Abstract
De-ubiquitinating enzymes (DUBs) can reverse the modifications catalyzed by ubiquitin ligases and as such are believed to be important regulators of a variety of cellular processes. Several members of this protein family have been associated with human cancers; however, there is little evidence for a direct link between deregulated de-ubiquitination and neoplastic transformation. Ubiquitin C-terminal hydrolase (UCH)-L1 is a DUB of unknown function that is overexpressed in several human cancers, but whether it has oncogenic properties has not been established. To address this issue, we generated mice that overexpress UCH-L1 under the control of a ubiquitous promoter. Here, we show that UCH-L1 transgenic mice are prone to malignancy, primarily lymphomas and lung tumors. Furthermore, UCH-L1 overexpression strongly accelerated lymphomagenesis in Eμ-myc transgenic mice. Aberrantly expressed UCH-L1 boosts signaling through the Akt pathway by downregulating the antagonistic phosphatase PHLPP1, an event that requires its de-ubiquitinase activity. These data provide the first in vivo evidence for DUB-driven oncogenesis and suggest that UCH-L1 hyperactivity deregulates normal Akt signaling.
Keywords: de-ubiquitinating enzymes, lymphoma, mouse model, Akt, PHLPP
Introduction
The ubiquitin (Ub)–proteasome system is involved in nearly all areas of cell biology and has long been implicated in cancer biology.1,2 The attachment of Ub to target proteins by Ub ligases leads to a variety of events, including target protein degradation, cellular relocalization or conformation-based changes in protein function. Counteracting the process of Ub attachment is a family of enzymes collectively referred to as de-ubiquitinating enzymes (DUBs) that consists of approximately 100 members.3 The DUB family is sub-classified, on the basis of sequence and structural clustering, into five groups, including the Ub C-terminal hydrolases (UCHs), the Ub-specific proteases (USPs), the ovarian tumor domain containing (OTU), the Josephin-domain containing and the jab1/MPN domain-associated metalloiso-peptidase class.3 It is believed that by reversing the ubiquitination status of targeted proteins, DUBs are important regulators of many cellular processes. Despite their emerging importance as regulatory molecules, the molecular targets for the vast majority of DUBs are not known.
Although the involvement of Ub ligases in cancer-related processes has been extensively reported, it is increasingly apparent that regulation of these processes by DUBs may be equally important, and that alterations may lead to malignancy. A few DUBs have been found to be directly mutated in human cancers,4–9 whereas many others are dysregulated.10 Mutations in the DUB CYLD were identified in patients with a skin tumor syndrome known as hereditary cylindromatosis. Although inactivation of CYLD does enhance the susceptibility to carcinogen-induced skin tumors, spontaneous tumors are not reported.9,11 Recently, inactivating mutations were identified in the DUB A20 (also known as TNFAIP3) in a variety of human B-cell lymphomas.4–7,12 Unfortunately, A20 knockout mice die at a very early age due to severe inflammation, making tumor studies impossible.13 It remains largely unproven in vivo, therefore, whether DUBs have a direct causal involvement in the pathogenesis of cancer. Furthermore, there is no conclusive evidence that DUBs can function as oncogenes in vivo. The enzyme Ub C-terminal hydrolase L1 (UCH-L1; PGP9.5) was among the first DUBs described, yet its molecular targets have been elusive.14 UCH-L1 is abundant in mammalian nervous tissue and in the testis, but it is not usually expressed in other somatic tissues.15 Although bearing the sequence signature of a DUB, UCH-L1 also has been shown to possess an unusual ligase activity, especially when present in high concentrations.16 This activity maps to residues that are distinct from the DUB catalytic cysteine (C90) and is greatly reduced in a mutant UCH-L1 (S18Y) that has been found to be protective for Parkinson’s disease.16
Several observations hint at a possible link between UCH-L1 overexpression and cancer. UCH-L1 was noted to be highly expressed in Burkitt’s lymphoma (BL) cell lines, where it was hypothesized to function non-oncogenically to alleviate proteotoxic stress.17 Using activity-based DUB probes, high levels of active UCH-L1 were also observed in cell lines derived from BL and chronic lymphocytic leukemia,18 but a role in malignant transformation was not established. Recently, UCH-L1 depletion was found to produce a modest reduction in the growth of BL cell lines, although it is not known whether its effect was on cell proliferation or cell death, or whether UCH-L1 expression can impact in vivo lymphoma behavior.19 Using transformed lung cancer cell lines, UCH-L1 expression was found to correlate with invasive and metastatic behavior.20 In these cells, UCH-L1 expression led to a slight increase in phosphorylation of Akt and mitogen-activated protein kinase family members, and this effect was required to enhance the migration behavior of these cells. It was not detailed in this study whether UCH-L1 affected phosphorylation of both of the required phospho-Akt residues (threonine 308 (T308) and serine 473 (S473)), whether downstream Akt targets are affected, or whether UCH-L1 catalytic activity is required to boost Akt signaling. In striking contrast to these studies that suggest a provocative role, several recent reports suggest that UCH-L1 expression acts as a tumor suppressor restricting the growth of malignant cells. Supporting this claim are data in which UCH-L1 promoter hypermethylation is observed in a variety of cancers.21–24 Re-expression of UCH-L1 in some cell lines derived from these cancers results in reduced proliferation and apoptosis. Similarly, incubation with the commercially available UCH-L1 inhibitor LDN57444 produced accelerated growth in a UCH-L1-expressing lung cancer cell line.25 Taken together, it remains unclear whether UCH-L1 is associated with the pathogenesis of human cancer or whether its expression is simply a cellular response to transformation.
