Abstract
Upon completion of synthesis of an Okazaki fragment, the lagging strand replicase must recycle to the next primer at the replication fork in under 0.1 second to sustain the physiological rate of DNA synthesis. We tested the collision model that posits that cycling is triggered by the polymerase encountering the 5′-end of the preceding Okazaki fragment. Probing with surface plasmon resonance, DNA polymerase III holoenzyme initiation complexes were formed on an immobilized gapped template. Initiation complexes exhibit a half-life of dissociation of approximately 15 minutes. Reduction of gap size to one nucleotide increased the rate of dissociation 2.5-fold and complete filling of the gap increased the off rate an additional three-fold (t½ ∼ 2 min). An exogenous primed template and ATP accelerated dissociation an additional four-fold in a reaction that required complete filling of the gap. Neither a 5′-triphosphate nor 5′-RNA terminated oligonucleotide downstream of the polymerase accelerated dissociation further. Thus, the rate of polymerase release upon gap completion and collision with a downstream Okazaki fragment is 1000-fold too slow to support an adequate rate of cycling and likely provides a backup mechanism to enable polymerase release when the other cycling signals are absent. Kinetic measurements indicate that addition of the last nucleotide to fill the gap is not the rate-limiting step for polymerase release and cycling. Modest (approximately 7 nucleotide) strand displacement is observed after the gap between model Okazaki fragments is filled. To determine the identity of the protein that senses gap filling to modulate affinity of the replicase for the template, we performed photo-crosslinking experiments with highly reactive and non-chemoselective diazirines. Only the α subunit cross-linked, indicating it serves as the sensor.
Introduction
Cellular chromosomal replicases from all branches of life are tripartite. They contain a polymerase (Pol III in bacteria and Pol δ and ε in eukaryotes), a sliding clamp processivity factor (β2 in bacteria and PCNA in eukaryotes), and a clamp loader (DnaX complex in bacteria and RFC in eukaryotes) 1–3. The E. coli replicase, the DNA polymerase III holoenzyme (Pol III HE), has the processivity required to replicate >150 kb 4,5, and perhaps the entire E. coli chromosome, without dissociation. Yet, the lagging strand polymerase must be able to efficiently cycle to the next primer synthesized at the replication fork upon the completion of each Okazaki fragment at a rate faster than Okazaki fragment production. The rate of replication fork progression in E. coli is about 600 nt/s at 30° C,2 approximately the rate of replicase progression on single-stranded DNA (ssDNA) templates.6 Thus, most of the time in an Okazaki fragment cycle is spent on elongation and little time (< 0.1 s) remains for the polymerase to release, bind the next primer, and begin synthesis. A processivity switch must be present to increase the off-rate of the lagging strand polymerase by several orders of magnitude.
Upon complete replication of primed single strand DNA, Pol III* (a complex of Pol III and DnaX complexes containing the τ form of DnaX that binds Pol III) dissociates from a duplex and leaves the β2 sliding clamp behind 7. There are two competing but non-exclusive models for the signal that throws the processivity switch. The first, the signaling model, proposes that a signal is provided by synthesis of a new primer at the replication fork that induces the lagging strand polymerase to dissociate, even if the Okazaki fragment has not been completed 8. The second, the collision model, originally proposed for T4 9 and then extended to the E. coli system 10, posits that the lagging strand polymerase replicates to the last nucleotide 10 or until the Okazaki fragment is nearly complete, signaling polymerase dissociation 11. A communication circuit that proceeds through the τ subunit has been proposed to sense the conversion of a gap to a nick, signaling release 10,12,13. τ was proposed to act by competing with a C-terminal site on Pol III for binding to β. However, other work has shown the essential site required for β interaction to be internal within the Pol III α subunit 14–16. Experiments designed to test whether the signaling or collision models are dominant have yielded equivocal results 17.
In the more fully characterized systems provided by the replication apparatus of bacteriophages T4 and T7, signaling through synthesis or the availability of a new primer appears to play an important role, with the collision pathway playing an apparent backup function 18–20. In T4, handoff of the nascent pentaribonucleotide primer occurs in a reaction that is facilitated by T4 ssDNA binding protein (T4 SSB) and the clamp and the clamp loader 21,22. A proposal was made that the T4 clamp loader/clamp interaction with a new primer might be the key signal required for release of the lagging strand polymerase 22.
In this report, we perform a kinetic assessment of the collision model on templates constructed such that elongating Pol III HE encounters an oligonucleotide hybridized to the template that mimics the 5′ terminus of the preceding Okazaki fragment on the lagging strand of a replication fork. We compared the rate of Pol III* release with the rate of addition of the last nucleotide. To determine candidate holoenzyme subunits that could serve as a sensor for completion of lagging strand synthesis, we identified the protein that contacts the template immediately ahead of the primer terminus and the 5′-end of the preceding Okazaki fragment.
Results
Most measurements assessing pathways that trigger Pol III* release and cycling have been performed using equilibrium binding measurements. While a decreased affinity of Pol III* for the template might be consistent with a role in accelerating release, the relevant issue is whether the lagging strand is triggered to release in less than 0.1 s upon collision with the preceding Okazaki fragment. To provide a method for quantitative assessment of the kinetics of Pol III* release, we set up a surface plasmon resonance (SPR) assay in which primers were placed on a surface-immobilized template a set number of nucleotides from an oligonucleotide that models the 5′-end of the preceding Okazaki fragment (Fig. 1a). Initiation complexes are mobile on DNA and unless bulky blocking agents flank the complex, it rapidly slides off the ends of the template 23. We placed biotin near the 3′-end of the template to permit attachment to a streptavidin-covered surface for SPR measurements. The other end of the construct was blocked by covalently attaching the 3′ terminus of the blocking oligonucleotide to His6-tagged O6-alkylguanine-DNA-transferase (SNAP tag)24. The sequence of the template downstream of the primer was designed to permit controlled advancement of the elongating Pol III HE by a subset of dNTPs.
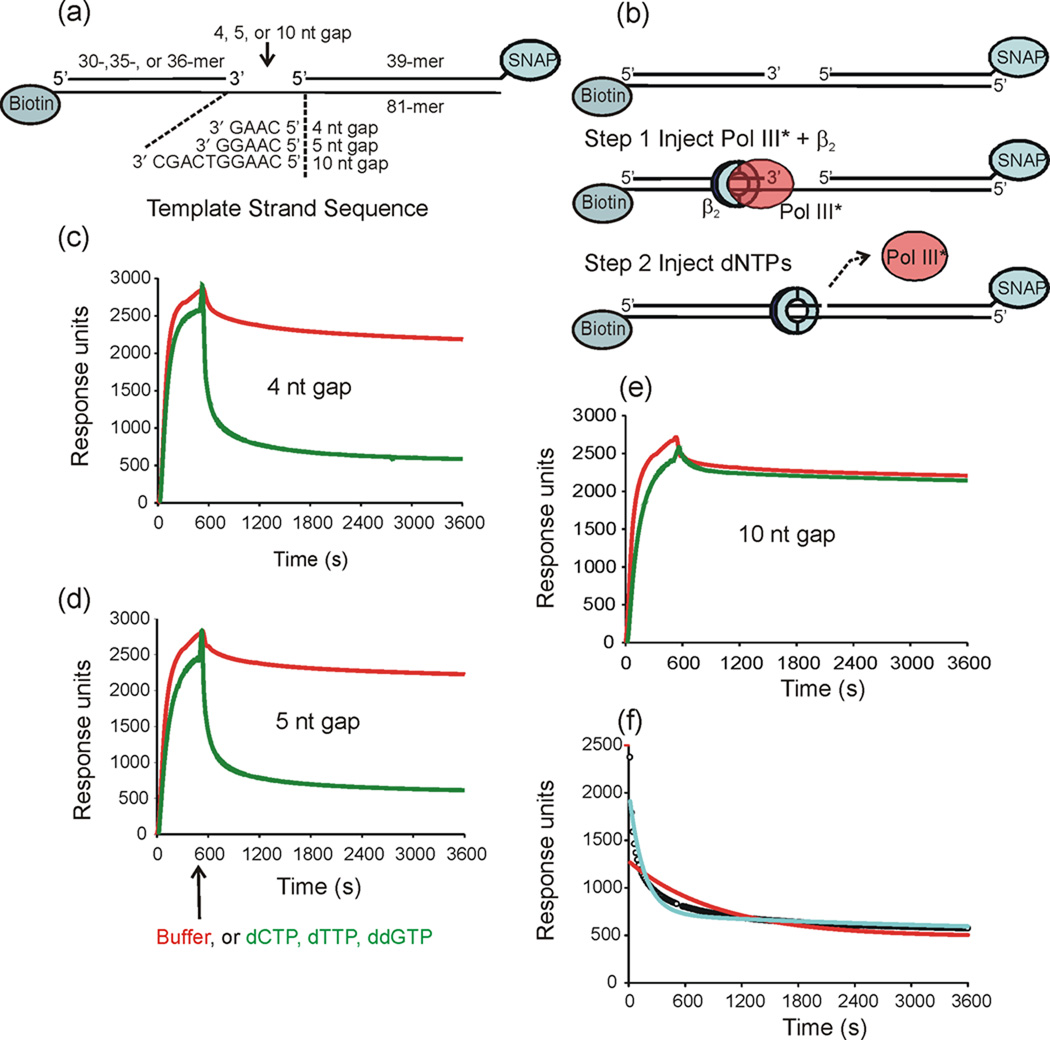
Fig. 1.
