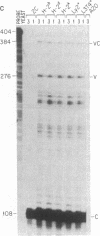
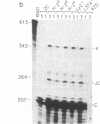
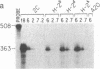
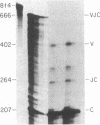
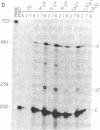
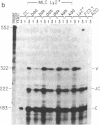
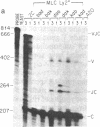
Abstract
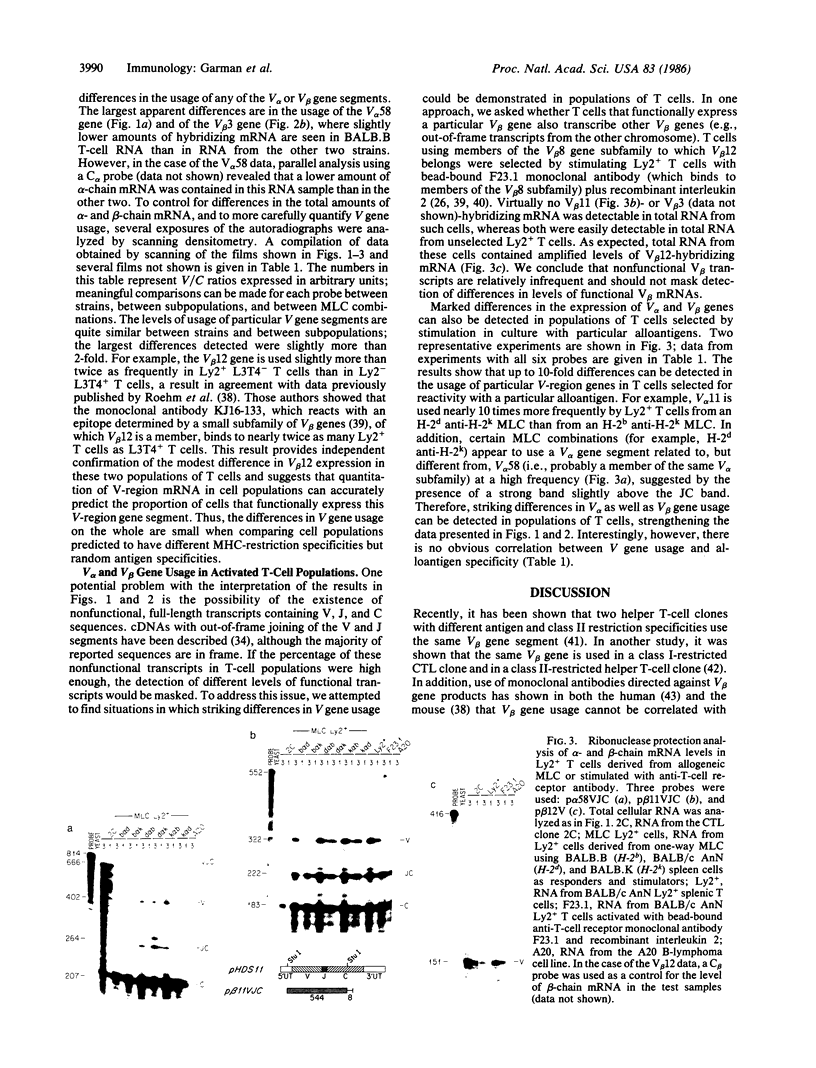
We have examined T-cell receptor alpha- and beta-chain variable (V) region gene usage in T-cell populations predicted to have different major histocompatibility complex-restriction specificities. Using a sensitive ribonuclease protection assay to measure T-cell receptor mRNA levels, we found no striking differences in the usage of three V alpha genes and three V beta genes in T-cell populations from three congeneic H-2-disparate strains of mice and between the mutually exclusive Ly2+ L3T4- and Ly2- L3T4+ T-cell subpopulations. These results suggest that major histocompatibility complex restriction cannot be explained by the differential usage of nonoverlapping V alpha or V beta gene pools. In contrast, striking but unpredictable differences were seen in V gene usage in populations of T cells selected by activation with particular alloantigens.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Acuto O., Campen T. J., Royer H. D., Hussey R. E., Poole C. B., Reinherz E. L. Molecular analysis of T cell receptor (Ti) variable region (V) gene expression. Evidence that a single Ti beta V gene family can be used in formation of V domains on phenotypically and functionally diverse T cell populations. J Exp Med. 1985 Jun 1;161(6):1326–1343. doi: 10.1084/jem.161.6.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison J. P., McIntyre B. W., Bloch D. Tumor-specific antigen of murine T-lymphoma defined with monoclonal antibody. J Immunol. 1982 Nov;129(5):2293–2300. [PubMed] [Google Scholar]
- Arden B., Klotz J. L., Siu G., Hood L. E. Diversity and structure of genes of the alpha family of mouse T-cell antigen receptor. 1985 Aug 29-Sep 4Nature. 316(6031):783–787. doi: 10.1038/316783a0. [DOI] [PubMed] [Google Scholar]
- Barth R. K., Kim B. S., Lan N. C., Hunkapiller T., Sobieck N., Winoto A., Gershenfeld H., Okada C., Hansburg D., Weissman I. L. The murine T-cell receptor uses a limited repertoire of expressed V beta gene segments. Nature. 1985 Aug 8;316(6028):517–523. doi: 10.1038/316517a0. [DOI] [PubMed] [Google Scholar]
- Behlke M. A., Spinella D. G., Chou H. S., Sha W., Hartl D. L., Loh D. Y. T-cell receptor beta-chain expression: dependence on relatively few variable region genes. Science. 1985 Aug 9;229(4713):566–570. doi: 10.1126/science.3875151. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bevan M. J., Fink P. J. The influence of thymus H-2 antigens on the specificity of maturing killer and helper cells. Immunol Rev. 1978;42:3–19. doi: 10.1111/j.1600-065x.1978.tb00256.x. [DOI] [PubMed] [Google Scholar]
- Bevan M. J. In a radiation chimaera, host H-2 antigens determine immune responsiveness of donor cytotoxic cells. Nature. 1977 Sep 29;269(5627):417–418. doi: 10.1038/269417a0. [DOI] [PubMed] [Google Scholar]
