer-1 cleaves pre-miRNAs to miRNAs, while Dicer-2 converts long dsRNA into small interfering RNAs (siRNAs), which direct RNA interference (RNAi), a distinct small RNA silencing pathway required for host defense against viral infection and somatic transposon mobilization, as well as gene silencing triggered by exogenous dsRNA [13, 14].
miRNA duplexes assemble into Argonaute proteins to form the precursor RNA-Induced Silencing Complex (pre-RISC), a process uncoupled from small RNA production [15]. In flies, miRNAs typically bind to Argonaute1 and siRNAs to Argonaute2 [16]. During RISC assembly one of the two strands of a miRNA duplex is selectively retained to form an active silencing complex. Strand selection is determined by the relative thermodynamic stability of the duplex ends, the identity of the 52 nucleotides, as well as the structure and length of the miRNA duplex [15]. In mature RISC, a single-stranded miRNA directs Ago1 to bind partially complementary sequences, typically within the 32 untranslated region (32 UTR) of mRNAs [1]. RISC-binding represses mRNA expression by accelerating its decay or inhibiting its translation [17].
Here, we report that more than one quarter of all miRNAs in Drosophila S2 cells are trimmed after their loading into Ago1, a process that can be recapitulated in cell extracts and that we can detect in vivo in flies. Trim ming of miRNAs is mediated by the Mg2+-dependent 32-to-52 exoribonuclease Knabber (CG9247), a member of the DEDD family of exonucleases. Knabber activity is required to trim Ago1-loaded miRNAs, and miRNA trimming enhances target RNA repression. Knabber is required for normal fly development. Our results show that the 32 ends of miRNAs are not simply defined by the RNase III enzymes Drosha and Dicer-1, but undergo exonucleolytic reshaping after their loading into Ago1. Thus, the previously described heterogeneity of miRNA 32 ends reflects mainly active trimming, rather than sloppy precursor processing.
Results
The 32 end of miR-34 is trimmed after its production by Dicer-1
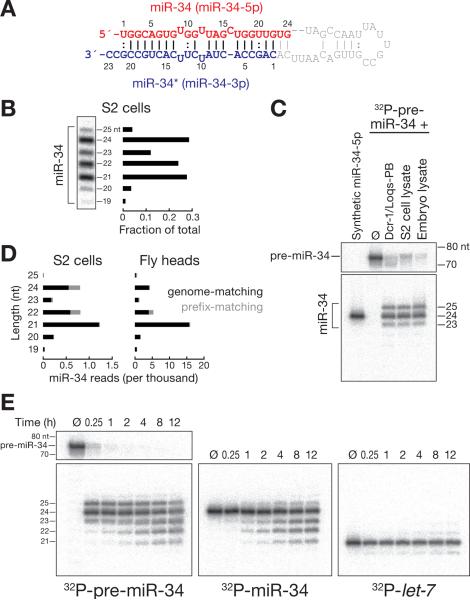
miRBase annotates miR-34 (miR-34-5p) as 24 nucleotide (nt) long, pairing to a 23 nt miR-34* strand (miR-34-3p) (Figure 1A), but high resolution Northern hybridization revealed additional, abundant 23, 22, and 21 nt miR-34 isoforms (Figures 1B). To test whether inaccurate processing of pre-miR-34 by Dicer-1 explains miR-34 heterogeneity, we incubated 52 32P-radiolabeled pre-miR-34 with purified, recombinant Dicer-1/ Loquacious PB, S2 cell lysate or 0–2 h Drosophila embryo lysate for 15 min (Figure 1C). In all three conditions, pre-miR-34 was rapidly converted to 24 nt (Dcr-1/ Loqs-PB: 61%; S2 cell lysate: 63%; Embryo lysate: 60 %), 25 nt (Dcr-1/ Loqs-PB: 25%; S2 cell lysate: 26%; Embryo lysate: 29%) and 23 nt (Dcr-1/ Loqs-PB: 13%; S2 cell lysate: 11%; Embryo lysate: 11%) products; we observed no isoforms shorter than 23 nt. Thus, the shorter isoforms of miR-34 are unlikely to reflect inaccurate processing of pre-miR-34 by Dicer-1.
Figure 1. miR-34 is trimmed after its production by Dicer-1.
(A) Structure of pre-miR-34. Red, miR-34 (24 nt); blue, miR-34* (23 nt). (B) miR-34 isoforms detected in total RNA from S2 cells by Northern hybridization. (C) 52 32P-radiolabeled pre-miR-34 was incubated with purified, recombinant Dicer-1/ Loquacious-PB heterodimer (Dcr-1/ Loqs-PB), S2 cell lysate or 0–2 hour embryo lysate. Products were resolved by denaturing polyacrylamide gel electrophoresis. (D) Abundance of miR-34 isoforms detected in fly heads and S2 cells by high throughput sequencing. Only reads with the annotated miR-34 52 ends were analyzed. Black, genome-matching; gray, prefix-matching reads. (E) 52 32P-radiolabeled pre-miR-34, 24 nt miR-34 or 21 nt let-7 RNA were incubated in 0–2 h embryo lysate. Products were resolved by denaturing polyacrylamide gel electrophoresis. See also Figure S1.