Pol III* does not release rapidly upon filling a gap. The dissociation of Pol III* from primed templates was measured using surface plasmon resonance. (a) Three primed templates containing 4-, 5- and 10-nt gaps were anchored to the surface of a streptavidin-coated sensor chip using biotin-labeled oligonucleotides. The 3′-end of the blocking oligonucleotide was covalently linked to the SNAP protein to prevent β2 from sliding off the end. (b) Initiation complexes were assembled onto the immobilized DNA primer-template by injecting Pol III*, β2 and ATP. A second injection contained either buffer or a solution containing 40µM dCTP, dTTP, and ddGTP (dNTPs) was made at 540 s. (c) (d) and (e) Injection of either buffer (red) or dNTPs (green) over the 4-nt gapped template 5-nt gapped template, and 10-nt gapped template, respectively. Dissociation was allowed to continue for three days. The data shown is only for the first h after injection. (f) Curve-fit analysis is shown for the for Pol III* dissociation in the presence of dNTPs over a 4-nt gapped template: data (open circles), curve fit to single exponential decay (red), curve fit to double exponential decay (cyan).
We observed stabile association of Pol III HE with each primer-template to form an initiation complex that dissociated with a half life of 10–15 min (Fig. 1c–e). Addition of the full complement of dNTPs required for conversion of the gap to a nick greatly increased the amount of the Pol III* released (Fig. 1c,d) and modestly (3- to 8-fold) increased the dissociation rate (Table I). However, the half life of polymerase release (∼2–5 min) is approximately 1000-fold slower than that expected to support the physiological rate of cycling upon completion of an Okazaki fragment. We observed that addition of any nucleotide, independent of whether the nucleotides present were able to fill the gap completely led to an increase in the amount of Pol III* that dissociates (amplitude column in Table I), but complete gap filling is needed to achieve the maximal rate.
Table 1.
Filling a gap completely is required to achieve the maximal rate of Pol III* release
| Injection | 4 nt gap template t1/2 [amplitude] |
Gap size |
5 nt gap template t1/2 [amplitude] |
Gap size |
|---|---|---|---|---|
| Buffer | 860 s [12%] | 4 | 620 s [10%] | 5 |
| dCTP | 390 s [69%] | 3 | 360 s [70%] | 3 |
| ddCTP | 660 s [25%] | 3 | 380 s [26%] | 4 |
| dCTP, dTTP | 210 s [62%] | 1 | 190 s [63%] | 1 |
| dCTP, ddTTP | 370 s [36%] | 2 | 350 s [37%] | 2 |
| dCTP, dTTP, dGTP | 290 s [55%] | 0 | 180 s [57%] | 0 |
| dCTP, dTTP, ddGTP | 110 s [64%] | 0 | 110 s [63%] | 0 |
Kinetic analysis was complicated by a second, more slowly dissociating component that required analysis using a double exponential decay equation (Fig 1f). The second component dissociated with a half-life of 1 – 5 h. This required each dissociation experiment to be run for up to 3 days to acquire a data set that allowed for complete decay (10 half lives) for an accurate curve fit. In the text of the main paper, we refer to the presumably biologically relevant fast dissociating component, and present the full data under Supplementary Data (Supplementary Tables S1–S3).
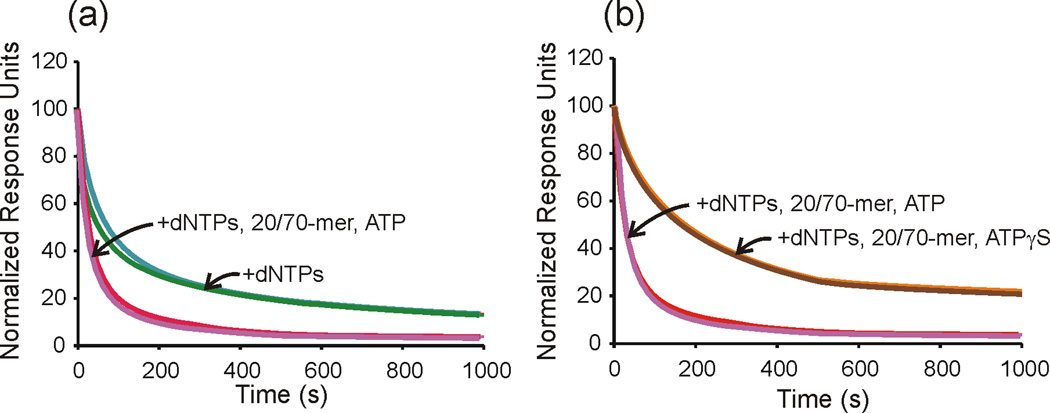
Recognizing that simple conversion of a gap to a nick did not provide an adequate rate of Pol III* release to achieve a physiologically relevant rate of cycling for Okazaki fragment synthesis, we sought external effectors that might stimulate dissociation. We found that the presence of ATP and an exogenous template-primer accelerates dissociation approximately four-fold (Fig. 2, Table 2). Raising the exogenous primed template concentration in the analyte solution did not further increase the rate. ATPγS will not substitute in this reaction, suggesting that ATP hydrolysis is required. The rate enhancement by these factors is only fully achieved when a gap has been filled. SSB and exogenous β2 are not required to accelerate Pol III release (Table 2).
Fig. 2.
An exogenous primed template and ATP accelerate Pol III* release from templates with filled gaps. The dissociation of Pol III* from primed template was measured using surface plasmon resonance following a second injection of dNTPs (40 µM dCTP, dTTP, ddGTP) alone or dNTPs plus additional components (+1 mM ATP or ATPγS, and 0.5 µM exogenous 20/70-mer). Data shown are normalized response units vs. time for the first 1000 s of dissociation. (a) Curves shown are for dNTPs alone with a 4-nt gapped template (green) and a 5-nt gapped template (teal) or for a dNTPs + 20/70-mer + ATP with 4-nt gapped template (magenta) and 5-nt gapped template (red). (B) The slowly hydrolyzed ATP analog ATPγS decreases the off-rate of Pol III* in the presence of dNTPs and exogenous 20/70-mer. Curves shown are for dNTPs + 20/70-mer + ATP with a 4-nt gapped template (magenta) and a 5-nt gapped template (red) or dNTPs + 20/70-mer + ATPγS with a 4-nt gapped template (brown) and 5-nt gapped template (dark orange). The curves for the 4 nt and 5 nt gapped templates are coincident and not resolved at most times shown.
Table 2.
