Abstract
The β amyloid (Aβ) peptide aggregates to form β-rich structures that are known to trigger Alzheimer’s disease. Experiments suggest that an α-helical intermediate precedes the formation of these aggregates. However, a description at the molecular level of the α-to-β transition has not been obtained. Because it has been proposed that the transition might be initiated in the amino-terminal region of Aβ, we studied the aggregation of the 28-residue amino-terminal fragment of Aβ (Aβ1–28) using molecular dynamics and a coarse-grained force field. Simulations starting from extended and helical conformations showed that oligomerization is initiated by formation of intermolecular β -sheets between the residues in the N-terminal regions. In simulations starting from the α-helical conformation, forcing residues 17–21 to remain in the initial (helical) conformation prevents aggregation but allows for the formation of dimers, indicating that oligomerization, initiated along the non-helical N-terminal regions, cannot progress without the α-to-β transition propagating along the chains.
Keywords: Amyloids, Aβ peptide, UNRES force field, molecular dynamics, Alzheimer’s disease
Introduction
The β amyloid (Aβ) peptide aggregates to form β -rich structures that evolve into fibrils. Both the fibrils and the pre-fibrillar aggregates have been identified as the cause of Alzheimer’s disease (AD).1,2 In its monomeric form, Aβ is not toxic; therefore, it is the transition from monomer to oligomer that marks the beginning of the disease. Understanding how aggregation is initiated and how it could be prevented is crucial for devising therapeutic agents to treat AD.
Experiments to obtain a structural description of Aβ monomers and the conformational changes leading to aggregation are extremely challenging because of the high propensity of Aβ to aggregate. To surmount this problem, organic solvents, such as trifluoroethanol (TFE),3–5 hexafluoroisopropanol (HFIP),6,7 or micellar solutions,8,9 that can stabilize the helical conformation, are often used. At very low concentrations of HFIP, Aβ1–42 does not aggregate and adopts conformations with a certain helical content.7 An α-helical intermediate has been detected during the aggregation of Aβ1–40 and Aβ1–42. 10 Simulations have also given important insight into the characterization of Aβ.11–20 The helix propensity11,12,21 as well as the aggregation of Aβ 13,15,17–19 have been studied by molecular dynamics (MD) simulations, and the tendency of Aβ1–40 monomers to adopt the α-helical conformation along their pathway towards fibril formation has been observed with this technique as well.18,19,21 The foregoing results support the hypothesis that an α-to-β transition precedes Aβ aggregation. However, it has not been possible to determine where and how this transition is initiated.
In this work, we use MD simulations and a coarse-grained united-residue (UNRES) model22–28 to study the aggregation of the Aβ1–28 fragment. Experiments have shown that Aβ1–28 forms fibrils with the cross-β structure characteristic of the full length Aβ,29 adopts a helical conformation in solution,11,30 and stabilization of a small helical fragment can prevent aggregation,31 all of which makes Aβ1–28 a good model for studying amyloid aggregation. Our results indicate that a helical intermediate precedes aggregation; however, starting the simulations from helical conformations does not speed up the process. Finally, stabilizing the helix between residues 17 and 21 prevents the formation of oligomers larger than dimers.
Methods
In the UNRES model,22–28 a polypeptide chain is represented as a sequence of α-carbon (Cα) atoms with united peptide groups (p) located halfway between the consecutive Cα atoms and united side chains (SC) attached to the Cα atoms. The force field has been defined as a potential of mean force (PMF) of a system of polypeptide chain(s) in water, where all degrees of freedom except the coordinates of the Cα atoms and SC centers have been averaged out.23,27 The effective energy function contains local and site-site interactions, as well as multibody terms, which have been obtained by decomposing the PMF into factors corresponding to clusters of interactions within and between coarse-grained sites. The SC-SC interaction potentials implicitly include the contribution from solvation.23,27 The force field was calibrated to reproduce the structure and thermodynamics of small model proteins;24,25,27,28 in this study, we used the version parameterized with the formin-binding WW domain (PDB ID: 1E0L).25
Systems formed by five monomers of Aβ1–28, initially separated by a 30 Å distance, were used to study aggregation. A total of 120 canonical MD trajectories were simulated. The temperature was held constant at 300 K with the Berendsen thermostat.32 Each of the 120 trajectories was 100 ns long. The monomers were confined within a given volume to define a 50 mM concentration. The 120 trajectories are divided into three groups of 40 trajectories each. In the first group, the monomers were initially in the extended conformation, while in the second and third group, they were initially in a fully helical conformation. For the first two groups, the monomers were allowed to interact freely. For the third group, the distance between the united peptide groups of residues 17 and 21 was restrained so that the α-helical conformation between these residues was preserved during the length of the simulation. A harmonic potential was used to restrain the distances. No restraints were imposed on the interactions between different monomers.
