Abstract
Facile reactivity of hydrazides and aldehydes was explored as potential coupling partners for incorporation into M(CO)3 (M = Re, 99mTc) based radiopharmaceuticals. Both ‘click, then chelate’ and ‘prelabel, then click’ synthetic routes produced identical products in high yields and lacked metal-hydrazide/-hydrazone interactions, highlighting the potential of this click strategy.
The benefits of click reactions are well known, and their applications have revolutionized numerous fields including bioconjugate chemistry, material chemistry and polymer sciences.1–4 However, the most widely used Huisgen 1,3-dipolar cycloaddition click reaction has restricted usefulness in biological applications due to the toxicity of copper in vivo.5 A sterically strained cyclooctyne group alleviates the toxicity issue in a copper-less click reaction. However, the installation of a bulky functional group can significantly impact the pharmacodynamics and biological efficacy of a clicked biomolecule. Alternatively, the non-aldol addition reaction between a carbonyl and a nucleophilic nitrogen as a click reaction bypasses both drawbacks mentioned above. Key benefits of this click strategy are the availability of amines for bioconjugation, rapid incorporation into molecular frameworks, and a simple resultant click product that minimally impacts the nature of the biomolecule.
Click reactions have become an integral feature in radio-pharmaceutical chemistry in designing ligands for metal complexation or for coupling pre-labeled radionuclide complexes with a targeting biomolecule.6–8 The latter approach reduces the exposure of biomolecules to harsh conditions (i.e., pH, temperature) required for complexation and improves the site-specific labeling of the metal. This “prelabel, then click” strategy allows efficient incorporation of radiometals into sensitive biomolecules which would be deactivated under standard radiolabeling conditions.
In this article, we pursue the non-aldol hydrazone click reaction between an aldehyde-containing biomolecule and a hydrazide bifunctional chelator for complexing the diagnostic molecular imaging radionuclide 99mTc (γ, 140 keV (89%), t1/2 = 6.0 h). The hydrazone moiety was selected for its high stability at physiological pH, and lability under strongly acidic and basic conditions as demonstrated by drug delivery agents in tumor targeting.9–11 To our knowledge, such a click approach with 99mTc has not yet been explored. Instead, the paradigm from the literature indicates that hydrazine precursors would be unavailable for hydrazone formation because they interact with 99mTc metal centers. For instance, 99mTcV oxo complexes react with hydrazines (i.e., 6-hydrazinonicotinic acid (HYNIC)) to form complexes through nitrogen coordination (Fig. 1, Route-1).12–17 Additionally, mixed observations of metal coordination or combined oxidation/decarbonylation of the metal are reported for hydrazines with the organometallic fac-(M(OH2)3(CO)3)+ (M = Re, 99mTc) complex.12,18 Such observations prompted our investigation of hydrazides as a clickable moiety for hydrazone generation. We find that the model hydrazide systems employed did indeed prefer hydrazone formation over interactions with the metal centers (Fig. 1, Route-2).
Fig. 1.
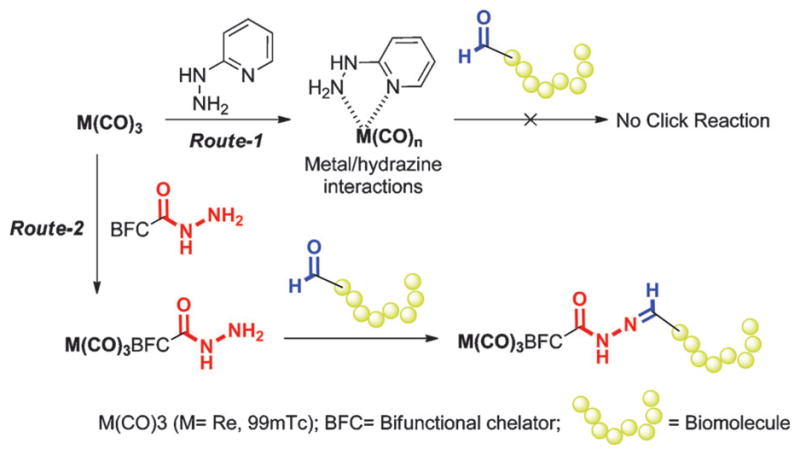
General hydrazine/hydrazide reaction scheme: Route-1:M-HYNIC type coordination and Route-2: Coupling with the aldehyde to give hydrazone.