Exogenous primed template and ATP stimulates Pol III* release from completed Okazaki Fragments.
| Injection | 4 nt gap template t1/2 [amplitude] |
5 nt gap template t1/2 [amplitude] |
|---|---|---|
| ATP | 990 s [60%] | 920 s [62%] |
| dNTPs | 110s [64%] | 110s [64%] |
| dNTPs, 20/70-mer, ATP | 30 s [86%] | 40 s [83%] |
| dNTPs, ATP | 60 s [75%] | 60 s [77%] |
| 20/70-mer, ATP | 330 s [52%] | 340 s [51%] |
| dNTPs, 20/-70-mer | 60 s [80%] | 70 s [77%] |
| dNTPs, 20/-70-mer, ATP, SSB4 | 50 s [77%] | 50 s [73%] |
| dNTPs, 20/-70-mer, ATP, SSB4, | 30 s [64%] | 30 s [59%] |
| β2 dNTPs, 20/-70-mer, ATPγS |
130 s [68%] | 130 s [67%] |
The blocking oligonucleotide used in our assay to mimic the preceding Okazaki fragment contained a 5′-hydroxyl group. The preceding fragment in cells would be expected to contain a 5′-triphosphate to be composed of RNA for ca. 10 nucleotides at the 5′-end. To determine whether either of these factors led to more rapid polymerase release, we had blocking oligonucleotides synthesized that contained either a 5′-triphosphate or a mixed RNA/DNA oligonucleotide with 12 RNA nucleotides at the 5′-end and a single 5′-phosphate. Neither of these modifications led to acceleration of release (Table 3). The experiments described so far were conducted using Pol III HE with the presumed physiological Pol III* composition: Pol III2-τ2γδδ′χ ψ1. To ensure that a dimeric polymerase was not the cause of slow release, we also prepared an assembly with a Pol III-τγ2δδ′χ ψ composition. Upon addition of dNTPs on the 4- and 5-nucleotide gapped templates, polymerase dissociated with a half life of 140 and 130s, respectively—similar to the 110s observed with the dimeric polymerase assembly. We observed a similar dissociation rate (t½ =140 s) upon the addition of dNTPs to allow gap filling for Pol III HE with a Pol III3-τ3γδδ′χ ψ1 composition.
Table 3.
RNA or a triphosphate on the 5′-terminus of the preceding Okazaki fragment do not contribute to the rate of Pol III* release
| Injection | 4 nt gap template t1/2 [amplitude] |
5 nt gap template t1/2 [amplitude] |
|---|---|---|
| 5′-OH DNA39 blocking oligo | 30 s [86%] | 40 s [83%] |
| 5′ PO4 RNA12/DNA27 blocking | 60 s [71%] | 70 s [68%] |
| oligo 5′ Tri-PO4 DNA39 blocking oligo |
80 s [51%] | 90 s [44%] |
The rate of polymerase release that we observe is much slower than the rate of less than or equal to (0.5 s−1; t1/2∼1.4 s) reported previously 10. However, re-analysis of the published data yielded a fit to a single exponential with a half-life of 14 s (0.05 s−1), too slow to support the rate of in vivo Okazaki fragment synthesis, albeit faster than the rates we observe in this work (Supplementary Fig. S1).
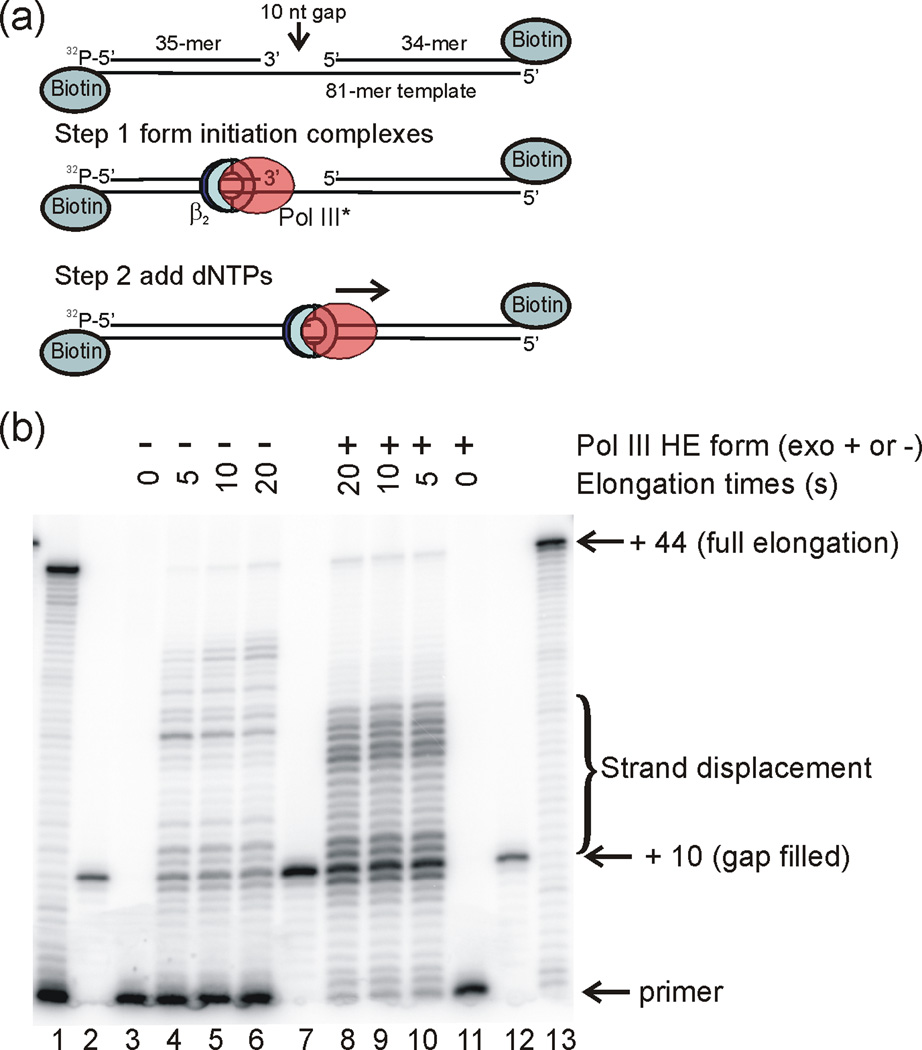
Our results are consistent with gap completion providing a signal for cycling, but with release slower than would be expected for cycling during Okazaki fragment synthesis. We wanted to determine whether the slow rate of polymerase release was due to a slow process after Okazaki fragment completion such as a conformational change or whether the rate of incorporation of the final nucleotide(s) to complete the fragment was slow. To enable these measurements, we constructed templates similar to those used for the SPR experiments with 5′-32P-labeled primers. This permitted monitoring the rate of primer elongation and gap completion using high resolution polyacrylamide gel electrophoresis. After formation of initiation complexes with Pol III HE on the primer-template, elongation reactions were initiated by addition of dNTPs. The majority (79%) of primer-templates are completely gap-filled before the first manually sampled time point could be taken (5 s), a time scale faster than Pol III* dissociation (Fig. 3). Thus, dissociation is not limited by the rate of gap filling. The nicked duplex represents 22% of all elongated products. Interestingly, most of the elongation product (57% for wild-type Pol III HE) corresponds to a modest level of displacement of the blocking oligonucleotide. The wild-type Pol III HE forms significant products up to 14 nt beyond the blocking oligonucleotide, and the average strand displacement is approximately 7 nucleotides.
Fig. 3.
Pol III HE rapidly fills a 10 nt gap and partially displaces a blocking oligonucleotide. (a) 32P-5′ end labeled primer 35-mer was hybridized to a template 81-mer with a 10 nt gap in front of blocking oligonucleotide 34-mer. This primer-template was incubated with Pol III HE and ATP for 60 s to form initiation complexes, followed by addition of dNTPs to initiate primer elongation for the 32P-labeled complexes. (b) Denaturing polyacrylamide gel analysis of the rate of nucleotide incorporation. The gel lanes were loaded with 1) unblocked primer-template fully extended by Pol III HE; 2) 32P-5′ end labeled 45-mer corresponding to a fully gap-filled product, annealed to the 81-mer template; 3) unextended 10 nt gap primer-template; 4) 10 nt gap primer-template after 5 s extension time; 5) 10 nt gap primer-template after 10 s extension time; 6) 10 nt gap primer-template after 20 s extension time; 7) 45-mer annealed to the 81-mer template (4-fold more sample than loaded than in lanes 2 and 12); 8) 10 nt gap primer-template after 20 s extension time; 9) 10 nt gap primer-template after 10 s extension time; 10) 10 nt gap primer-template after 5 s extension time; 11) unextended 10 nt gap primer-template; 12) 45-mer annealed to the 81-mer template; 13) unblocked primer-template fully extended by Pol III HE. The samples in lanes 1 and 4 – 6 were extended by exonuclease-deficient Pol III HE. The samples in Lanes 8 – 10 and 13 were extended by wild-type Pol III HE.