Monomer simulations were carried out using replica exchange MD (REMD). A total of 150 trajectories were simulated with temperatures ranging from 300 K to 480 K. Each trajectory was 20 ns long, and exchanges were attempted every 20,000 steps. Between exchanges, the temperature was held constant with the Berendsen thermostat.32 The α-helix and β -sheet conformational fractions were calculated as the fraction of snapshots, from all snapshots at 300 K, in which a residue was found in an α-helix or a β -sheet conformation.
Results and discussion
An α-helical intermediate precedes Aβ1–28 aggregation
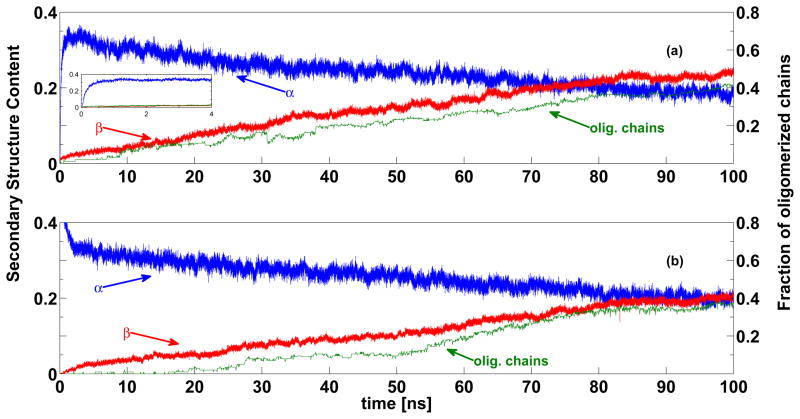
We simulated 40 trajectories of a system of 5 monomers, initially in an extended conformation, confined within a given volume and allowed to interact freely. During the simulation, we computed the fraction of residues in the α-helix and β -sheet conformations, as well as the fraction of chains in the oligomeric state (i.e., forming a dimer or higher order oligomer). The average values (over all trajectories) are shown in Figure 1a. At the beginning of the simulation, the α-helical content grows rapidly reaching almost 40 %, but as the simulation progresses, the helical content slowly decreases, while the β -content increases, indicating a transition from α to β. This transition is accompanied by the oligomerization of the initially free monomers (Figure 1a, green line). We conclude that the α-helical conformations are accessible to Aβ monomers, and that the monomers rarely adopt fibril-like conformations. However, these transient fibril-like conformations can be stabilized by the interactions with other monomers.
Figure 1.
Fraction of residues with α-helical (blue) and β -sheet (red) conformations, and the fraction of chains that have formed dimers or higher order oligomers (green). The values were obtained by averaging over 40 canonical MD trajectories of 5-chain systems with the chains initially in the extended (a) or helical (b) conformation. The inset on panel (a), shows the fast growth of the α-helical content during the first 4ns of simulation.
The α → β transition has been observed in experiments of aggregation of Aβ1–40 and Aβ1–42, with the α-helical content present in the monomeric form and the β content arising together with aggregation. Klimov and Thirumalai21 have also reported an α → β transition during all-atom simulations of the Aβ16–22 fragment, with the β content growing only when the peptides are allowed to interact with each other. Therefore, the oligomerization scenario proposed here might be shared by the different Aβ species.
Facilitating the formation of the α-helical intermediate does not speed up aggregation
Experiments have shown that stabilizing helical conformations, up to a certain degree, accelerates fibril formation,33 which suggests that the helical intermediate might facilitate the aggregation process.10,33 To explore this possibility, we carried out simulations under the same conditions as those illustrated in Figure 1a, except that the monomers were initially in a completely α-helical conformation. Figure 1b shows the fraction of residues in α-helix and β -sheet conformation and the fraction of chains in oligomeric state as a function of simulation time. The behavior is not very different from that of the previous simulations (shown in panel a). Here as well, as the monomers start to associate and oligomerize, the helical content decreases and the β content increases. However, in panel b, the β content increases slightly slower than in panel a. This suggests that the formation of the helical structures does not speed up aggregation, and that the peptides need a longer time to transition to β -sheet conformations when started in a fully helical conformation.