Sloppy Drosha cleavage of pri-miR-34 might also generate the shorter miR-34 isoforms. Such inaccurate Drosha processing would be expected to generate heterogeneous 52 ends for miRNAs, like miR-34, that derive from the 52 arm of their pre-miRNA. We analyzed small RNA high throughput sequencing data from fly heads for reads mapping to the miR-34 genomic locus. Of those reads mapping to the miR-34 locus, 98.5% began at the annotated 52 end of miR-34; for miR-34-mapping reads bound to Ago1, 99.0% shared this same, unique 52 end (Figures S1A and S1B). Similarly, 98.8% of all miR-34 reads in total small RNA datasets from S2 cells shared this 52 end (Figure S1C). We conclude that neither Dicer-1 nor Drosha contribute to the observed heterogeneity in miR-34 length.
Despite the uniformity of the 52 end of miR-34 in the high throughput sequencing datasets, miR-34 reads showed a length heterogeneity similar to that detected by Northern hybridization (Figures 1B), with 21, 22 and 24 nt isoforms accounting for most of the miR-34 reads in both S2 cells and fly heads (Figures 1D and S1). We concluded that the observed miR-34 length heterogeneity reflects 32 heterogeneity generated after dicing by 32 trimming. Supporting this idea, incubation of 52 32P-radiolabeled pre-miR-34 or a mature miR-34/ miR-34* duplex in 0–2 h embryo lysate produced 21 to 22 nt isoforms (Figure 1E). In contrast, let-7, a 21 nt miRNA that has been extensively used to study canonical miRNA biogenesis, function and turnover, was not shortened when incubated in embryo lysate.
miRNA trimming requires Ago1
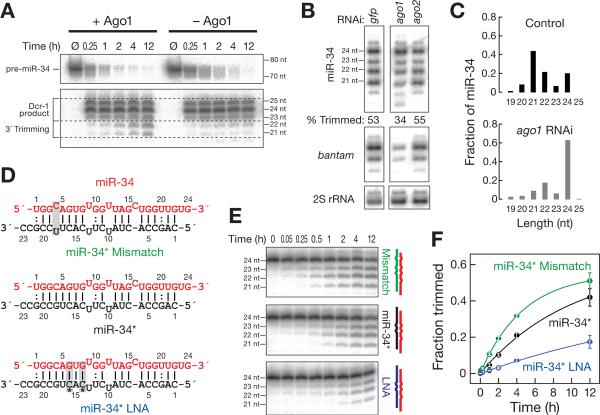
Trimming of miR-34 might occur immediately after its production by Dicer-1 when miR-34 is still bound to miRNA*, after loading of the miR-34/ miR-34* duplex into Ago1 to generate pre-RISC, or following the eviction of miR-34* from pre-RISC to create miR-34-guided Ago1-RISC. To distinguish among these possibilities, we monitored pre-miR-34 processing and miRNA trimming in 0–2 h embryo lysate immunodepleted of Ago1 (Figure 2A). Although pre-miR-34 was efficiently converted into miR-34 in the absence of Ago1, the resulting 23–25 nt Dcr-1 products were not trimmed. In contrast, the miR-34 cleaved from pre-miR-34 was trimmed in lysate containing Ago1 (Figure 2A). Similarly, the fraction of trimmed miR-34 decreased in S2 cells depleted of Ago1 by RNAi, compared to the control, when measured by both Northern hybridization (Figure 2B) and high throughput sequencing (Figure 2C). RNAi depletion of Ago2—the Argonaute protein that binds small interfering RNAs in the RNA interference pathway [18]—had no effect on the amount of trimmed miR-34. We conclude that trimming of miR-34 requires Ago1, presumably because miR-34 trimming occurs after loading into Ago1.
Figure 2. Trimming of miR-34 requires Ago1 and is limited by the removal of the miRNA* strand.
(A) 52 32P-radiolabeled pre-miR-34 was incubated in 0–2 h embryo lysate or lysate immunodepleted of Ago1. Products were resolved by denaturing polyacrylamide gel electrophoresis. (B) DsRNA-triggered RNAi targeting Ago1, but not Ago2, decreased trimming of miR-34, compared to treatment with a control dsRNA targeting GFP. “Trimmed” indicates the fraction of all miR-34 corresponding to 21 and 22 nt isoforms. The bantam miRNA and 2S rRNA served as controls. (C) The fraction of long miR-34 isoforms, measured by high throughput sequencing, increased when S2 cells were depleted of Ago1 by RNAi. Only isoforms with the annotated miR-34 52 end were analyzed. The abundance of miR-34 in the two libraries was 3499 ppm (control) and 4506 ppm (ago1 RNAi). (D, E) Synthetic duplexes of 52 32P-radiolabeled miR-34 (red) paired to variants of miR-34* (D) were incubated in 0–2 h embryo lysate (E), and the products analyzed by denaturing polyacrylamide gel electrophoresis. (F) Mean ± standard deviation for three independent replicates of the experiment in (E). See also Figure S2.
miRNA* strand dissociation limits the rate of miRNA trimming
A key step in the assembly of mature Ago1-RISC is the removal of the miRNA* strand from the Ago1-bound, miRNA/ miRNA* duplex, a process that converts pre-RISC to RISC. Mismatches between the miRNA seed sequence and the corresponding nucleotides in the miRNA* promote maturation of pre-Ago1-RISC [19–21]. We performed in vitro trimming assays using three miR-34/ miR-34* duplexes that differ in the strength of pairing of the miR-34 seed sequence to the seed match in miR-34* (Figure 2D). One duplex contained a mismatch within the miR-34 seed sequence. A second duplex, included two locked nucleic acid (LNA) ribose modifications within the seed match of miR-34*; LNA modifications increase the strength of base pairing by favoring the C32 endo ribose conformation found in RNA helices. None of the modifications within the miR-34* strand are predicted to alter the relative thermodynamic stability of the miR-34 versus miR-34* 52 ends, and therefore preserve the preference to load miR-34 rather than miR-34* into Ago1. The mismatch miR-34* more than doubled the rate of miR-34 trimming (kobs = 5.8 × 10−5 nM/ sec), compared to the canonical miR-34* (kobs = 2.7 × 10−5 nM/ sec). In contrast, the miR-34* containing LNA modifications more than halved the rate of trimming (kobs = 1.1 × 10−5 nM/ sec; Figures 2E and 2F).