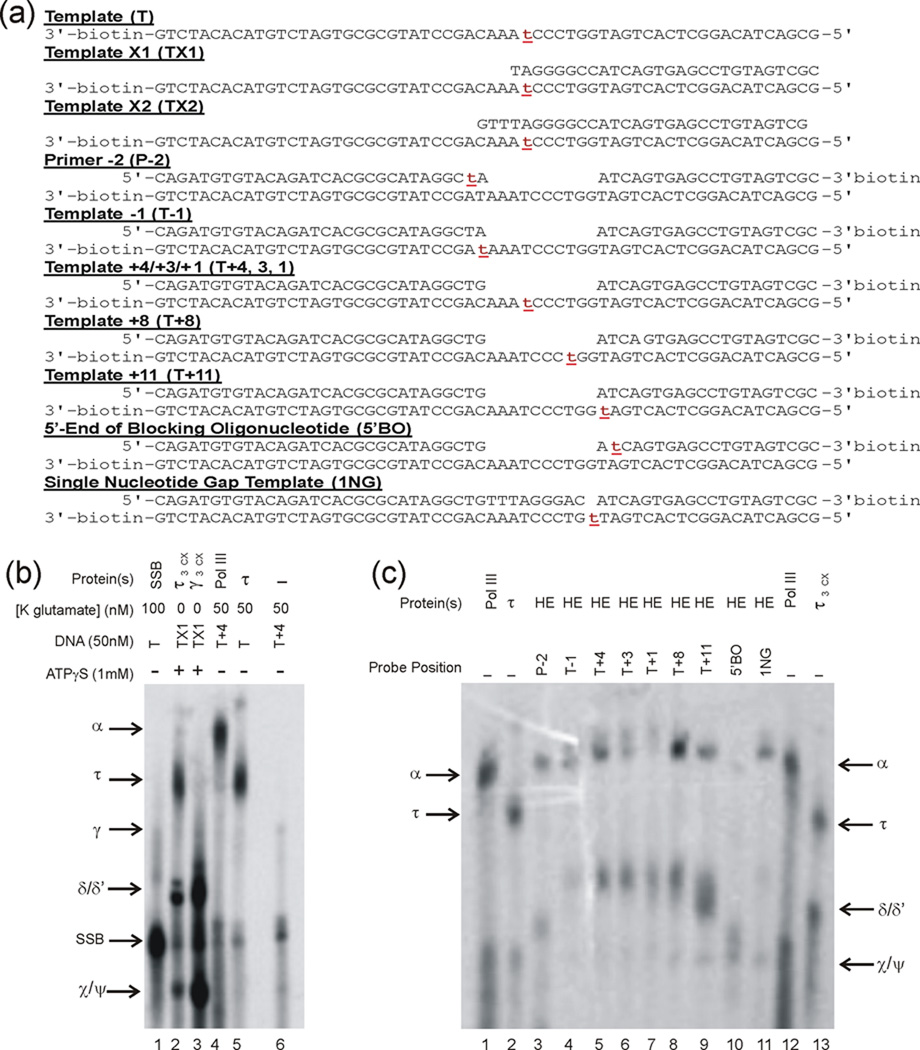
Previous models have proposed roles for either τ or α to serve as the sensor for conversion of a gap to a nick upon completion of Okazaki fragment synthesis, serving as a trigger for accelerated polymerase release 10,11,25,26. To determine the proteins that contact the template and blocking oligonucleotide immediately ahead of the primer terminus, we performed cross-linking experiments. A phenyldiazirine, linked to the 5-position of thymidylate was placed at a unique position in a series of templates that mimicked those used in the SPR experiments (Fig. 4a). We chose diazirines because, when irradiated, they generate carbenes that efficiently insert into C–H bonds of all amino acids 27. This permits interpretation of negative results with certainty—if a protein does not crosslink, it is not bound to the template position to which the diazirine is linked.
Fig. 4.
The Pol III α subunit and not τ is positioned to serve as the sensor for the completion of Okazaki fragment synthesis (a) DNA constructs used to determine Pol III HE contacts with model templates. The position of the phenyldiazirine photo-crosslinker is indicated by red lowercase t. The oligonucleotide containing the photocrosslinker was labeled with 32P on the 5′-end prior to annealing. Template T+4 also served as Templates T+3 and T+1 by the addition of 10 µM ddTTP or dTTP, respectively. (b) Denaturing polyacrylamide gel analysis of Pol III HE subunit standards. Generation of lanes 1–3 is described in Supplementary Fig. S4. Pol III (0.62 µM final concentration) used to generate lane 4 and τ (2 µM) used to generated lane 5. (c) Identification of protein contacts with model templates. Photo-crosslinking took place within initiation complexes formed with the designated model templates. The gel lanes were loaded with approximately equal counts of radioactivity. Lanes 1, 2, 12, and 13 are identical to lanes 4, 5, 4 and 2, respectively, in panel b.
To enable assignment of crosslinked proteins by their electrophoretic mobility, we forced template-protein contacts with high concentrations of single or simple subsets of Pol III HE subunits to generate standards (Supplementary Fig. S4 and Fig. 4b). Control experiments were performed with the phenyldiazirine placed at the penultimate (−2) position at the 3′-end of the primer terminus and at the template position opposite the 3′-primer terminus. As expected from previous work25,28, α was observed to crosslink. Cross-linking at template positions one, three, four and eight nucleotides ahead of the primer and at the template position opposite the 5′-nucleotide of the blocking oligonucleotide led to the cross-linking of the α subunit of Pol III and not τ (Fig 4c). We also observed a crosslink to single-stranded template positions that may correspond to either δ or δ′. In a control experiment, where the material in lane 8 of Fig 4c was mixed with lane 2, the τ and δ/δ′ crosslinks were clearly distinguishable (data not shown). These species may represent two binding states—one with Pol III bound to the primer terminus and another with DnaX complex bound. Crosslinks to α were also observed to the nucleotide in the gap for templates containing a single nucleotide gap (Fig 4c). When phenyldiazirine was placed at the penultimate 5′-position of the blocking oligonucleotide, a very low yield of crosslinks to α was observed, consistent with the strand displacement observed upon completion of synthesis (Fig.3). This result suggests that displaced blocking oligonucleotide does not maintain contact with Pol III. Thus, α and not τ is the only protein positioned to function as the sensor for gap completion.
Discussion
To permit a kinetic evaluation of the rate of polymerase release upon completing Okazaki fragment synthesis, we immobilized model templates on a surface, assembled initiation complexes, and monitored polymerase release upon nucleotide addition by SPR. We found that conversion of a gap to a nick accelerates release, but the rate is far too slow to support the physiological rate of Okazaki fragment synthesis. The differences from other published models likely arises from their derivation from equilibrium measurements and ours from kinetic determinations.
In a search for factors that accelerate collision-induced dissociation, we found that a mixture of ATP and an exogenous primed template accelerates dissociation four-fold. ATPγS will not substitute in this reaction, suggesting that ATP hydrolysis is required. These findings partially reconcile previous models and observations pertaining to the requirement for gap completion 10, the availability of a new template primer 8, and the requirement for τ 10. A series of switches may exist, all of which must be accommodated for maximal cycling. Gap completion may put the Pol III HE in a state in which a signal can be received from the associated τ-containing DnaX complex that involves binding of a new primer template and ATP hydrolysis. Additional acceleration might be derived from other factors or their arrangement/ interaction at the fork. Or, the collision mechanism might only be a backup, with the primary signal provided by events associated with new primer generation. The Pol III HE is involved in mismatch repair and other repair reactions where large stretches of DNA need to be copied 29. The collision mechanism might provide a means for dissociating Pol III* once it has completed its task in repair reactions. Previous reports describe that gaps need to be completely filled 10 or only mostly filled 11 before polymerase release is triggered. These measurements were made primarily by equilibrium binding measurements. Our kinetic experiments support the conclusion that gaps need to be completely filled before the maximal rate of release is induced, but this rate is too slow to support chromosomal replication.
Our experiments were performed on a DNA substrate that mimicked the gap in the final stages of Okazaki fragment synthesis. True replication forks contain additional interactions, including two coupled leading and lagging strand polymerases that are bound to the replicative helicase. It is possible that the polymerase could switch to a state where it could more rapidly dissociate in the presence of these additional potential allosteric effectors. Nevertheless, our substrates are similar to those used to initially formulate the collision hypothesis.