This behavior might seem to contradict the experimental results10,33 that showed that conditions stabilizing α-helical conformations can speed up fibril formation. However, these experiments10,33 also showed that, if the helical state is made too stable, fibril formation does not occur. Therefore, it is the presence of partially folded, helix-containing conformers that facilitates fibril formation.33 Our results are consistent with this hypothesis because they suggest that, by temporarily preventing the C-terminal region from participating in the oligomerization process, the α-helical intermediate facilitates an ordered association of the peptides, from the N-terminal towards the C-terminal regions, ultimately accelerating the formation of the fibrils. But the helices need to be partially unfolded for the monomers to associate.
Stabilization of the 17–21 helical region prevents aggregation
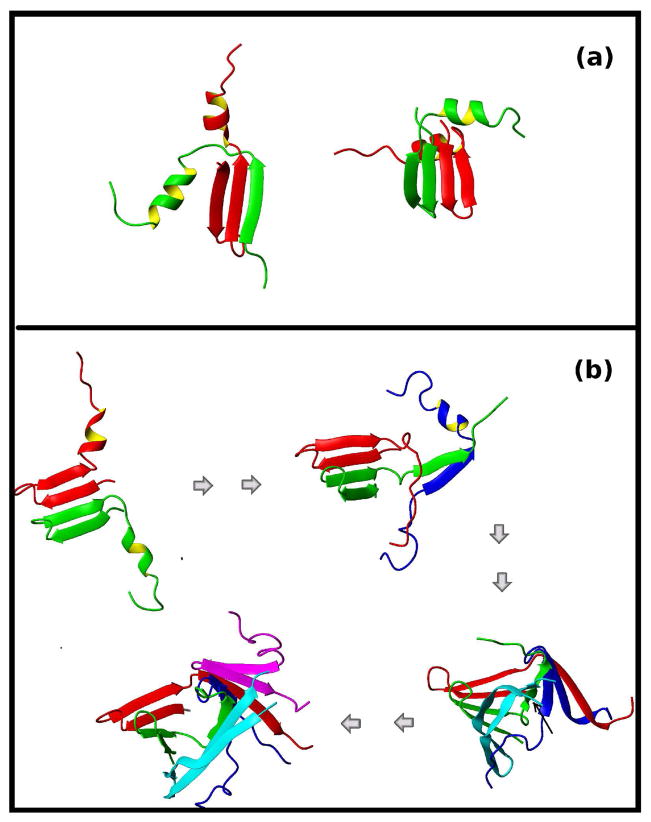
The fact that only oligomeric forms of Aβ can cause neuronal death, has encouraged the study of compounds that can prevent aggregation. Kapurniotu et al.31 studied the aggregation of wild type Aβ1–28, the mutant Aβ1–28(L17K,A21D), and a peptide similar to Aβ1–28(L17K,A21D) that had a lactam-bridge between the side chains of K17 and D21 (cyclo17,21-[Lys17,Asp21]Aβ1–28). The goal of this experiment was to design a peptide that was similar to Aβ1–28 but would not aggregate. By bridging residues Lys17 and Asp21, the helix along this region was stabilized, and although the unbridged Aβ1–28(L17K,A21D) formed fibrils in vitro, the lactam-bridged cyclo 17,21-[Lys17,Asp21]Aβ1–28 did not. To explore the origin of such behavior, we carried out simulations on the wild type protein with residues Leu and Ala at positions 17 and 21, respectively, under the conditions described in the previous paragraph (free monomers with initial helical conformation), but with the distance between L17 and A21 restrained to the initial distance. This restraint was meant to mimic the experiments by Kapurniotu et al.31 and prevent the helix between residues L17 and A21 from unfolding. In agreement with the experiments, we did not detect β -rich aggregates in these simulations. We did observe the formation of dimers through the association of the N-terminal β -hairpins at residues 1 to 12. However, some of these dimers had a short life and, in none of the trajectories, did the association progress to higher orders. Two examples of the most stable dimers are shown in Figure 2a. In both examples, the chains form intermolecular β -sheets involving residues 1 to 12 in the N-terminal half of the peptide. As we will show in the following paragraph, dimers of this type were usually found on the pathway to oligomerization for the unrestrained simulations. But here, by preserving the helical region between L17 to A21, we prevented the formation of larger complexes. Interestingly, in a recent study of a number of potential anti AD drugs,34 the ability to stabilize the helical conformation along the L17-A21 region of Aβ1–42 was shown to correlate with the effectiveness of the drugs, supporting the scenario proposed by our simulations.