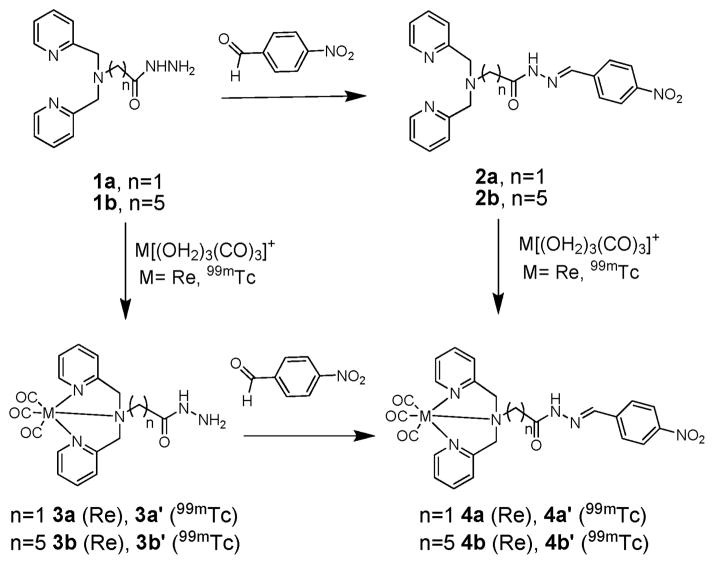
The focus of this investigation was to prove the concept of the hydrazide/hydrazone click reaction with a model aldehyde for application with M(CO)3 + and to show its viable incorporation into a biomolecule-related framework. To minimize possible coordination of the hydrazide or hydrazone groups, a well-known and efficient tridentate ligand, dipicolylamine (DPA), was utilized for selective coordination of M(CO)3 +.19 The clickable, bifunctional chelator was generated by functionalizing the central amine of DPA with a hydrazide moiety. We expected this to avoid undesirable interactions seen between hydrazines and 99mTc metal centers while maintaining sufficient reactivity for hydrazone formation. Our approach involved two parallel synthetic routes to determine the influence of the hydrazide and hydrazone products in the complexation of M(CO)3 + (Scheme 1). Route 1: the DPA-linked hydrazide ligand was reacted with an aldehyde to yield the corresponding hydrazone clicked ligands followed by complexation with M(CO)3 +. Route 2: the DPA-hydrazide ligand was complexed directly with M(CO)3 + followed by hydrazone formation with an aldehyde. Ideally, both routes would lead to the same complex, indicating that neither the hydrazide in the prelabeled approach nor the clicked hydrazone moiety interact with the metal centers.
Scheme 1.
Parallel synthesis approaches Route 1 (1 → 2 → 4, click, then chelate) and Route 2 (1 → 3 → 4, prelabel, then click) for the formation of M(CO)3 +, (M=Re, 99mTc) DPA-hydrazone complexes.
Two variations of the DPA-hydrazide ligands, 1a (n = 1) and 1b (n = 5), were prepared in two steps with reasonable overall yields: (1) alkylation of DPA with the corresponding 2-bromoacetic acid and 6-bromohexanoic acid ethyl esters in the presence of triethylamine; (2) conversion of the ester to the hydrazide by the addition of 35% hydrazine in ethanol.20 These two different linkers permitted the investigation of the potential electrochemical or coordination interactions of the hydrazide group with the M(CO)3 centers based on the proximity of these groups. Linker 1a is more likely to coordinate the metal center (5 or 6 member coordination ring) than the longer linker 1b.
In Route 1 (click, then chelate), the clicked hydrazone analogues were prepared by the condensation of the hydrazide (1a, 1b) with p-nitrobenzaldehyde (PNB) as a model system. The reaction proceeded smoothly to completion at 50–55 °C, 30–45 min at pH 4.0 in an ethanol/water mixture to form the hydrazone DPA ligands, 2a (81%) and 2b (62%). The clicked hydrazone ligands (2a, 2b) were complexed with fac-Re(OH2)3(CO)3 + in methanol/water at pH 8 (dropwise addition of saturated NaHCO3 solution) at 55 °C for 2–3 h to yield a single product 4a (45%) and 4b (57%), respectively. Confirmation of the DPA coordination of 2a and 2b in the Re(CO)3 complexes were elucidated from X-ray crystallographic and NMR data.† The X-ray structure of 4a clearly delineates the DPA coordination to Re(CO)3, while the hydrazone is orientated away from the metal center (Fig. S5†).22 1H NMR data of 4a and 4b also confirmed the DPA coordination of Re(CO)3 indicated by the HA,B splitting for the methylene protons adjacent to the pyridine typically observed upon complexation. The minimal shift of the hydrazone (CH) (7.86 ppm) upon complexation also confirmed the non-coordination of the hydrazone with the Re center.