Assembly of miRNA/ miRNA* duplexes into pre-RISC proceeds normally at 15°C, but low temperature slows the transition from pre-RISC to mature RISC [19]. For all of the three miR-34/ miR-34* duplexes, incubation at 15°C further reduced the fraction of miR-34 that was trimmed (Figure S2A). The rate of destruction of a miRNA* strand reflects the rate at which the miRNA and miRNA* strands dissociate. Mismatches between miR-34 and miR-34* accelerated the rate of destruction of miR-34*, whereas the addition of LNA modifications to miR-34* slowed the decay of miRNA*, compared to an unmodified miR-34* RNA (Figure S2B). Thus, miR-34* modifications that accelerate RISC assembly also accelerated trimming, whereas modifications that slow RISC assembly also slowed trimming. Our results suggest that miR-34 is first loaded into Ago1 as a 24 nt RNA and is only converted into shorter isoforms after miR-34* is removed from pre-RISC. The majority of 24 nt miR-34 likely corresponds to miR-34 bound to miR-34* in pre-RISC, since the 24 nt isoform, unlike the 21–23 nt isoforms, is not susceptible to target RNA-directed destruction, a process that requires extensive base pairing between the small RNA and its RNA target [22, 23].
The 32-to-52 exoribonuclease Knabber trims miR-34
To identify the exoribonuclease that trims miR-34, we performed a candidate RNAi screen in S2 cells using long double-stranded RNA targeting genes with sequence similarity to known or suspected exoribonucleases. Our screen included Drosophila homologs of exonucleases previously implicated in small RNA silencing pathways, such as the small RNA degrading nucleases (SDN) of plants [24], Enhancer of RNAi-1 (Eri-1) [25] and Mut-7 in C. elegans [26], as well as components of the general cellular RNA decay machinery such as RRP4, a core component of the exosome, the SKI-2 ortholog Twister, and the general 52-to-32 exonuclease Pacman (XRN1) [27]. RRP4, Twister and Pacman were previously proposed to degrade the mRNA products generated by RNAi [28] and Xrn-1 was implicated in miRNA turnover[29, 30]. The miRNA bantam, which does not undergo detectable trimming served as a control for general destabilization of miRNAs.
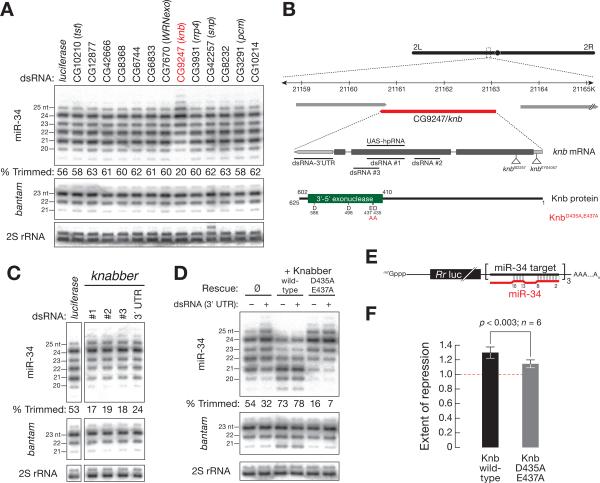
Among the exonucleases we tested, only depletion of CG9247 decreased the fraction of trimmed miR-34 (fraction trimmed = 20%), compared to control RNAi (fraction of miR-34 trimmed = 56%; Figure 3A). We observed a similar loss of miR-34 trimming for two additional, non-overlapping dsRNAs targeting different regions within the second exon and the 32 untranslated region of CG9247 (Figures 3B and 3C). In all cases trimming of miR-34 was reduced by more than half. To reflect its role in 32 shortening of miRNAs, we named CG9247, knabber (knb), German for “nibbler.”
Figure 3. The 32-to-52 exoribonuclease Knabber (CG9247) trims miR-34, enhancing miR-34 function in S2 cells.
(A) S2 cells were transfected with dsRNA against a panel of predicted exonucleases and the effect on miR-34 length analyzed by high resolution Northern hybridization. bantam and 2S rRNA served as controls. The fraction of miR-34 trimmed to 21–22 nt is indicated below each lane. (B) The predicted structure of the knabber (CG9247) gene, mRNA, and protein. (C) S2 cells were transfected with three dsRNAs targeting the second exon or the 32 UTR of knabber as indicated in (B). All four dsRNAs decreased miR-34 trimming, relative to a control dsRNA targeting firefly luciferase. bantam and 2S rRNA served as controls. (D) S2 cells stably expressing wild -type or D435A,E437A mutant Knabber were transfected with dsRNA targeting the 32 UTR of endogenous knabber, and the effect on miR-34 trimming measured. bantam and 2S rRNA served as controls. (E) Reporter construct used in (F). The three miR-34 binding sites pair with miR-34 nucleotides 2–8 and 13–15, mimicking typical animal miRNA binding sites [54]. Rr luc, Renilla reniformis luciferase. (F) Knabber trimming of miR-34 enhances miRNA function. Repression by miR-34 in S2 cells expressing wild-type or D435A,E437A mutant Knabber was measured by blocking miR-34 using a 22-O-methyl-modified anti-miRNA oligonucleotide and measuring the increase in Rr luciferase expression compared to a control oligonucleotide targeting let-7, a miRNA not normally expressed in S2 cells. See also Figure S3.