Measurement of the rate of gap filling indicates that the reaction is complete before the first time point (5 s) was taken. Thus, the rate of gap filling is fast relative to the rate of polymerase dissociation. For a majority of primer-templates, filling of the gap is followed by a strand displacement of approximately 7 nucleotides on average. This value is less than the processivity of 300 nucleotides observed in a strand displacement reaction that benefitted from a stabilizing interaction of χ with SSB 30. Comparison of these results suggests that the χ-SSB interaction increases the processivity of strand displacement approximately 40-fold. The fact that most of the product observed involves strand displacement argues against exact gap filling to form a nick providing the signal for Pol III* release. If the signaling model is dominant, many Okazaki fragments may end before the gap between their terminus and the primer, for the following Okazaki fragment, is filled. However, if the lagging strand polymerase is sufficiently fast relative to the leading strand polymerase, complete gap filling will occur. In that case, strand displacement is likely to occur. DNA polymerase I is recognized as an enzyme that fills any remaining gaps between Okazaki fragments and removes RNA primers with a duplex-specific 5′-3′ exonuclease activity, providing a nick that can be sealed by DNA ligase 31, 32. If there is a portion of the product that arises from strand displacement, another repair enzyme would be required to excise the displaced single-stranded flap, analogous to the function of the flap endonuclease, FEN1, in eukaryotes 33.
We applied diazirine-based photo-crosslinking to identify which proteins contact a uniquely reactive position within model templates. The advantages of the method include high yields, irradiation at wavelengths (350 nm) far removed from chromophores present in proteins or nucleic acids, and the ability of the carbene formed upon irradiation to insert into all amino acids. The efficiency and lack of chemical specificity permit negative results to be interpreted with confidence. Using this method, we have identified that α, and not τ, contacts the template and blocking oligonucleotide in front of the primer, making it the candidate for sensing conversion of a gap to a nick, presumably triggering change to a conformation that more rapidly releases from DNA.
In a structure of Pol III α complexed to DNA, an OB fold was located close to the primer terminus 25. Because OB folds commonly bind to ssDNA, a proposal was made that the OB fold could be part of the sensing network 25, 26. Consistent with this hypothesis, the ssDNA-binding portion of Pol III was localized to a C-terminal region of α that contains the OB-fold element 34. A test of the importance of the OB-fold motif was made using a mutant in which three basic residues located in the β1-β2 loop were changed to serine 11. No ssDNA binding was observed in the mutant, indicating diminution in affinity. However, even the wild-type polymerase bound ssDNA extremely weakly, near the limit of detection in the assays used (KD ∼8 µM), so a modest decrease in affinity would appear to be a null result. The processivity of the mutant polymerase was decreased by the β1-β2 loop mutations, an effect that was rescued by the presence of the τ complex 11. The latter observation would seem to suggest that, although the OB fold contributes to ssDNA affinity and processivity, it is not the processivity sensor, or at least the residues mutated are not the key interactors. The OB fold might act in concert with other binding changes as part of a more complex signaling network.
In our view, the entire polymerase active site is likely the processivity switch. Steitz and colleagues 25 have elegantly demonstrated a conformational change induced by substrate binding in which several elements move, one of the results of which is to place the β2 binding domain in a position where it can productively interact with the β2 clamp on DNA. Follow-on studies with a Gram-positive polymerase suggest this observation may be general 35,36. The geometry and spatial constraints around the active site when the exiting template is double-stranded might make insertion of the last nucleotide and ensuing strand displacement energetically unfavorable. Thus, these products may have decreased affinity for the active site, triggering a reversal of the conformational changes that occurred upon primer-template and dNTP binding, causing the β2 binding domain to be pulled away, and switching the polymerase to a low-processivity mode. The presence of a polymerase domain that is not bound to DNA serves to decrease the affinity of the C-terminal domains of Pol III α for β2 14 which is consistent with this model.
Our results demonstrate that the collision of the Pol III HE with the downstream elongated primer of the preceding Okazaki fragment is inadequate to support the rapid release required for cycling to the next primer at the replication fork. The only alternative explanation in the literature is provided by the signaling model, originally proposed by Ken Marians8. We have shown that providing a competing primer in the presence of ATP only modestly stimulates dissociation. So, the function of primase in the signaling model must be more than just providing a competing primer. One possibility is that primase, upon primer synthesis, sends a signal to the Pol III HE through the replicative helicase to which both are bound. Another possibility is that the competing primer is provided in a special context created by the molecular interactions at the replication fork.
Methodologies have been developed in the T4 and T7 systems18–20 that might enable a more rigorous test of the signaling mechanism in E. coli. If the signaling model proves correct, an understanding of the specific reaction steps and molecular interactions that throw the processivity switch and direct the replicase to the next primer will be the next challenge. This remains a critical deficit in our understanding of all DNA replication systems.
Materials and Methods
DNA templates
The templates diagrammed in Fig. 1 were assembled from the following gel-purified oligonucleotides: template 81-mer- 5′ GCT AAT GAA TTC CCG GTT CTT GAC TAC ATT ACT CTT GAT CAA GGT CAG CCA GCC TAT GCG CGT GAT CTG TAC ACC GTT C biotinT T (biotinT represents T with biotin attached, via a linker, to the C5 position); primer 30-mer- 5′ GAA CGG TGT ACA GAT CAC GCG CAT AGG CTG (to make 10 nt gapped template); primer 35-mer- 5′ GAA CGG TGT ACA GAT CAC GCG CAT AGG CTG GCT GA (to make 5 nt gapped template); primer 36-mer- 5′ GAA CGG TGT ACA GAT CAC GCG CAT AGG CTG GCT GAC (to make 4 nt gapped template); blocking oligonucleotide 39-mer- 5′ ATC AAG AGT AAT GTA GTC AAG AAC CGG GAA TTC ATT AGC-SNAP; blocking oligonucleotide 39-mer (5′-phosphate RNA12/DNA27)- 5′-PO4rArUrC rArArG rArGrU rArArU GTA GTC AAG AAC CGG GAA TTC ATT AGC-SNAP; blocking oligonucleotide 39-mer (5′ triphosphate)- 5′ triphosphate-ATC AAG AGT AAT GTA GTC AAG AAC CGG GAA TTC ATT AGC C-SNAP. The exogenous primed template used to stimulate polymerase release (Table 3) was assembled by annealing a 20-mer primer (5′-TTG TTC AGA TGA AGG CGC AT) to a 70-mer template (5′-TAA AAA AAA AAA AAA AAA AAG GAT TAC TGG ATC CGA AGG TCA GCC AGC CTA TGC GCC TTC ATC TGA ACA A).
For experiments reported in Fig. 3, in addition to oligonucleotides described in this subsection, the following oligonucleotides were synthesized: blocking oligonucleotide 34-mer- 5′ GAG TAA TGT AGT CAA GAA CCG GGA ATT CAT TAG C-biotin and extended primer 45-mer- 5′ GAA CGG TGT ACA GAT CAC GCG CAT AGG CTG GCT GAC CTT GAT CAA.
Preparation of the SNAP-conjugated blocking oligonucleotides
A method24 for attaching the Hexa-His tagged O6-Alkylguanine-DNA-Transferase (SNAP-tag, New England Biolabs) to a DNA oligomer was adapted for our purposes. The blocking oligonucleotides (PAGE-purified from Integrated DNA Technologies or Biosynthesis, Inc. for the 5′-triphosphate oligonucleotide) contained 3′-O-CH2-CH2-SH blocked by formation of a disulfide bond. Disulfide-blocked oligonucleotide (35 nmol) was reduced by incubation (1 h) at room temperature with 10 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) (Pierce) in 500 µl TE buffer (10 mM Tris-HCl, 1 mM EDTA (pH 8.0)) (Supplementary Fig. S2). The solution was applied to a NAP5 (0.5 ml) gel filtration column (GE Healthcare) with TE as the running buffer to remove TCEP and the 3-carbon thiol linker (Supplementary Fig. S2). The most concentrated fractions, determined spectrophotometrically, were combined giving a total 800 µl. 300 µl of O6-benzylguanine-maleimide (BG-maleimide; New England Biolabs) (2.5 mM in dimethyl formamide (DMF)) was added to the 800 µl of reduced 39-mer in a total volume of 1.1 ml and the reaction mixture was allowed to proceed at room temperature (1 h) before application to a 2.5 ml NAP25 column to remove unreacted BG-maleimide. To verify attachment of the O6-benzylguanine to the 3′end of the 39-mer, product (12,738 Da) was analyzed on a 9% denaturing polyacrylamide gel using the unmodified 39-mer (12,316 Da) for comparison. Greater than 90% of the oligonucleotide was modified (Supplementary Fig. S3). Unreacted/unmodified oligomer was removed in the next step.