In Route 2 (prelabel, then click), the DPA-hydrazide ligands (1a, 1b) were complexed with Re(CO)3. 1a was refluxed in methanol with Re(CO)5OTf and 1b reacted with fac-Re(CO)3(H2O)3 + at 55 °C 30 min at pH 8 in a methanol/water solution to yield 3a (46%) and 3b (87%), respectively. 1H NMR of 3a and 3b shows characteristic downfield shifts and an HA,B type splitting of the methylene protons of DPA indicative of Re(CO)3 + coordination. Whereas, the methylene protons adjacent to the hydrazide group do not shift significantly from the free ligand as would be observed upon metal coordination. This data supports the non-coordination of hydrazide and the non-HYNIC behavior of these molecules. The characteristic IR pattern of the fac-Re(CO)3 also confirmed that oxidative decarbonylation to a Re(CO)2 species was not observed under these conditions.
In the second step, the Re(CO)3DPA-hydrazide complexes (3a, 3b) were condensed with PNB at pH ~ 4 for 2–4 h at 50 °C in ethanol/water to yield the Re(CO)3DPA-hydrazone complexes 4a (93%) and 4b (62%), respectively. The analytical data collected of 4a and 4b in Route 2 was identical to the results observed in Route 1. The reactivity of 3a and 3b towards PNB was comparable in reaction conditions and yields observed with the ligands (2a, 2b). In both Re(CO)3DPA-hydrazide variations, the hydrazide exhibits preferential condensation with the aldehyde over coordination. Re(CO)3DPA complexation does not appear to impact the reactivity, through either electronic or coordination effects, of the hydrazone formation. The lability of the hydrazone linkage was explored, and the hydrazone bond is stable at pH 3–12 and shows no decomposition on heating for over 10 h at 90 °C.
Radiolabeling studies with fac-99mTc(OH2)3(CO)3 + were conducted to evaluate the efficiency and efficacy of Routes 1 and 2 for comparison to the Re analogs. In Route 1, the clicked DPA-hydrazone ligands (2a, 2b) in various concentrations (10−4–10−5 M) were incubated for 30 min at 90 °C with fac-99mTc(OH2)3(CO)3 + to generate the corresponding 99mTc(CO)3 complexes 4a′ (97%) and 4b′ (91%) in excellent yields. A single chromatographic peak for 4a′ (21.93 min) and 4b′ (21.68 min) was observed in radio (γ) HPLC that correlated with the tr of the Re analogs 4a (21.76 min) and 4b (21.82 min). In Route 2, the DPA-hydrazide ligands (1a, 1b) were complexed with fac-99mTc(OH2)3(CO)3 + at 50 °C to generate fac-99m-Tc(CO)3DPA-hydrazide complexes 3a′ (46%) and 3b′ (78%) in good yield. Observed as single peak on HPLC, the tr’s (3a′, 16.98 min and 3b′, 17.95 min) of 99mTc species correlated with the Re analogs (3a, 16.85 min and 3b, 17.89 min). The hydrazone condensation of 3a′ and 3b′ with PNB at pH 4 was found to be analogous to the Re complexes (3a, 3b) by comparison of retention times by analytical HPLC. Comparative HPLC results for compounds 3a (and 3a′) and 4a (and 4a′) can be seen in Fig. S2†. To determine the optimal coupling conditions, several variables such as temperature (60/90 °C), reaction time (30/60 min), and [PNB] (10−4–10−6 M), were independently evaluated (Table S1). Increased temperature and reaction times were found to modestly improve the condensation yields. However, PNB concentration had a significant effect in the hydrazone formation. Excellent yields of 71–91% of the hydrazone product were observed at 10−4 M PNB, while at lower concentrations the product yields declined even with increased temperature and reaction time.