(We note that RNAi depletion of snipper (snp; CG42257) decreased full length 2S rRNA and caused the accumulation of higher molecular weight isoforms of 2S rRNA, suggesting that Snipper plays a previously unknown role in the maturation of 2S rRNA, which is generated by the processing of 5.8S rRNA in flies.)
Knabber homologs include Mut-7 in C. elegans and EXD3 in humans (Figure S3A). mut-7, which was one of the very first genes discovered to act in the RNAi pathway, is required for transposon silencing, RNAi, and cosuppression in worms [26, 31–33], but no role for mut-7 in miRNA biogenesis has been reported. Like Mut-7 and EXD3, Knabber belongs to the DEDD family of exoribonucleases, which are part of a larger superfamily that includes DNA exonucleases as well as the proof-reading domains of many DNA polymerases [34]. DEDD exonucleases contain three characteristic sequence motifs (Figures S3A), which include four invariant acidic amino acids (DEDD) (Figure 3B) [34, 35]. The structure of DNA polymerase suggests that these four amino acids organize two divalent metal ions at the catalytic center [36]. Consistent with the view that Knabber is a metal-dependent DEDD exoribonuclease, miR-34 trimming in fly lysate was inhibited by EDTA; adding additional Mg2+ rescued the inhibition (Figure S3B) [37, 38].
We changed two of the four invariant amino acids of the Knabber DEDD motif to alanine (D435A and E437A; Figure 3B), mutations predicted to block exonuclease activity, then reintroduced wild -type or mutant knabber open reading frame into S2 cells depleted of endogenous knabber using dsRNA targeting its 32 UTR (dsRNA 32 UTR; Figure 3B). knabber cDNA expression was driven by the constitutive Actin5C promoter (Figure S3C). In these experiments, depletion of endogenous knabber in control S2 cells decreased the fraction of trimmed miR-34 from 54% to 32% (Figure 3D); the presence of a stable, wild -type Knabber transgene enhanced miR-34 trimming (73% trimmed), even after depletion of endogenous knabber (78% trimmed miR-34; Figure 3D). Enhanced miR-34 trimming likely reflects the greater abundance of Knabber protein in the stable transgenic cell line, since knabber mRNA levels were ~100-times higher than in control S2 cells (data not shown). In contrast, expression of the D435A, E437A mutant Knabber protein reduced miR-34 trimming. The fraction of trimmed miR-34 decreased to 16% when transgenic, D435A, E437A mutant Knabber was expressed along with endogenous Knabber. The fraction of trimmed miR-34 decreased to 7% when D435A, E437A mutant Knabber was expressed and endogenous Knabber was depleted by RNAi.
In cultured Drosophila S2 cells, trimming of miR-34 by Knabber enhanced its target mRNA silencing activity. We compared the repression of a miR-34-regulated Renilla reniformis luciferase reporter in S2 cells stably expressing wild -type Knabber to cell expressing D435A,E437A mutant Knabber. S2 cells expressing transgenic wild -type Knabber produced mostly the 21 nt miR-34 isoform, whereas S2 cells stably expressing mutant Knabber produce predominantly the 24 nt miR-34 isoform (Figure 3D). For each cell line, we compared the level of reporter expression when the cells were transfected with a control anti-miRNA 22-O-methyl oligonucleotide to the reporter expression when the cells were transfected with an anti-miR-34 22-O-methyl oligonucleotide. The ratio of anti-miR-34 to control indicated the extent of repression. We observed significantly (p-value = 0.003, n = 6) greater repression of the miR-34 reporter in the cells expressing wild -type Knabber, compared to those expressing the mutant protein, indicating that trimming of miR-34 to shorter isoforms enhances its activity. We conclude that trimming of long miRNAs by the Mg2+-dependent, 32-to-52 exoribonuclease Knabber enhances miRNA function.
Knabber trims many miRNAs
To assess the role of Knabber in the production of other miRNAs, we sequenced 18-30 nt small RNAs from S2 cells treated with knabber dsRNA and from S2 cells treated with a control dsRNA. S2 cells produce 36 distinct miRNAs that were detected at >200 parts per million (ppm) in our high throughput sequencing. Among the isoforms of these 36 miRNA, we detected a small but statistically significant increase in the overall mean length of miRNAs when Knabber was depleted: 21.96 nt in the control versus 22.11 nt in knabber(RNAi) (p-value = 3.9 × 10−5, Wilcoxon signed rank test). If all of miR-34 were 22 nt long in the control and became 24 nt in the knabber dsRNA-treated cells, the mean miRNA length would be expected to increase by 0.056. Thus, a 0.15 increase in mean length suggests that miR-34 is not the only miRNA trimmed by Knabber in S2 cells.