The BG-modified-39-mer (33 nmol) was covalently attached to SNAP by incubating 1.5 fold excess of oligonucleotide over SNAP in (50 mM sodium phosphate (pH 8.0), 300 mM NaCl, 20% glycerol, 1 mM dithiothreitol, 5 mM imidazole) for 1 h at room temperature with gentle rotation of the tube in a final volume of 2 ml. The oligonucleotide-SNAP conjugate was applied to a 0.75 ml Ni2+-NTA column (Qiagen) pre-equilibrated with wash buffer (50 mM sodium phosphate (pH 8.0), 300 mM NaCl, 20% glycerol, 1 mM dithiothreitol, 5 mM imidazole). All subsequent column procedures were performed at 0–4 °C. The column was washed with 10 column volumes of wash buffer (50 mM sodium phosphate (pH 8.0), 300 nM NaCl, 20% glycerol, 1 mM dithiothreitol, 5 mM imidazole) to remove unreacted oligonucleotide. Protein was eluted with elution buffer (50mM sodium phosphate (pH 8.0), 300 nM NaCl, 20% glycerol, 1 mM dithiothreitol, 400 mM imidazole). Eluted fractions containing the protein samples were detected by the Bradford method (Pierce). Analysis of the SNAP-tag-39-mer, monitored on a 4–20% gradient SDS-PAGE gel from its molecular weight shift (unmodified SNAP-tag, 22.2 kDa; modified SNAP-tag-39-mer, 34.9 kDa), indicated that approximately 90% of the SNAP-tag protein that eluted off the column contained a covalently attached 39-mer oligonucleotide (Supplementary Fig. S3). The protein/oligo conjugate was frozen in small aliquots by flash freezing in liquid nitrogen.
DNA polymerase III holoenzyme components
Protein components were purified as described for Pol III37, ε38, τ 39, γ 39, δ 40, δ′40, χ 41, ψ 41, β2 42, and SSB4 43. Four different DnaX protein complexes with stoichiometries of τ2γδδ′χψ, τγ2δδ′χψ, τ3δδ′χψ, and γ3δδ′χψ were purified as described 44,45. Unless otherwise noted, the ε subunit used in our experiments was mutated at D12A and E14A to eliminate endogenous 3′-5′ exonuclease activity which degrades the primer46.
For experiments using Pol III* exhibiting a τ2γ stoichiometry, complexes were assembled from purified sub-complexes of Pol III (αεθ; 40 nM final) and DnaX complex (τ2γδδ′χψ; 20 nM final). For experiments using Pol III* exhibiting γ3 stoichiometry, complexes were assembled from purified sub-complexes of Pol III (αεθ; 20 nM final) and DnaX complex (γ3δδ′χψ; 20 nM final). For experiments using Pol III* exhibiting a τγ2 stoichiometry, the Pol III* was isolated and purified first by cation exchange chromatography over a Mono-S column 37, 45, 47. DnaX complex (τγ2δδ′χψ; 2nmol) was incubated with Pol III (αε D12A E14A, θ; 10 nmol) for 15 min at room temperature. The incubated mixture was applied to a NAP25 2.5 ml (GE Healthcare) gel filtration column to exchange the buffer (25 mM Hepes (pH 7.5), 5% glycerol, 25 mM NaCl) prior to running the mixture over a 1 ml Mono-S column at 4 °C. The column was washed with 4 column volumes of wash buffer (25 mM Hepes (pH 7.5), 5% glycerol, 25 mM NaCl). Fractions (0.5 ml) were collected after application of a 150 mM NaCl step (in 25 mM Hepes (pH 7.5), 5% glycerol). The Pol III* fractions eluted approximately five column volumes after the 150 mM NaCl step. Purified Pol III* was injected over the Sensor chip SA at a final concentration of 50 nM.
Surface Plasmon Resonance
A BIAcore 3000 instrument was used to quantify and measure release of Pol III from immobilized oligonucleotide templates. A flow rate of 5 µl/ min in KCl-EDB buffer (50 mM Hepes (pH 7.5), 125 mM KCl, 10 mM magnesium acetate, 0.005% tween-20, 10 µM TCEP) at 25 °C was used for all reactions. All buffers were filtered and degassed before use. The Sensor Chip SA (BIAcore), a sensor chip pre-immobilized with streptavidin for capture of biotinylated ligands, was preconditioned with 3 one min injections of 5 µl of 1 M NaCl/ 50 mM NaOH prior to attachment of DNA primer-templates. Three different DNA primer-templates were immobilized. The 81-mer template annealed to the 30-mer primer was attached to flowcell 1 of the Sensor Chip SA. The template 81-mer annealed to the 35-mer, and the template 81-mer annealed to 36-mer were attached to flowcells 2 and 3, respectively. Flowcell 4 was not derivatized and was used as a control for background subtraction.
DNA primer-templates were assembled in two steps. First, duplex DNA containing the template oligonucleotide 81-mer (500 pmol) and the binding oligonucleotide 30-mer (or 35-mer, or 36-mer) (5 nmol) was annealed in vitro in (10 mM Tris-HCl (pH 8.0), 300 mM NaCl, 30 mM sodium citrate) in a volume of 250 µl. The sample was heated to 95 °C for 10 min and allowed to cool slowly to 23 °C at 1 °C/ min. The sample was diluted 1000-fold to a concentration of 2 fmol/µl with respect to the 81-mer, in KCl-EDB buffer and aliquoted. To prepare the flowcell with the DNA primer-template, a total of ca. 50 µl of the annealed DNA primer-template was injected over the surface of an individual flow-cell on the sensor chip SA which resulted typically in an increase of ca. 270–300 response units (RU). Second, SNAP-tag-39-mer was diluted to a final concentration of 80 fmol/ µl in KCl-EDB buffer and 300 µl was injected over all 4 flowcells simultaneously. The SNAP-tag-39-mer does not bind to the underivatized flowcell. Complete annealing to the DNA templates on the remaining flowcells was observed for the SNAP-tag-39-mer by determining the binding signal after injection. Since the output obtained from the BIAcore is directly proportional to the mass bound, stoichiometries could be calculated. Complete annealing was observed for the SNAP-tag-39-mer.
Pol III* complexes were injected over the DNA primer-template at concentrations determined empirically for each individual assembly. To determine the optimum concentration for binding, a series of different concentrations ranging from 5 nM to 80 nM Pol III* complex was injected. Concentrations were chosen that gave maximum stoichiometry of binding. In a typical experiment, Pol III*, β2 (300 nM), and ATP (1 mM) was injected over all four flowcells of the SA sensor chip simultaneously in a 45 µl injection. The ‘KINJECT’ command was used for measuring dissociation of Pol III complexes in the presence of buffer alone. Data was collected for 3 days following the injection to allow the dissociation of complexes to come to baseline. The long dissociation time was required to gather data for 10 half lives for the slow, non physiological second phase. Having a complete time course for the slow phase proved essential for a proper kinetic fit for accurate determination of the fast phase. To measure the dissociation of Pol III* in the presence of dNTPs and/or other additional components, the ‘COINJECT’ command was used. The ‘COINJECT’ command introduces a second sample injection (45µl was used) immediately upon completion of the first. The following components were used either individually, or in combination, to determine the requirements for full recycling: dNTPs (40 µM of each, final), ddNTPs (40 µM final), ATP (1 mM final), ATPγS (1 mM final), β2 (0.5 µM final), SSB4 (0.5 µM final), 20-mer/70-mer (0.5 µM final). All were diluted in KCl-EDB running buffer. Pol III* failed to bind to DNA primer templates using control reactions that lacked β2 or ATP in the first injection demonstrating dependence on both for binding.
The rate of Pol III*/β2 dissociation from DNA primer-templates was determined using non-linear regression analysis using SigmaPlot software. The dissociation data was then fit to several models for exponential decay. All models tested included an offset value that defined the point at which the decay curve leveled out. To help assess whether a term was necessary to generate a good fit, a ‘dependency’ value for each parameter in the fitting equation was determined. The simplest model that best fit our data was double exponential decay.