A one-pot synthesis was also carried out and it was proven that addition of PNB to 3a (or 3b) without purification also yielded 4a (or 4b). X-Ray quality crystals of 4a were isolated by this method via repeated crystallization from methanol (Fig. S5†). Labeling studies with 99mTc were also performed to confirm the results from the Re studies. Details can be found in supplementary information Table S1.†
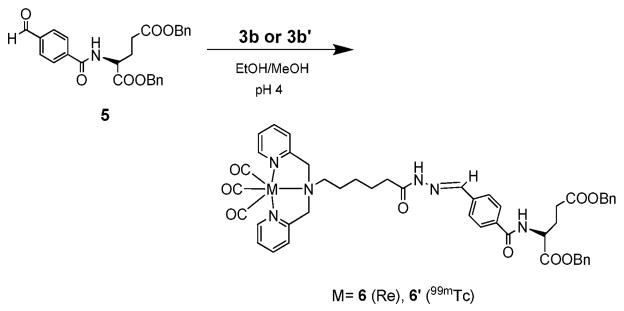
The successful results obtained with PNB as a model provided an important proof of concept for the Re/99mTc(CO)3DPA-hydrazide/hydrazone click reaction and indicated its viable application within a biomolecule-related framework. A model system using glutamic acid was investigated to establish a potential route for incorporation of the hydrazide/hydrazone click reaction into a biomolecule having free amines (Scheme 2). 4-carboxybenzaldehyde provides a unique motif to couple to free amines under standard peptide conditions, while maintaining efficient reactivity with metal-hydrazides to yield a single product.
Scheme 2.
Glutamate model system for the hydrazide/hydrazone Route 2 (1b → 3b → 6b, prelabel, then click).
Compound 5 was synthesized by coupling 4-carboxybenzal-dehyde with H2N-Glu(OBn)(OBn) in the presence of EDC and HoBt.21 Parallel synthetic approaches (Route 1 and 2) were investigated with 5 under similar conditions (pH 4, 30–60 min) as the PNB system. In Route 1 “click, then chelate”, the clicked ligand (7b) was complexed with fac-Re(OH2)3(CO)3 + to yield 6 (66%). In Route 2 “prelabel, then click”, 6 was also obtained in good yield (61%) by the coupling of 3b with 5 at pH 4 in 40 min. Characterization of the products from both routes confirmed the identical nature of the compounds. 99mTc(OH2)3(CO)3 + studies performed according to the PNB conditions confirmed product formation analogous to the Re compounds. A single peak with a tr matching the Re analog 6 and similar labeling yields as 3b′ were obtained (60/90 °C, 10−4 M 5, 60/90 min; 70–80%).
In conclusion, we have presented here a novel hydrazide/hydrazone click reaction which highlights the potential application for labeling biomolecules with 99mTc(CO)3 as a fast and efficient reaction. Either Route 1 or 2 takes advantage of the strong chelator DPA to yield single, well-defined products that differ considerably in behavior from previous reports of decarbonylation, HYNIC, or hydrazone coordination to the metal center. The inclusion of the hydrazide/hydrazone system into a peptide framework reported in this work can be readily extended to a number of biomolecules for labeling with 99mTc(CO)3.
Supplementary Material
Acknowledgments
This research was funded in part by the Washington State LSDF (08-012374880), Office of Science (BER), U.S. DOE (DE-FG02-08-ER64672), NSF’s REU program at WSU (#0851502) and NIH Biotechnology Training Program at WSU (T32 GM008336) and NIH (5R01CA1406170).
Footnotes
Electronic supplementary information (ESI) available. CCDC 829258. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c1cc15451f
Contributor Information
Leonard R. MacGillivray, Email: len-macgillivray@uiowa.edu.
Paul D. Benny, Email: bennyp@wsu.edu.