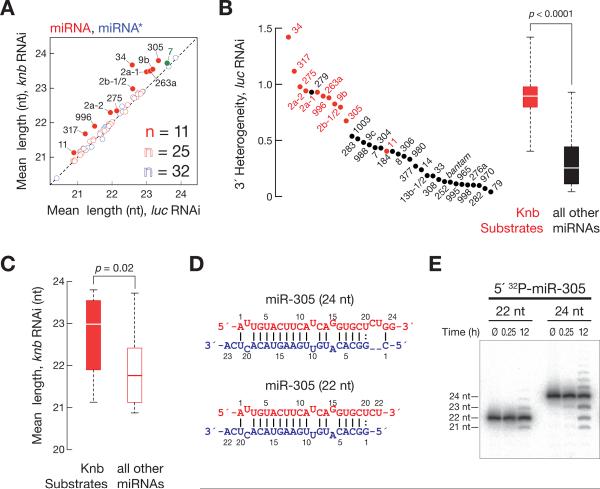
In fact, of the 36 abundantly expressed S2 cell miRNAs, 28 increased in mean length. Of these, 13 increased by more than 0.1 nt, and 9 by more than 0.33 nt. We used a chi-square test to assess the significance of the change in the distributions of isoform lengths in the knabber (RNAi) S2 cells for each miRNA (Supplemental Table 1). An increase of ~0.2 nt in mean length was the smallest change we could corroborate by Northern hybridization, an admittedly less sensitive method than high throughput sequencing. Using the 0.2 nt mean length increase as a conservative threshold, 11 S2 cell miRNAs correspond to Knabber substrates (red filled circles, Figures 4A and S4A). Thus, ≥30% of S2 cell miRNAs are trimmed by Knabber after their production by Dicer-1.
Figure 4. Knabber trims a quarter of all miRNAs in S2 cells.
(A) Analysis of mean miRNA and miRNA* length in S2 cells transfected with dsRNA targeting knabber or a control dsRNA targeting firefly luciferase. miRNA, red; miRNA*, blue; filled circles indicate miRNAs with a significant increase in mean length. In S2 cells, miR-7 (green) does not match our conservative criteria for Knabber substrates, but in flies, miR-7 is trimmed by Knabber (Figure S5C). (B) Knabber trimming explains miRNA 32 heterogeneity. 32 heterogeneity was determined for all S2 cell miRNAs that were more abundant than 200 ppm in high throughput sequencing data. Red, the 11 Knabber substrates identified in this study. Boxplots illustrate 32 heterogeneity of Knabber substrate miRNAs (red) versus all other miRNAs (black). P-value was determined using the Mann-Whitney U test. (C) The mean length of Knabber substrate miRNAs is longer than non-Knabber substrate miRNAs in S2 cells treated with knabber dsRNA. P-value was determined using the Mann-Whitney U test. (D, E) Synthetic miRNA/ miRNA* duplexes comprising a 24 or 22 nt 52 32P-radiolabeled miR-305 RNA and the corresponding miRNA* strand (D) were incubated in embryo lysate, and the products analyzed by denaturing polyacrylamide gel electrophoresis (E). See also Figure S4 and Tables S1 and 2.
Knabber substrates included both miRNAs derived from the 52 arm of their pre-miRNA (4 miRNAs) and miRNAs derived from the 32 arm of their premiRNA (7 miRNAs). miRNAs trimmed by Knabber account for most of the previously identified 32 heterogeneity of S2 cell miRNAs, because Knabber-substrates exhibit significantly higher 32 heterogeneity than non-substrate miRNAs (p<0.0001, Mann-Whitney U-test, Figure 4B). In contrast, 52 heterogeneity, which is generally low because of the purification process associated with Argonaute loading [39], was unaffected by the depletion of Knabber by RNAi (Figure S4B).
The 11 Knabber substrate miRNAs were significantly longer in S2 cells treated with knabber dsRNA than non-Knabber substrate miRNAs: the median of the mean lengths was 23.0 nt for Knabber substrates versus 21.8 nt for all others (p-value = 0.02, Mann-Whitney U test, Figure 4C). In contrast to Knabber substrate miRNAs, the length of endogenous siRNAs did not change after depletion of Knabber by RNAi, suggesting that Ago2-bound small RNAs are not Knabber substrates (Figure S4C). We also analyzed the effect of Knabber depletion on the length of the 32 miRNA* strands for which we detected >10 ppm by high throughput sequencing. The overall miRNA* mean length changed from 22.00 to 22.02 nt (p-value = 0.04, Wilcoxon signed rank test), but only two miRNA* strands showed a significant increase in the knabber dsRNA-treated S2 cells when analyzed using the chi-square test; neither of the two miRNA* strands increased more than 0.1 nt (Supplemental Table 1). Consistent with the proposal that miRNA trimming occurs after miRNA* strands depart from pre-RISC, those miRNA* strands whose miRNAs were Knabber substrates did not change significantly in length when compared to all other miRNA*s (Figure S4D).