Kinetics of filling the gap within model templates
A primer-template system similar to the surface plasmon resonance experiments was used, with an identical primer 35-mer and template 81-mer. Blocking oligonucleotide 34-mer was used to generate a 10 nt gap on the template. The primer 35-mer was 5′-32P end-labeled using T4 polynucleotide kinase. The construct was annealed by combining 200 nM 5′-32P-primer 35-mer, 250 nM template 81-mer, and 300 nM blocking oligonucleotide 34-mer (to ensure complete capture of the primer by a fully blocked template) and heating to 95 °C for 10 min and cooling to 23 °C at 1 °C/ min. An unblocked control sample was prepared by the same procedure but omitting the blocking oligonucleotide 34-mer. A marker for the expected product resulting from primer extension by 10 nt to fully fill the gap was prepared by annealing 200 nM 5′-32P-extended primer 45-mer with 250 nM template 81-mer.
Primer elongation assays were conducted at ambient temperature in a buffer containing 50 mM Hepes (pH 7.5), 100 mM potassium glutamate, 10 mM magnesium acetate, 0.20 mg/mL bovine serum albumin, 10 mM dithiothreitol, 2.5% (v/v) glycerol, and 0.02% (v/v) Nonidet-P40 detergent. Initiation complexes were generated by pre-incubating 10 nM of the 32P-labeled 10 nt gap containing primed template with 30 nM streptavidin, then combining this mixture with 25 nM τ2γ DnaX complex, 25 nM Pol III, 100 nM β2, 200 µM ATP, and 1 µM hexokinase and incubating for 60 s. For experiments with wild-type Pol III, this initiation complex formation reaction included 20 µM dATP, enabling the polymerase to replace the 3′-terminal nucleotide of the primer 35-mer if digested by the 3′-5′ exonuclease. Initiation complex formation was terminated and primer elongation was initiated by adding an equal volume of 0.25 mg/mL activated calf thymus DNA (a cold trap for unassociated Pol III HE), 10 mM glucose (to activate the hexokinase ATPase activity), and 40 µM dNTPs 48. The total volume of each gap filling reaction was 40 µl. All concentrations above are the final values after initiating gap filling. Primer elongation reactions were quenched after varying reaction times with 40 µl 50 mM EDTA. To ensure a high yield of the full length product, the primer elongation reaction for the unblocked primer-template was conducted for 30 s in the absence of calf thymus DNA and glucose. All other control DNA samples were not exposed to Pol III HE. All reactions and control samples were ethanol precipitated, and the air-dried pellets were each dissolved in 40 µl 75% formamide and 17 µl loaded onto 10% polyacrylamide/8.0 M urea/12% formamide sequencing gels to resolve the products. The gels were visualized using a Storm 840 Phosphor Imager and analyzed using ImageQuant 5.2.
Attachment of phenyldiazirine to oligonucleotides
In the absence of ambient light, 4-[3-(trifluoromethy)-diazirin-3-yl]benzoate N-hydroxysucinimide (custom synthesized by BioSynthesis, Inc.) was dissolved in anhydrous DMF (0.5 M final concentration). A 4.4 µmol sample was combined with 20 nmol of oligonucleotide (Integrated DNA Technologies) containing amino-modified C2 dT from Glen Research in a 0.1 M sodium tetraborate buffer (pH 8.5) (final volume 50 µl). This reaction mixture was incubated in the dark at room temperature overnight. The derivatized product was HPLC purified using a Waters XBridge OST C18 column (4.6 × 50 mm, pore size 2.5 µm). The sample was applied in 0.1 M triethylammonium acetate (pH 7.0) at a flow rate of 1.0 ml/min and eluted with a gradient changing to acetonitrile at 1.54 %/min. The column was run at 60 °C and absorbance was monitored at 265 nm and 350 nm. Approximately 15 nmol of photoreactive oligonucleotide was recovered. The presence of photocrosslinker in the oligonucleotide was verified by observing peaks at 260 nm and 350 nm and by a shift in chromatographic mobility from the unreacted oligonucleotide.
Photo-crosslinking oligonucleotides
Oligonucleotides (Fig. 4a) were combined with proteins and nucleotides at room temperature in a buffer containing 50 mM Hepes (pH 7.5), 10 mM magnesium acetate, 10 mM dithiothreitol, 20% (v/v) glycerol, 0.02% (v/v) Nonidet-P40 detergent, and 100 mM potassium glutamate (unless otherwise indicated) in siliconized Pyrex tubes covered in Parafilm and irradiated at 350 nm in a Rayonet photochemical reactor for 1 h at room temperature. The samples were quenched in 2X SDS-PAGE sample buffer (0.25 M Tris-HCl (pH 6.8), 5% SDS, 0.02% β-mercaptoethanol, and 0.2% glycerol). Samples were analyzed by 4–11% gradient SDS-PAGE containing 4 M urea.
For experiments in Fig. 4c, initiation complexes were generated by pre-incubating 4 nM of the 32P-labeled primed template (Fig. 4a) with 40 nM streptavidin, then combining with 16 nM τ3 complex, 16 nM Pol III, 200 nM β2, and 0.8 mM ATP prior to irradiation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 GM060273. We thank Anna Wieczorek for preparation of proteins and Diane Hager for assistance in figure preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McHenry CS. DNA replicases from a bacterial perspective. Annu Rev Biochem. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- 2.Breier AM, Weier HU, Cozzarelli NR. Independence of replisomes in Escherichia coli chromosomal replication. Proc Natl Acad Sci U S A. 2005;102:3942–3947. doi: 10.1073/pnas.0500812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- 4.Mok M, Marians KJ. Formation of rolling-circle molecules during ϕX174 complementary strand DNA replication. J Biol Chem. 1987;262:2304–2309. [PubMed] [Google Scholar]
- 5.Mok M, Marians KJ. The Escherichia coli preprimosome and DNA B helicase can form replication forks that move at the same rate. J Biol Chem. 1987;262:16644–16654. [PubMed] [Google Scholar]
- 6.Johanson KO, McHenry CS. The β subunit of the DNA polymerase III holoenzyme becomes inaccessible to antibody after formation of an initiation complex with primed DNA. J Biol Chem. 1982;257:12310–12315. [PubMed] [Google Scholar]
- 7.Stukenberg PT, Turner J, O'Donnell ME. An Explanation for Lagging Strand Replication: Polymerase Hopping among DNA Sliding Clamps. Cell. 1994;78:877–887. doi: 10.1016/s0092-8674(94)90662-9. [DOI] [PubMed] [Google Scholar]
- 8.Wu CA, Zechner EL, Reems JA, McHenry CS, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork: V. Primase action regulates the cycle of Okazaki fragment synthesis. J Biol Chem. 1992;267:4074–4083. [PubMed] [Google Scholar]
- 9.Alberts BM, Barry J, Bedinger P, Formosa T, Jongeneel CV, Kreuzer KN. Studies on DNA replication in the bacteriophage T4 in vitro system. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):655–668. doi: 10.1101/sqb.1983.047.01.077. [DOI] [PubMed] [Google Scholar]
- 10.Leu FP, Georgescu R, O'Donnell ME. Mechanism of the E. coli τ processivity switch during lagging-strand synthesis. Mol Cell. 2003;11:315–327. doi: 10.1016/s1097-2765(03)00042-x. [DOI] [PubMed] [Google Scholar]
- 11.Georgescu RE, Kurth I, Yao NY, Stewart J, Yurieva O, O'Donnell M. Mechanism of polymerase collision release from sliding clamps on the lagging strand. EMBO J. 2009;28:2981–2991. doi: 10.1038/emboj.2009.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez de Saro FJ, Georgescu RE, O'Donnell ME. A peptide switch regulates DNA polymerase processivity. Proc Natl Acad Sci U S A. 2003;100:14689–14694. doi: 10.1073/pnas.2435454100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez de Saro FJ, Georgescu RE, Goodman MF, O'Donnell ME. Competitive processivity-clamp usage by DNA polymerases during DNA replication and repair. EMBO J. 2003;22:6408–6418. doi: 10.1093/emboj/cdg603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DR, McHenry CS. Identification of the β-binding domain of the α subunit of Escherichia coli polymerase III holoenzyme. J Biol Chem. 1996;271:20699–20704. doi: 10.1074/jbc.271.34.20699. [DOI] [PubMed] [Google Scholar]