Notes and references
- 1.Hein CD, Liu XM, Wang D. Pharm Res. 2008;25:2216–2230. doi: 10.1007/s11095-008-9616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lutz JF, Zarafshani Z. Adv Drug Delivery Rev. 2008;60:958–970. doi: 10.1016/j.addr.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Martin ME, Parameswarappa SG, O’Dorisio MS, Pigge FC, Schultz MK. Bioorg Med Chem Lett. 2010;20:4805–4807. doi: 10.1016/j.bmcl.2010.06.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR. Proc Natl Acad Sci U S A. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheiber F, Schmidt MM, Dringen R. Neurochem Int. 2010;57:314–322. doi: 10.1016/j.neuint.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Mamat C, Ramenda T, Wuest FR. Mini-Rev Org Chem. 2009;6:21–34. [Google Scholar]
- 7.Struthers H, Mindt TL, Schibli R. Dalton Trans. 2010;39:675–696. doi: 10.1039/b912608b. [DOI] [PubMed] [Google Scholar]
- 8.Waengler C, Schirrmacher R, Bartenstein P, Waengler B. Curr Med Chem. 2010;17:1092–1116. doi: 10.2174/092986710790820615. [DOI] [PubMed] [Google Scholar]
- 9.Dirksen A, Dawson PE. Bioconjugate Chem. 2008;19:2543–2548. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coessens V, Schacht E, Domurado D. J Controlled Release. 1996;38:141–50. [Google Scholar]
- 11.Barbour NP, Paborji M, Alexander TC, Coppola WP, Bogardus JB. Pharm Res. 1995;12:215–22. doi: 10.1023/a:1016274825322. [DOI] [PubMed] [Google Scholar]
- 12.Alberto R, Cowley AR, Dilworth JR, Donnelly PS, Pratt J. Dalton Trans. 2004:2610–2611. doi: 10.1039/B407708C. [DOI] [PubMed] [Google Scholar]
- 13.Alberto R, Schibli R, Schubiger AP, Abram U, Pietzsch HJ, Johannsen B. J Am Chem Soc. 1999;121:6076–6077. [Google Scholar]
- 14.Banerjee SR, Maresca KP, Stephenson KA, Valliant JF, Babich JW, Graham WA, Barzana M, Dong Q, Fischman AJ, Zubieta J. Bioconjugate Chem. 2005;16:885–902. doi: 10.1021/bc050040y. [DOI] [PubMed] [Google Scholar]
- 15.Edwards DS, Liu S, Harris AR, Poirier MJ, Ewels BA. Bioconjugate Chem. 1999;10:803–7. doi: 10.1021/bc990022e. [DOI] [PubMed] [Google Scholar]
- 16.Harris TD, Sworin M, Williams N, Rajopadhye M, Damphousse PR, Glowacka D, Poirier MJ, Yu K. Bioconjugate Chem. 1999;10:808–814. doi: 10.1021/bc9900237. [DOI] [PubMed] [Google Scholar]
- 17.Meszaros LK, Dose A, Biagini SCG, Blower PJ. Inorg Chim Acta. 2010;363:1059–1069. [Google Scholar]
- 18.Barbazan P, Hagenbach A, Oehlke E, Abram U, Carballo R, Rodriguez-Hermida S, Vazquez-Lopez EM. Eur J Inorg Chem. 2010:4622–4630. [Google Scholar]
- 19.Bartholomae M, Valliant J, Maresca KP, Babich J, Zubieta J. Chem Commun. 2009:493–512. doi: 10.1039/b814903h. [DOI] [PubMed] [Google Scholar]
- 20.Begum A, Aparna AV, Sireesha B, Devi CS, Raghavaiah P. Indian J Chem, Section B: Org Chem Including Med Chem. 2009;48B:1565–1570. [Google Scholar]
- 21.Poulsen SA. J Am Soc Mass Spectrom. 2006;17:1074–1080. doi: 10.1016/j.jasms.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 22.Crystal data for 4a, C34H29F3N6O9ReS, Mr = 940.89 gmol−1, triclinic, space group P 1̄, a = 10.9711(12) Å, b = 11.6456(13) Å, c = 15.6740(17) Å, α = 92.749(5), β = 109.576(5), γ = 101.608(5), V = 1833.7(3) Å3, T = 125(2) K, Z = 2, μ(MoKα) = 3.447 mm−1, 12 482 reflections measured, 6421 independent reflections (Rint = 0.0165). The final R1 values were 0.023 (I > 2σ(I)). The final wR(F2) values were 0.0572 (I > 2σ(I)). The final R1 values were 0.0242 (all data). The final wR(F2) values were 0.0578 (all data). The goodness of fit on F2 was 1.046. CCDC deposition number: 829258.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.