Figure 2.
(a) Two examples of dimers formed during simulations with residues 17 to 21 restrained to remain in the helical conformation. Different colors are used for each chain. Restrained residues L17 and A21 as well as the C-terminal residues K28 are indicated. (b) Snapshots from a representative trajectory illustrate the process of aggregation. Different colors are used for different chains. Oligomerization starts with the formation of a dimer, which acts as a seed and triggers further aggregation. The C-terminal residue (K28) is marked for each new chain.
Aggregation is initiated by formation of intermolecular β-sheets involving the residues in the N-terminal region
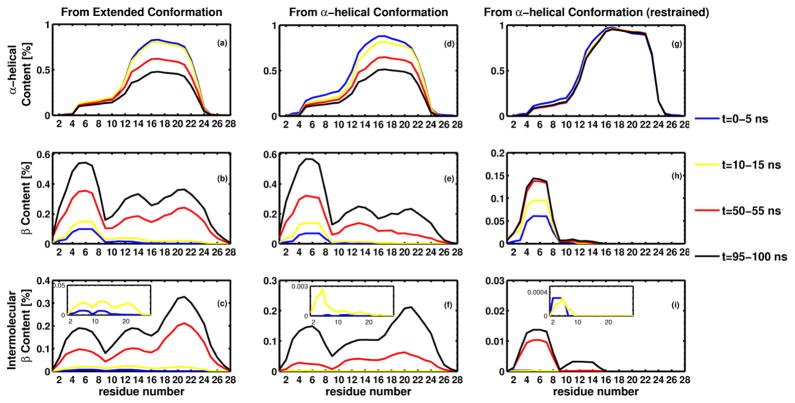
To explore the aggregation process further, we followed the conformations adopted by each residue along the chains for all the trajectories described above. For each type of simulation (initially extended, initially helical, and initially helical with the distance between L17 and A21 restrained), we calculated the fraction of time, during 5 ns-long intervals, that each residue remained in the α-helix or β -sheet conformation, and whether the β -sheet was intermolecular. Figure 3 shows these values, averaged over all chains and all trajectories, for the three different simulations. For clarity, only four time intervals are shown. It should be noticed that, because the helices form very quickly, the first interval (0–5 ns in blue color), which includes the initially extended conformation, also contains conformations with a high α-helical content. Therefore in Figure 3a, it might seem as if the initial conformation already had a certain α-helical content, but this is a consequence of averaging over the first 5-ns interval during which helices appear rapidly.
Figure 3.
Secondary structure information for the three different types of simulation: free monomers initially in: the extended conformation (a to c), helical conformation (d to f) and helical conformation with residues 17 to 21 restrained to the initial helical conformation (g to i). For each type of simulation, the fraction of time during a 5 ns interval that each residue remains in α-helical or β -sheet conformation was computed and averaged over all chains and trajectories. Four intervals are shown, 0–5 ns (blue), 10–15 ns (yellow), 50–55 ns (red) and 95–100 ns (black). Panels a, d and g show the values corresponding to α-helical content, panels b, e and h, the total β -sheet content, and panels c, f and i, the intermolecular β -sheet content.
For all simulations, the helical content (panels a, d and g) is higher between residues 12 and 24, and for the unrestrained simulations (panels a and d), it decreases during the length of the simulation. It can be observed that, initially, the regions adopting β-sheet conformations (panels b, e and h) extend from residue 2 to residue 10 and, to a lesser degree, from residue 11 to residue 15. Intermolecular β-sheets (panels c, f and i) are also initially formed between residues in the N-terminal half of the peptides. For the unrestrained systems, as the simulation progresses, the total and intermolecular β-sheet content propagates towards the C-termini. This cannot happen for the restrained systems because residues 17 to 21 are forced to remain in the α-helical conformation, and therefore we see only a growth in the total and intermolecular β-sheet content between residues 2 an 17. The small fraction of intermolecular β-sheets (panel i) is produced by the formation of dimers such as those shown in Figure 2a.