- Chien Y., Becker D. M., Lindsten T., Okamura M., Cohen D. I., Davis M. M. A third type of murine T-cell receptor gene. Nature. 1984 Nov 1;312(5989):31–35. doi: 10.1038/312031a0. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Crispe I. N., Bevan M. J., Staerz U. D. Selective activation of Lyt 2+ precursor T cells by ligation of the antigen receptor. Nature. 1985 Oct 17;317(6038):627–629. doi: 10.1038/317627a0. [DOI] [PubMed] [Google Scholar]
- Dialynas D. P., Wilde D. B., Marrack P., Pierres A., Wall K. A., Havran W., Otten G., Loken M. R., Pierres M., Kappler J. Characterization of the murine antigenic determinant, designated L3T4a, recognized by monoclonal antibody GK1.5: expression of L3T4a by functional T cell clones appears to correlate primarily with class II MHC antigen-reactivity. Immunol Rev. 1983;74:29–56. doi: 10.1111/j.1600-065x.1983.tb01083.x. [DOI] [PubMed] [Google Scholar]
- Goverman J., Minard K., Shastri N., Hunkapiller T., Hansburg D., Sercarz E., Hood L. Rearranged beta T cell receptor genes in a helper T cell clone specific for lysozyme: no correlation between V beta and MHC restriction. Cell. 1985 Apr;40(4):859–867. doi: 10.1016/0092-8674(85)90345-9. [DOI] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Haskins K., Kubo R., White J., Pigeon M., Kappler J., Marrack P. The major histocompatibility complex-restricted antigen receptor on T cells. I. Isolation with a monoclonal antibody. J Exp Med. 1983 Apr 1;157(4):1149–1169. doi: 10.1084/jem.157.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayday A. C., Diamond D. J., Tanigawa G., Heilig J. S., Folsom V., Saito H., Tonegawa S. Unusual organization and diversity of T-cell receptor alpha-chain genes. 1985 Aug 29-Sep 4Nature. 316(6031):828–832. doi: 10.1038/316828a0. [DOI] [PubMed] [Google Scholar]
- Hedrick S. M., Cohen D. I., Nielsen E. A., Davis M. M. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984 Mar 8;308(5955):149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- Katz D. H., Hamaoka T., Dorf M. E., Maurer P. H., Benacerraf B. Cell interactions between histoincompatible T and B lymphocytes. IV. Involvement of the immune response (Ir) gene in the control of lymphocyte interactions in responses controlled by the gene. J Exp Med. 1973 Sep 1;138(3):734–739. doi: 10.1084/jem.138.3.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindred B., Shreffler D. C. H-2 dependence of co-operation between T and B cells in vivo. J Immunol. 1972 Nov;109(5):940–943. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuer S. C., Fitzgerald K. A., Hussey R. E., Hodgdon J. C., Schlossman S. F., Reinherz E. L. Clonotypic structures involved in antigen-specific human T cell function. Relationship to the T3 molecular complex. J Exp Med. 1983 Feb 1;157(2):705–719. doi: 10.1084/jem.157.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozato K., Mayer N., Sachs D. H. Hybridoma cell lines secreting monoclonal antibodies to mouse H-2 and Ia antigens. J Immunol. 1980 Feb;124(2):533–540. [PubMed] [Google Scholar]
- Raulet D. H., Garman R. D., Saito H., Tonegawa S. Developmental regulation of T-cell receptor gene expression. Nature. 1985 Mar 7;314(6006):103–107. doi: 10.1038/314103a0. [DOI] [PubMed] [Google Scholar]
- Raulet D. H., Gottlieb P. D., Bevan M. J. Fractionation of lymphocyte populations with monoclonal antibodies specific for LYT-2.2 and LYT-3.1. J Immunol. 1980 Sep;125(3):1136–1143. [PubMed] [Google Scholar]