What destines miRNAs for trimming by Knabber? Perhaps many Knabber substrate miRNAs are initially prod uced by Dicer-1 as long isoforms that are trimmed to a more typical miRNA length. To test this idea, we incubated synthetic miR-305/ miR-305* duplexes (Figure 4C) in Drosophila embryo lysate and monitored their trimming. In vivo in flies, miR-305 is efficiently trimmed (Fig. S5A, adult males). Moreover, miR-305 is abundantly expressed and efficiently trimmed in 0–2h embryos: among the 3,668 ppm miR-305 reads detected in the total small RNAs of 0–2h embryos, 23 nt (5%) and 24 nt (14%) miR-305 isoforms represent just 19% of all miR-305 reads, whereas the shorter, trimmed 21 nt (45%) and 22 nt (32%) isoforms represent 77% of all miR-305 reads. When a 24 nt 52 32P-radiolabeled miR-305, paired to a 23 nt miR-305* strand, was incubated overnight in embryo lysate, 17% was trimmed to shorter isoforms: 10% accumulated as 23 nt, 5% as 22 nt, and 2% as 21 nt (Figure 4D). In contrast, only 2% of a duplex comprising the 22 nt isoform of miR-305 paired to a 22 nt miR-305* strand was converted to a 21 nt form; no species shorter than 21 nt were detectable. We conclude that miRNA trimming is triggered, at least in part, by the length of the miRNA, with ~24 nt miRNAs being converted by Knabber into the 21–22 nt length, which is more typical for miRNAs at steady-state.
Knabber trims miRNAs in vivo
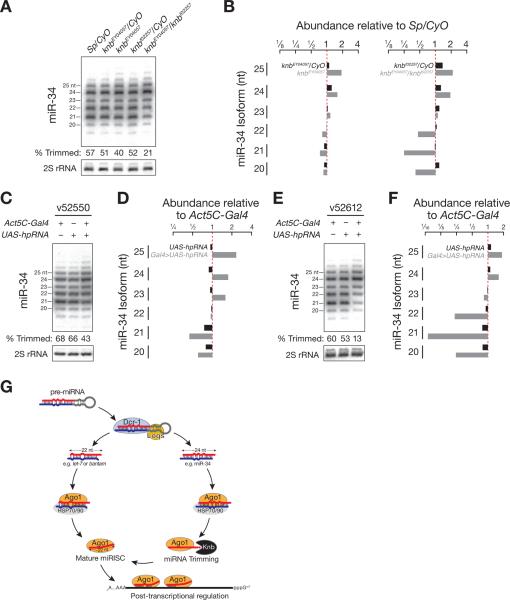
To test the role of Knabber in vivo, we obtained two publicly available Drosophila strains bearing a transposon insertion in knabber: knabberEY04057, corresponding to a p-element insertion in the 52 UTR of knabber and knabberf02257, corresponding to a piggyBac insertion in the first exon of knabber (Figure 3B). knabberEY04057 was homozygous viable and showed no change in knabber mRNA abundance compared to Oregon R or w1118 control flies (Figure S5B). However, our preliminary data suggests that the knabber mRNAs in knabberEY04057 originate within the p-element (data not shown) and may therefore not produce wild -type levels of Knabber protein. knabberf02257 was homozygous lethal, and knabberf02257 heterozygotes produced 38 ± 24% of the amount of knabber mRNA present in w1118 control flies (p = 0.008, Figure S5B). The fraction of miR-34 that was trimmed was reduced, albeit slightly, in both mutants: 57% of miR-34 was trimmed in Sp/ CyO control flies, whereas 51% was trimmed in knabberEY04057/ CyO and 52% was trimmed in knabberf02257/ CyO (Figure 5A). Trimmed miR-34 accounted for only 40% of all miR-34 isoforms in knabberEY04057 homozygotes, and in knabberEY04057/ knabberf02257 trans-heterozygotes just 21% of miR-34 was trimmed (Figure 5A). We conclude that trimming of miR-34 requires Knabber in vivo. Similarly, the two knabber mutations recapitulated the effect on eleven miRNAs identified as Knabber substrates in S2 cells (Figure S5C).
Figure 5. miRNA trimming requires Knabber in vivo.
(A, C, and E) High resolution Northern hybridization of miR-34 from 3–5 day-old male flies. 2S rRNA served as a loading control. (B, D and F) Abundance of miR-34 isoforms in flies carrying a knabber mutant allele (B) or in which Knabber was depleted by RNAi (D and F). miR-34 isoform abundance was measured relative to the indicated controls. (G) A model for miRNA trimming by Knabber. See also Figure S5.
knabberf02257 likely corresponds to a strong allele, but this m utation is not homozygous viable. To test whether loss of Knabber affects fly development, we used RNAi to deplete Knabber in vivo [40]. When driven by an Actin5C-Gal4 driver, a UAS-hpRNA transgene on the second chromosome (UAS-hpRNAv52550) reduced the fraction of miR-34 that was trimmed to 43% of all miR-34 isoforms, compared to 68% in flies expressing the Act5C-Gal4 driver alone or to 66% in the flies carrying only the UAS-hpRNA transgene. An insertion of the same hpRNA construct on the third chromosome (hpRNAv52612), reduced the fraction of miR-34 that was trimmed to 13% of all miR-34 isoforms, compared to 60% in flies carrying only the Actin5C-Gal4 driver or 53% in flies bearing only the UAS-hpRNA transgene (Figure 5B, right). Notably, 29% (69 of 239) of the flies expressing UAS-hpRNAv52612, the RNAi transgene with the stronger effect on miR-34 trimming, failed to eclose from their puparia. Only 5% (16 of 327) of the Actin5C-Gal4/ CyO; Dr/ TM3,Sb control flies and only 2% (5 of 298) of the +; UAS-hpRNAiv52612 control flies died as pupae. Although miRNAs regulate fly development and Knabber acts on miRNAs, we currently cannot exclude the possibility that this pupal lethality reflects a requirement for Knabber in processing substrates other than miRNAs.