In order to illustrate how the aggregation process is initiated, in Figure 2b, we show snapshots along one of the trajectories started from the extended conformation. Oligomerization begins with the formation of a dimer, with two β-hairpins along the N-terminal regions associating to form an intermolecular β-sheet, and the rest of the residues retaining certain helical structure. It should be noted that this dimer, which will act as a seed for the formation of a larger aggregate, shares certain similarities with those shown in panel a (those that did not produce higher order oligomers). However, in the example shown in Figure 2b, the helices are free to unwind, and in the second snapshot, the C-terminal region of the red dimer forms an intermolecular β-sheet with a third chain. In the following snapshots two more chains are added to the complex, finally forming a pentamer with no α-helical conformation. Simulations of Aβ1–28 dimers with the OPEP force field 35 also showed that the N-terminal regions form β -hairpins that can participate in intermolecular β-sheets, supporting the scenario described here.
In the structural models of the fibrils formed by Aβ1–40 and Aβ1–42, 36,37 the monomers adopt a conformation with a turn around residues 26–27 that permits the formation of a stabilizing salt bridge between Asp 23 and Lys 28. The N-terminal region (residues 1–8 in Aβ1–40 and 1–17 in Aβ1–42) is disordered and does not contribute to stabilize the fibrils. Therefore, it is surprising that the same N-terminal region plays such an important role in Aβ1–28 aggregation. This apparent discrepancy can be explained by comparing our results with those obtained for different Aβ species. Recent μs long REMD all-atom simulations of Aβ1–42 in explicit solvent, 20 showed that a β -turn about residues 7–8 forms in ≈ 50% of the ensemble conformations, with the residues around the turn (3–6 and 10–13) often forming a β -hairpin. The 26–27 turn is not found as often, but when it is found, it can be stabilized by a β -sheet between residues 16–21 and 29–36 with the peptide adopting a fibril-like conformation. This work also showed that Aβ1–42 can sample diverse conformations that often combine small α-helices and β -sheets, and the residues that were found to form an α-helix in one conformation might participate in a β -sheet, a turn, or a random coil in another conformation. MD simulations of Aβ1–42 with a different force field also showed that residues 3–5 and 7–11 form a β -hairpin that later forms a β -sheet with residues 33–37, allowing residues 24–27 to form a turn.14 In a different study, Reddy et al.16 used all-atom MD simulations to study the formation of the Asp 23-Lys 28 salt bridge on Aβ10–35. The authors concluded that the conformations containing the salt bridge were rarely sampled, and that hydrophobic interactions between the residues at N- and C-terminal regions stabilized these conformations. Based on the aforementioned results, we conclude that, because Aβ1–28 lacks residues 29–36, the 26–27 turn together with Asp 23-Lys 28 salt bridge cannot be stabilized. Therefore, in the fibrils formed by Aβ1–28, the monomers might not have the 26–27 turn and the Asp 23-Lys 28 salt bridge. The aggregation mechanism might also be different from that of Aβ1–40 and Aβ1–42 in which other competent conformations can initiate the process. However, given the high tendency of the N-terminal residues of Aβ1–42 to form a β -hairpin (as shown in20 and14), it is possible that this region could initiate aggregation in this peptide as well, allowing the formation of a nucleus that can serve as a platform for new monomers to bind and adopt the fibril-like conformation. Results from our previous work18 indicate that Aβ1–40 monomers do not adopt the fibril-like conformation prior to binding, but that a conformational change occurs when locking themselves into the fibril. Therefore, even when the N-terminal region can form a β -hairpin, because of the high diversity of conformations adopted by all regions in the peptides, the same residues could be disordered upon reorganization into the fibril.