- Roehm N., Herron L., Cambier J., DiGuisto D., Haskins K., Kappler J., Marrack P. The major histocompatibility complex-restricted antigen receptor on T cells: distribution on thymus and peripheral T cells. Cell. 1984 Sep;38(2):577–584. doi: 10.1016/0092-8674(84)90512-9. [DOI] [PubMed] [Google Scholar]
- Rupp F., Acha-Orbea H., Hengartner H., Zinkernagel R., Joho R. Identical V beta T-cell receptor genes used in alloreactive cytotoxic and antigen plus I-A specific helper T cells. 1985 May 30-Jun 5Nature. 315(6018):425–427. doi: 10.1038/315425a0. [DOI] [PubMed] [Google Scholar]
- Saito H., Kranz D. M., Takagaki Y., Hayday A. C., Eisen H. N., Tonegawa S. A third rearranged and expressed gene in a clone of cytotoxic T lymphocytes. Nature. 1984 Nov 1;312(5989):36–40. doi: 10.1038/312036a0. [DOI] [PubMed] [Google Scholar]
- Saito H., Kranz D. M., Takagaki Y., Hayday A. C., Eisen H. N., Tonegawa S. Complete primary structure of a heterodimeric T-cell receptor deduced from cDNA sequences. 1984 Jun 28-Jul 4Nature. 309(5971):757–762. doi: 10.1038/309757a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer G. M. Cell-mediated cytotoxicity to trinitrophenyl-modified syngeneic lymphocytes. Eur J Immunol. 1974 Aug;4(8):527–533. doi: 10.1002/eji.1830040802. [DOI] [PubMed] [Google Scholar]
- Sim G. K., Augustin A. A. V beta gene polymorphism and a major polyclonal T cell receptor idiotype. Cell. 1985 Aug;42(1):89–92. doi: 10.1016/s0092-8674(85)80104-5. [DOI] [PubMed] [Google Scholar]
- Staerz U. D., Rammensee H. G., Benedetto J. D., Bevan M. J. Characterization of a murine monoclonal antibody specific for an allotypic determinant on T cell antigen receptor. J Immunol. 1985 Jun;134(6):3994–4000. [PubMed] [Google Scholar]
- Symington F. W., Sprent J. A monoclonal antibody detecting an Ia specificity mapping in the I-A or I-E subregion. Immunogenetics. 1981;14(1-2):53–61. doi: 10.1007/BF00344299. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Winoto A., Mjolsness S., Hood L. Genomic organization of the genes encoding mouse T-cell receptor alpha-chain. 1985 Aug 29-Sep 4Nature. 316(6031):832–836. doi: 10.1038/316832a0. [DOI] [PubMed] [Google Scholar]
- Yanagi Y., Yoshikai Y., Leggett K., Clark S. P., Aleksander I., Mak T. W. A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature. 1984 Mar 8;308(5955):145–149. doi: 10.1038/308145a0. [DOI] [PubMed] [Google Scholar]
- Yoshikai Y., Clark S. P., Taylor S., Sohn U., Wilson B. I., Minden M. D., Mak T. W. Organization and sequences of the variable, joining and constant region genes of the human T-cell receptor alpha-chain. 1985 Aug 29-Sep 4Nature. 316(6031):837–840. doi: 10.1038/316837a0. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkernagel R. M., Callahan G. N., Althage A., Cooper S., Klein P. A., Klein J. On the thymus in the differentiation of "H-2 self-recognition" by T cells: evidence for dual recognition? J Exp Med. 1978 Mar 1;147(3):882–896. doi: 10.1084/jem.147.3.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkernagel R. M., Doherty P. C. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974 Apr 19;248(5450):701–702. doi: 10.1038/248701a0. [DOI] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]