Discussion
miRNA 32 heterogeneity [39, 41, 42] has been attributed to inaccurate processing by Dicer or Drosha. Our data suggest that much of the 32 diversity of miRNAs reflects their trimming by a novel processing step catalyzed by the 32-to-52 exoribonuclease Knabber. Figure 5G presents a revised model for the production of mature miRNAs from pre-miRNAs in flies. First, Dicer-1 converts premiRNAs to miRNA/ miRNA* duplexes. These are then sorted between Ago1 and Ago2 to generate Ago1- and Ago-2 pre-RISC complexes, with Ago1 selecting ≥22 nt miRNAs that begin with an unpaired U or A and containing an unpaired region centered on position 9 [19–21, 23, 43–47]. The Ago1 sorting process helps restrict the diversity of 52 ends of miRNAs. Next, the miRNA* strand dissociates from pre-RISC to produce RISC. We imagine that the 32 ends of “long” miRNAs bound to Ago1 are available for trimming by Knabber because they spend less time bound to the Ago1 PAZ domain than do 22 nt miRNAs. Once Knabber has shortened a long miRNA to 22 nt, its 32 end can bind the PAZ domain, protecting it from further trimming. For miR-34, we observed that trimming enhanced miRNA activity.
Depletion of Knabber in S2 cells (Figures 3A and 3C) and in flies (Figures 5A, C, E and S5) resulted in the appearance of higher molecular weight species, reminiscent of tailed small RNAs rather than bona-fide Dicer-products. Such non-templated addition of nucleotides to the 32 ends of mature miRNAs has been implicated in miRNA turnover in plants [48] and animals [22, 23, 49] and may indicate that Knabber-substrate miRNAs are marked for decay when not properly trimmed. In contrast to Ago1-bound miRNAs, Ago2-bound small RNAs would be protected from Knabber, perhaps because of their shorter mean length of ~21 nt (Figure S4C) or because they are 22-O-methyl modified by Hen1 [50–52].
This model does not invoke specific recruitment of Knabber to Ago1-RISC and is consistent with our preliminary experiments, in which we were unable to detect epitope-tagged, over-expressed Knabber bound to immunoprecipitated Ago1 (data not shown). However, such a simple model cannot explain why some trimmed miRNAs do not accumulate isoforms longer than 22 nt even after Knabber was depleted by RNAi (e.g., miR-11; Figure S4A), suggesting that miRNA length alone does not define a Knabber substrate. Perhaps additional proteins help recruit Knabber to Ago1-RISC for some miRNAs. A requirement for Knabber co-factors could also explain why Knabber trims miR-7 in flies but not in S2 cells (Figures 4 and S5A).
Is miRNA-trimming conserved in other organisms? Small RNAs in the human cervical carcinoma cell line HeLa exhibit an overall miRNA 32 heterogeneity similar to that observed for fly miRNAs (Figures S5E and S5F). Several human miRNAs with high 32 heterogeneity show a length distribution in HeLa cells reminiscent of Knabber-substrates in flies (Figure S5G). Perhaps a human homolog of fly Knabber processes these miRNAs. The C. elegans homolog of Knabber, Mut-7, is required for the accumulation of the 22G RNAs that direct worm Piwi proteins to represses transposon expression [26, 53]. We do not yet know if Knabber functions in the analogous piRNA pathway in flies or if Mut-7 has a yet undiscovered role in miRNA maturation in worms.
Experimental Procedures
Pre-miRNA Processing and Trimming Assays
Pre-miR-34 was transcribed with T7 RNA polymerase using a double-stranded DNA oligonucleotide template (see Supplemental Experimental Procedures), dephosphorylated with Calf Intestinal Phosphatase (New England Biolabs, Ipswich, MA, USA), and 52 32P-radiolabeled with T4 Polynucleotide Kinase (New England Biolabs). Pre-miR-34 (2 nM) was incubated with recombinant Dicer-1/ Loquacious PB (5 nM) or S2 cell or 0–2 h embryo lysate for 15 min at 25°C in a typical RNAi reaction [55]. Ago1 immunodepletion was as described [20].
For miRNA trimming, 52 32P-radiolabeled RNAs (2 nM) were incubated with 0-2 h embryo lysate as described [55], except that RNase inhibitor was omitted. Products were resolved by electrophoresis through a 15% denaturing polyacrylamide sequencing gel. Gels were dried, exposed to storage phosphor screens (Fuji, Tokyo, Japan) and quantified using ImageGauge 4.22 (Science Lab 2003, Fuji).
To analyze miRNA trimming for synthetic 52 32P-radiolabeled 24 nt miR-34, all isoforms shorter than 24 nt were considered to be trimmed. When pre-miR-34 was used as a substrate, we considered only isoforms shorter than 23 nt to be trimmed, because Dicer-1 produces 23, 24, and 25 nt miR-34 isoforms from premiR-34, so only isoforms shorter than 23 nt could be unambiguously considered to be trimmed. Similarly, we only considered isoforms shorter than 23 nt to be trimmed for Northern hybridization experiments. The fraction of miR-34 trimmed was defined as the sum of trimmed isoforms divided by the sum of all isoforms.