The N-terminal hairpin is not driven by the oligomerization process
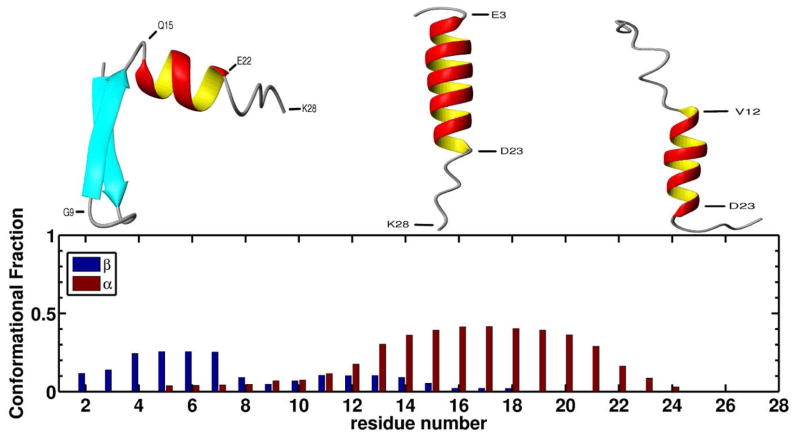
As in the example shown in Figure 2b, oligomerization was often initiated by the association of two β -hairpins from different chains, forming an intermolecular β -sheet. In order to assess whether the formation of the hairpins was promoted by the interactions between the monomers or whether it was a conformation that the monomers would adopt in isolation, we carried out REMD simulations of isolated monomers. Figure 4 shows the α-helix and β -sheet conformational fractions for each residue (see Methods). As can be seen from the Figure, residues 13 to 21 have a high tendency to adopt α-helical conformations while the residues in the N-terminal region (3 to 7 and to a lesser degree 11 to 14) tend to adopt β -sheet conformations. Residues 5 to 18 have the ability to adopt either α-helix or β -sheet conformations. Three snapshots, representative of the typical conformations found during the simulations are shown in Figure 4. The different snapshots show the capability of the N-terminal half of the peptide to form a β -hairpin (left) or an α-helix (center), and how the N-terminal α-helix can partially unwind (right). Therefore, the N-terminal β -hairpin is not driven by protein-protein interactions. However, when a β -hairpin is formed, upon interaction with other chains, it can be stabilized and serve as a seed for oligomerization, as in the example shown in Figure 2b.
Figure 4.
(top) Three representative snapshots of the typical conformations found during REMD simulations of isolated monomers. The three snapshots show residues 15 to 23 forming an α-helix, and illustrate the ability of the N-terminal region to form a β -hairpin (left) or an α-helix (center) that can partially unwind (right). (bottom) The α-helix (red) and β -sheet (blue) conformational fractions for each residue, calculated from all snapshots at 300 K.
According to the NMR model of Zagorski and Barrow,4 Aβ1–28 can adopt two possible motifs: one in which residues 2–27 form an α-helix, and another one in which two α-helices are formed along residues 2–7 and 10–27. Our simulations produced less structured peptides, with residues 1–8 and 25–28 very rarely adopting an α-helical conformation (see Figure 4). This difference is expected because the NMR studies4 were carried out in a mixture of water and TFE-d3, and TFE is known to stabilize α-helical conformations.
Raman spectroscopy experiments indicate that Aβ1–28 exists in a mixture of polyprolineII (PPII) and β -strand conformations in acidic solution.38 We did not observe PPII conformations in our simulations. However, in the present UNRES,22 the side-chain - side-chain interaction potentials are statistical potentials determined from the Protein Data Bank.39 Therefore, for charged amino-acid side chains, they correspond to the pH at which the proteins from the database were crystallized, i. e., at the physiological pH in most cases. Consequently, all acidic and basic residues are modeled in their ionized state, while the acidic residues are expected to be protonated at acidic pH. We are now replacing the statistical potential with physics-based potentials,40,41 which will include pH dependence for acidic and basic residues.
It should be noted that recent studies20,42 have shown that the AMBER43 force field with the new ff99SB parameters,44 can underestimate the PPII content especially for Ala residues.