- 15.Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci U S A. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dohrmann PR, McHenry CS. A bipartite polymerase-processivity factor interaction: Only the internal β binding site of the α subunit is required for processive replication by the DNA polymerase III holoenzyme. J Mol Biol. 2005;350:228–239. doi: 10.1016/j.jmb.2005.04.065. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Marians KJ. Two distinct triggers for cycling of the lagging strand polymerase at the replication fork. J Biol Chem. 2000;275:34757–34765. doi: 10.1074/jbc.M006556200. [DOI] [PubMed] [Google Scholar]
- 18.Hamdan SM, Loparo JJ, Takahashi M, Richardson CC, van Oijen AM. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature. 457:336–339. doi: 10.1038/nature07512. (9 A.D.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 20.Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel SS. Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature. 2009;462:940–943. doi: 10.1038/nature08611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manosas M, Spiering MM, Zhuang Z, Benkovic SJ, Croquette V. Coupling DNA unwinding activity with primer synthesis in the bacteriophage T4 primosome. Nat Chem Biol. 2009;5:904–912. doi: 10.1038/nchembio.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Nelson SW, Benkovic SJ. The control mechanism for lagging strand polymerase recycling during bacteriophage T4 DNA replication. Mol Cell. 2006;21:153–164. doi: 10.1016/j.molcel.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 23.Kaboord BF, Benkovic SJ. Rapid assembly of the bacteriophage T4 core replication complex on a linear primer/template construct. Proc Natl Acad Sci U S A. 1993;90:10881–10885. doi: 10.1073/pnas.90.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jongsma MA, Litjens RH. Self-assembling protein arrays on DNA chips by auto-labeling fusion proteins with a single DNA address. Proteomics. 2006;6:2650–2655. doi: 10.1002/pmic.200500654. [DOI] [PubMed] [Google Scholar]
- 25.Wing RA, Bailey S, Steitz TA. Insights into the replisome from the structure of a ternary complex of the DNA polymerase III α-subunit. J Mol Biol. 2008;382:859–869. doi: 10.1016/j.jmb.2008.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamers MH, Georgescu RE, Lee SG, O'Donnell M, Kuriyan J. Crystal structure of the catalytic α subunit of E. coli replicative DNA polymerase III. Cell. 2006;126:881–892. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 27.Tate JJ, Persinger J, Bartholomew B. Survey of Four Different Photoreactive Moieties for DNA Photoaffinity Labeling of Yeast RNA Polymerase III Transcription Complexes. Nucleic Acids Res. 1998;26:1421–1426. doi: 10.1093/nar/26.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reems JA, Wood S, McHenry CS. Escherichia coli DNA polymerase III holoenzyme subunits α, β and γ directly contact the primer template. J Biol Chem. 1995;270:5606–5613. doi: 10.1074/jbc.270.10.5606. [DOI] [PubMed] [Google Scholar]
- 29.Modrich P. Methyl-directed DNA mismatch correction. J Biol Chem. 1989;264:6597–6600. [PubMed] [Google Scholar]
- 30.Yuan Q, McHenry CS. Strand displacement by DNA polymerase III occurs through a τ-ψ-χ link to SSB coating the lagging strand template. J Biol Chem. 2009;284:31672–31679. doi: 10.1074/jbc.M109.050740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kornberg A. DNA Replication. United Kingdom: W. H. Freeman and Company; 1992. DNA Polymerase II of E. coli; p. 167. [Google Scholar]
- 32.Konrad EB, Lehman IR. A Conditioned Lethal Mutant of Escherichia-coli K-12 Defective in the 5′ → 3′ Exonuclease Associated with DNA Polymerase I. Proc Natl Acad Sci U S A. 1974;71:2048–2051. doi: 10.1073/pnas.71.5.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Kao HI, Bambara RA. Flap Endonuclease 1: A Central Component of DNA Metabolism. Annu Rev Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 34.McCauley MJ, Shokri L, Sefcikova J, Venclovas C, Beuning PJ, Williams MC. Distinct double- and single-stranded DNA binding of E. coli replicative DNA polymerase III α subunit. ACS Chem Biol. 2008;3:577–587. doi: 10.1021/cb8001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans RJ, Davies DR, Bullard JM, Christensen J, Green LS, Guiles JW, Pata JD, Ribble WK, Janjic N, Jarvis TC. Structure of polC reveals unique DNA binding and fidelity determinants. Proc Natl Acad Sci U S A. 2008;105:20695–20700. doi: 10.1073/pnas.0809989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wing RA. Structural studies of the prokaryotic replisome. Yale University; 2010. p. 170. [Google Scholar]
- 37.Kim DR, McHenry CS. In vivo assembly of overproduced DNA polymerase III: Overproduction, purification, and characterization of the α, α–ε, and α–ε–θ subunits. J Biol Chem. 1996;271:20681–20689. doi: 10.1074/jbc.271.34.20681. [DOI] [PubMed] [Google Scholar]
- 38.Wieczorek A, McHenry CS. The NH(2)-terminal php domain of the α subunit of the E. coli replicase binds the ε proofreading subunit. J Biol Chem. 2006;281:12561–12567. doi: 10.1074/jbc.M513844200. [DOI] [PubMed] [Google Scholar]
- 39.Dallmann HG, Thimmig RL, McHenry CS. DnaX complex of Escherichia coli DNA polymerase III holoenzyme: Central role of τ in initiation complex assembly and in determining the functional asymmetry of holoenzyme. J Biol Chem. 1995;270:29555–29562. [PubMed] [Google Scholar]
- 40.Song MS, Pham PT, Olson M, Carter JR, Franden MA, Schaaper RM, McHenry CS. The δ and δ′ subunits of the DNA polymerase III holoenzyme are essential for initiation complex formation and processive elongation. J Biol Chem. 2001;276:35165–35175. doi: 10.1074/jbc.M100389200. [DOI] [PubMed] [Google Scholar]
- 41.Olson MW, Dallmann HG, McHenry CS. DnaX-complex of Escherichia coli DNA polymerase III holoenzyme: The χψ complex functions by increasing the affinity of τ and γ for δ–δ′ to a physiologically relevant range. J Biol Chem. 1995;270:29570–29577. [PubMed] [Google Scholar]
- 42.Johanson KO, Haynes TE, McHenry CS. Chemical Characterization and Purification of the β Subunit of the DNA Polymerase III Holoenzyme from an Overproducing Strain. J Biol Chem. 1986;261:11460–11465. [PubMed] [Google Scholar]
- 43.Griep MA, McHenry CS. Glutamate Overcomes the Salt Inhibition of DNA Polymerase III Holoenzyme. J Biol Chem. 1989;264:11294–11301. [PubMed] [Google Scholar]
- 44.Pritchard AE, Dallmann HG, McHenry CS. In vivo assembly of the τ-complex of the DNA polymerase III holoenzyme expressed from a five-gene artificial operon: Cleavage of the τ-complex to form a mixed γ–τ-complex by the OmpT protease. J Biol Chem. 1996;271:10291–10298. doi: 10.1074/jbc.271.17.10291. [DOI] [PubMed] [Google Scholar]
- 45.Glover BP, McHenry CS. The DnaX-binding subunits δ′ and ψ are bound to γ and not τ in the DNA polymerase III holoenzyme. J Biol Chem. 2000;275:3017–3020. doi: 10.1074/jbc.275.5.3017. [DOI] [PubMed] [Google Scholar]
- 46.Fijalkowska IJ, Schapper RM. Mutants in the Exo I Motif of Escherichia coli dnaQ: Defective Proofreading and Inviability Due to Error Catastrophe. Proc Natl Acad Sci U S A. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pritchard AE, Dallmann HG, Glover BP, McHenry CS. A novel assembly mechanism for the DNA polymerase III holoenzyme DnaX complex: association of δδ′ with DnaX(4) forms DnaX(3)δδ′. EMBO J. 2000;19:6536–6545. doi: 10.1093/emboj/19.23.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Downey CD, Crooke E, McHenry CS. Polymerase Chaperoning and Multiple ATPase Sites Enable the E. coli DNA Polymerase III Holoenzyme to Rapidly Form Initiation Complexes. J Mol Biol. 2011;412:340–353. doi: 10.1016/j.jmb.2011.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.