RNAi in S2 cells
Regions targeted by double-stranded RNA were from [Ref. 40]. DNA templates for in vitro transcription were amplified from genomic DNA or cDNA from Oregon R flies by PCR using primers incorporating the T7 promoter sequence (see Supplemental Experimental Procedures). After isopropanol precipitation, PCR products were used as templates for transcription by T7 RNA polymerase. DsRNA products were purified using MEGA clear RNA purification kit (Ambion, Austin, TX, USA). S2 cells were transfected on day 1 and day 4 with 20 μg dsRNA using Dharmafect4 (Dharmacon, Lafayette, CO, USA), and then total RNA was extracted on day 7 using the mirVana kit (Ambion). In Figure 3D, only one round of dsRNA transfection was performed.
Reporter assay
S2 cells stably expressing wild-type or mutant Knabber were seeded in 24-well plates at 1.0×106 cells/ ml and transfected immediately after seeding using DharmaFECT Duo (Dharmacon) and 500 ng per well psiCHECK-2 bearing three sites partially complementary to miR-34 in the 3' UTR of Rr luciferase, together with 20 nM 2-O-methyl-modified oligonucleotide complementary to miR-34 or let-7. Rr and Photinus pyralis luciferase activities were measured 72 h later. Six biological replicates were used to compare the repression conferred by miR-34 for the two cell lines; error was propagated by standard methods. P-values were determined using Student's t-test.
Bioinformatics Analyses and Statistics
Insert extraction, mapping and filtering was as described [22], except that after removing the 3 adaptor and 5 barcode, only inserts longer than 18 nt were analyzed. 5 and 3 heterogeneity was determined as described [39]. Briefly, for each miRNA the heterogeneity of the termini of its isoforms was calculated as the mean of the absolute values of the distance between the 52 or 32 extremity of an individual read and the most abundant 52 or 32 end for that miRNA. For 52 heterogeneity, all isoforms of a miRNA were examined. For 32 heterogeneity, only the most abundant 52 isoforms (i.e., that with the annotated seed sequence) were evaluated.
Supplementary Material
Highlights
More than one quarter of all miRNAs in flies are trimmed after their production by Dicer-1
miRNA trimming occurs after loading into Ago1 and is limited by miRNA* strand removal
The 32 to 52 exoribonuclease Knabber trims miRNAs, enhancing miRNA function.
Knabber is required for normal fly development.
Acknowledgements
We thank Ryuya Fukunaga for providing recombinant Dicer-1/ Loqs PB, Gwen Farley for technical assistance, Alicia Boucher for help with fly husbandry and members of the Zamore lab for support, discussions, and comments on the manuscript. This work was supported by NIH grants GM62868 and GM65236 to P.D.Z. and an EMBO long-term fellowship (ALTF 522-2008) and an Austrian Science Fund (FWF) Erwin Schrödinger-Auslandsstipendium (J2832-B09) to S.L.A. P.D.Z. is a member of the scientific advisory board of Regulus Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 5.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 6.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and Mechanisms Related to RNA Interference Regulate Expression of the Small Temporal RNAs that Control C. elegans Developmental Timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 10.Hutvágner G, McLachlan J, Pasquinelli AE, Balint É, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 11.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/ miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 14.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 19.Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 20.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ameres SL, Hung JH, Xu J, Weng Z, Zamore PD. Target RNA-directed tailing and trimming purifies the sorting of endo-siRNAs between the two Drosophila Argonaute proteins. RNA. 2011;17:54–63. doi: 10.1261/rna.2498411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 26.Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 27.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 28.Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chatterjee S, Fasler M, Bussing I, Grosshans H. Target-mediated protection of endogenous microRNAs in C. elegans. Dev Cell. 2011;20:388–396. doi: 10.1016/j.devcel.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 32.Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- 33.Ketting RF, Plasterk RH. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- 34.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. N ucleic Acids Research. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernad A, Blanco L, Lázaro JM, Martín G, Salas M. A conserved 32–52 exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 36.Brautigam CA, Sun S, Piccirilli JA, Steitz TA. Structures of normal single-stranded DNA and deoxyribo-3-S-phosphorothiolates bound to the 32–52 exonucleolytic active site of DNA polymerase I from Escherichia coli. Biochemistry. 1999;38:696–704. doi: 10.1021/bi981537g. [DOI] [PubMed] [Google Scholar]
- 37.Deutscher MP, Marlor CW. Purification and characterization of Escherichia coli RNase T. J Biol Chem. 1985;260:7067–7071. [PubMed] [Google Scholar]
- 38.Cudny H, Zaniewski R, Deutscher MP. Escherichia coli RNase D. Purification and structural characterization of a putative processing nuclease. The Journal of biological chemistry. 1981;256:5627–5632. [PubMed] [Google Scholar]
- 39.Seitz H, Ghildiyal M, Zamore PD. Argonau te loading improves the 5′ precision of both MicroRNAs and their miRNA* strands in flies. Curr Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 41.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naive, effector and memory CD8 T cells. PLoS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 43.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36:445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010 doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 46.Seitz H, Tushir JS, Zamore PD. A 5′-uridine amplifies miRNA/ miRNA* asymmetry in Drosophila by promoting RNA-induced silencing complex formation. Silence. 2011;2:1–10. doi: 10.1186/1758-907X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baccarini A, Chauhan H, Gardner TJ, Jayaprakash AD, Sachidanandam R, Brown BD. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr Biol. 2011;21:369–376. doi: 10.1016/j.cub.2011.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 52.Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu W, Shirayama M, Conte D, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haley B, Tang G, Zamore PD. In vitro analysis of RNA interference in Drosophila melanogaster. Methods. 2003;30:330–336. doi: 10.1016/s1046-2023(03)00052-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.