Conclusions
The results of our UNRES simulations of the Aβ1–28 peptide system have demonstrated that the N-terminal half of the peptide has a propensity to form a β -hairpin in the monomeric state, while the C-terminal half readily forms an α-helix. This property of the peptide seems to determine the mechanism of aggregation. Oligomerization starts from the N-terminus by association of the N-terminal β -hairpins. Once these events have taken place and the two α-helices (one from each molecule) come close to each other, they interact and unfold. Then, the next monomer joins in, initially associating with one of the unfolded helices of the dimer by its β -hairpin segment (Figure 2b). The β -hairpins eventually straighten to form longer intermolecular β -sheet segments. Consistent with experiments,31 our simulations starting from the α-helical conformation and stabilizing the helix between residues 17 and 21 showed that preserving this helical region inhibits the propagation of association (Figure 2a and the last column of three graphs in Figure 3). From these simulations, it follows that association is stopped at the dimer stage because the α-helical segments cannot unfold and join the β -sheets. Another fact supporting the oligomerization scenario outlined above is that, by starting the simulations from fully α-helical conformations, the simulated oligomerization slows down; the reason for this is that the α-helix in the N-terminal segment has to unfold and convert into a β -hairpin to initiate dimer formation. However, helix formation in the C-terminal half of the monomeric peptide can facilitate association as inferred from experiments10,33 because it forces association to start in an ordered way from the N-terminal half, preventing the formation of irregular portions of strands that are difficult to be joined by another molecule.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM-14312), the National Science Foundation (MCB10-19767), and by the CCT Graduate Assistantship Program from Louisiana State University. This research was conducted by using the resources of (a) our 616-processor Beowulf cluster at Baker Laboratory of Chemistry, Cornell University, (b) the National Science Foundation Terascale Computing System at the Pittsburgh Supercomputer Center, and (c) the Informatics Center of the Metropolitan Academic Network (IC MAN) in Gdańsk.
References
- 1.Lorenzo A, Yuan M, Zhang Z, Paganetti PA, Sturchler-Pierrat C, Staufenbiel M, Mautino J, Vigo FS, Sommer B, Yankner BA. Nat Neurosci. 2000;3:460–464. doi: 10.1038/74833. [DOI] [PubMed] [Google Scholar]
- 2.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 3.Barrow CJ, Yasuda A, Kenny PTM, Zagorski MG. J Mol Biol. 1992;225:1075–1093. doi: 10.1016/0022-2836(92)90106-t. [DOI] [PubMed] [Google Scholar]
- 4.Zagorski MG, Barrow CJ. Biochemistry. 1992;31:5621–5631. doi: 10.1021/bi00139a028. [DOI] [PubMed] [Google Scholar]
- 5.Sticht H, Bayer P, Willbold D, Dames S, Hilbich C, Beyreuther K, Frank RW, Rösch P. Eur J Biochem. 1995;233:293–298. doi: 10.1111/j.1432-1033.1995.293_1.x. [DOI] [PubMed] [Google Scholar]
- 6.Crescenzi O, Tomaselli S, Guerrini R, Salvadori S, D’Ursi AM, Temussi PA, Picone D. Eur J Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 7.Tomaselli S, Esposito V, Vangone P, van Nuland NAJ, Bonvin AM, Guerrini R, Tancredi T, Temussi PA, Picone D. ChemBioChem. 2006;7:257–267. doi: 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- 8.Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ. Biochemistry. 1998;37:11064–11077. doi: 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- 9.Shao H, Jao S, Ma K, Zagorski MG. J Mol Biol. 1999;285:755–773. doi: 10.1006/jmbi.1998.2348. [DOI] [PubMed] [Google Scholar]
- 10.Kirkitadze MD, Condron MM, Teplow DB. J Mol Biol. 2001;312:1103–1119. doi: 10.1006/jmbi.2001.4970. [DOI] [PubMed] [Google Scholar]
- 11.Kirshenbaum K, Daggett V. Biochemistry. 1995;34:7629–7639. doi: 10.1021/bi00023a009. [DOI] [PubMed] [Google Scholar]
- 12.Khandogin J, Brooks CL., III Proc Natl Acad Sci U S A. 2007;104:16880–16885. doi: 10.1073/pnas.0703832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fawzi NL, Okabe Y, Yap EH, Head-Gordon T. J Mol Biol. 2007;365:535–550. doi: 10.1016/j.jmb.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raffa DF, Rauk A. J Phys Chem B. 2007;111:3789–3799. doi: 10.1021/jp0689621. [DOI] [PubMed] [Google Scholar]
- 15.O’Brien EP, Okamoto Y, Straub JE, Brooks BR, Thirumalai D. J Phys Chem B. 2009;113:14421–14430. doi: 10.1021/jp9050098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy G, Straub JE, Thirumalai D. J Phys Chem B. 2009;113:1162–1172. doi: 10.1021/jp808914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy G, Straub JE, Thirumalai D. Proc Natl Acad Sci USA. 2009;106:11948–11953. doi: 10.1073/pnas.0902473106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rojas A, Liwo A, Browne D, Scheraga HA. J Mol Biol. 2010;404:537–552. doi: 10.1016/j.jmb.2010.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee C, Ham S. J Comput Chem. 2011;32:349–355. doi: 10.1002/jcc.21628. [DOI] [PubMed] [Google Scholar]
- 20.Ball KA, Phillips AH, Nerenberg PS, Fawzi NL, Wemmer DE, Head-Gordon T. Biochemistry. 2011;50:7612–7628. doi: 10.1021/bi200732x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klimov DK, Thirumalai D. Structure. 2003;11:295–307. doi: 10.1016/s0969-2126(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 22.Liwo A, Ołdziej S, Pincus MR, Wawak RJ, Rackovsky S, Scheraga HA. J Comput Chem. 1997;18:849–873. [Google Scholar]
- 23.Liwo A, Czaplewski C, Pillardy J, Scheraga HA. J Chem Phys. 2001;115:2323–2347. [Google Scholar]
- 24.Ołdziej S, Ła̧giewka J, Liwo A, Czaplewski C, Chinchio M, Nanias M, Scheraga HA. J Phys Chem B. 2004;108:16950–16959. [Google Scholar]
- 25.Liwo A, Khalili M, Czaplewski C, Kalinowski S, Ołdziej S, Wachucik K, Scheraga H. J Phys Chem B. 2007;111:260–285. doi: 10.1021/jp065380a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rojas AV, Liwo A, Scheraga HA. J Phys Chem B. 2007;111:293–309. doi: 10.1021/jp065810x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liwo A, Czaplewski C, Ołdziej S, Rojas AV, KaŸmierkiewicz R, Makowski M, Murarka RK, Scheraga HA. In: Coarse-Graining of Condensed Phase and Biomolecular Systems. Voth G, editor. Taylor & Francis; 2008. pp. 107–122. [Google Scholar]
- 28.He Y, Xiao Y, Liwo A, Scheraga HA. J Comput Chem. 2009;30:2127–2135. doi: 10.1002/jcc.21215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikros E, Benaki D, Humpfer E, Spraul M, Loukas S, Stassinopoulou CI, Pelecanou M. Angew Chem Int Ed Engl. 2001;40:3603–3605. doi: 10.1002/1521-3773(20011001)40:19<3603::AID-ANIE3603>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 30.Talafous J, Marcinowski KJ, Klopman G, Zagorski MG. Biochemistry. 1994;33:7788–7796. doi: 10.1021/bi00191a006. [DOI] [PubMed] [Google Scholar]
- 31.Kapurniotu A, Buck A, Weber M, Schmauder A, Hirsch T, Bernhagen J, Tatarek-Nossol M. Chem Biol. 2003;10:149–159. doi: 10.1016/s1074-5521(03)00022-x. [DOI] [PubMed] [Google Scholar]
- 32.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 33.Fezoui Y, Teplow DB. J Biol Chem. 2002;277:36948–36954. doi: 10.1074/jbc.M204168200. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Liu R, Lam KS, Jin L-W, Duan Y. Biophys J. 2011;100:1076–1082. doi: 10.1016/j.bpj.2010.12.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong X, Chen W, Mousseau N, Derreumaux P. J Chem Phys. 2008;128:125108. doi: 10.1063/1.2890033. [DOI] [PubMed] [Google Scholar]
- 36.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petkova AT, Yau W-M, Tycko R. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eker F, Griebenow K, Schweitzer-Stenner R. Biochemistry. 2004;43:6893–6898. doi: 10.1021/bi049542+. [DOI] [PubMed] [Google Scholar]
- 39.Bernstein FC, Koetzle TF, Williams GJB, Meyer EFJ, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 40.Makowski M, Liwo A, Sobolewski E, Scheraga HA. J Phys Chem B. 2011;115:6119–6129. doi: 10.1021/jp111258p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makowski M, Liwo A, Scheraga HA. J Phys Chem B. 2011;115:6130–6137. doi: 10.1021/jp111259e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nerenberg PS, Head-Gordon T. Journal of Chemical Theory and Computation. 2011;7:1220–1230. doi: 10.1021/ct2000183. [DOI] [PubMed] [Google Scholar]
- 43.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Wang B, Hayik S, Roitberg A, Seabra G, et al. AMBER 11. University of California; San Francisco: 2010. [Google Scholar]
- 44.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]