A central question, therefore, is whether UCH-L1 has oncogenic properties that can by itself drive malignant transformation in vivo or whether it acts as a ‘non-oncogene’26 that establishes a favorable intra-cellular environment supporting proliferation without itself providing the oncogenic stimuli. To address these issues, we generated a novel transgenic mouse model with enforced UCH-L1 expression under the control of a ubiquitous promoter. These animals have a striking tumor-prone phenotype with the development of lymphomas and lung tumors. These data therefore provide compelling evidence that UCH-L1 is an oncogene and that DUBs can act oncogenically in vivo. Moreover, when this transgene was combined with the Eμ-myc mouse lymphoma model, we observed dramatically accelerated lymphomagenesis associated with increased proliferation and reduced apoptosis of lymphoid tissues, and reduced apoptosis in established lymphomatous masses. Exploring the underlying mechanism, we found that aberrant UCH-L1 expression leads to a dramatic increase in Akt signaling in vitro and in vivo by decreasing the levels of PHLPP1: a phosphatase that reverses Akt phosphorylation. These effects require UCH-L1 de-ubiquitinase activity, as shown by the ability of wild-type—but not catalytic mutant—UCH-L1 to rescue RNAi-induced cell death in vitro. These data suggest that UCH-L1 is a potent oncogene and provide a new insight into the mechanism by which this enzyme affects the Akt signaling pathway.
Materials and methods
Generation of UCH-L1 and UCH-L1/Eμ-myc double-transgenic mice
Uchl1 transgenic mice were generated as described in the Supplementary methods. Male Eμ-myc mice (a kind gift from Dr Richard Bram, Mayo Clinic) were bred with female Uch11 mice to generate cohorts consisting of Eμ-myc, Uchl1 and Eμ-myc/Uchl1 double-transgenic mice. The breeding was accomplished with two breeding boxes—one for each clone of Uch11 to minimize variations related to differences in genetic background.
Collection and analysis of tissues and tumors
The paraffin-embedded human BL samples were de-identified residual diagnostic samples and use was approved by the institutional review board of the Mayo Clinic and the University of Utah. De-identified samples for qRT-PCR were obtained from the Mayo Clinic Cancer Center and were collected with the approval of the Mayo Clinic institutional review board. B-cell lymphoma tissue microarray was obtained from US Biomax. Mice for the spontaneous tumor study were humanely killed at between 16–18 months of age and examined with a dissection microscope to screen all major organs for overt tumors. Eμ-myc and Uchl1/Eμ-myc mice were killed when it was apparent that lymphoma was present. Fisher’s exact test was used to compare tumor incidence proportions across the genotypes for mice that developed tumors. Four- or six-week-old mice were humanely killed and lymph nodes dissected for analysis. Tumors and tissues were processed by standard procedures for histopathology. The BrdU proliferation assay was performed as described.27 The percentage of BrdU-positive cells was determined by counting the total and BrdU-positive nuclei in 10 non-overlapping fields at ×40 magnification (n=3 mice per genotype). TUNEL staining was performed according to the manufacturer (Roche).
Cell lines, immunoblotting and reagents
The cell lines KMS-11, KMS-12, KMS-18, KMS-28 (kindly provided by Takemi Otsuki), and HeLa cells were cultured in DMEM supplemented with 10% FCS. SDS-PAGE, electrophoretic transfer and immunoblotting were performed according to standard procedures. The source of all antibodies is provided in the Supplementary methods. For the assessment of Akt phosphorylation, cells were incubated in serum free medium overnight before the addition of interleukin-6 (10 ng/ ml) or IGF-1 (50 ng/ml) for the indicated times. MG132 was used where indicated at a final concentration of 20 μg/ml. Cell numbers and viability were determined in triplicate by trypan blue exclusion and the MTS assay (Promega, Madion, WI, USA), respectively.
RNA interference
The sequences of all shRNA constructs are detailed in the Supplementary methods. Indicated pre-designed pGIPZ and pTRIPZ miR-30-based lentiviral constructs were used to generate lentivirus with the Trans-Lentiviral Packaging System (Open Biosystems, Huntsville, AL, USA). Cells were transduced using a 50/50 mix of lentivirus-containing culture supernatant and fresh complete DMEM with 8 μg/ml (final concentration) polybrene for 48 h. Following 48 h (96 h following the first addition of virus), cells were selected with puromycin (2 μg/ml) for 48 h. Experiments using pGIPZ were performed between days 3–5 following transduction and cultures were discarded after 8 days to prevent selection of knockdown-resistant clones. Experiments performed with the tet-inducible pTRIPZ system were identical to the above, except that the experiments were performed 3–5 days after shRNA induction with 1 μg/ml doxycycline.
Quantitative real-time PCR
After obtaining the approval of the research protocol by the University of Utah Institutional Review Board, formalin-fixed, paraffin-embedded (FFPE) BL samples were retrieved from the archives of Primary Children’s Medical Center at the University of Utah. All diagnostic material on each case was reviewed (RRM and SLP) to confirm the diagnosis and to select blocks with at least 80% tumor cells, good preservation and minimal necrosis. Five 5-μm sections were cut into microfuge tubes, and total RNA was isolated using the RecoverAll Total Nucleic Acid Isolation Kit for FFPE Tissues (Applied Biosystems/Ambion, Austin, TX, USA). cDNA was generated from 500 ng of total RNA using the High Capacity cDNA Reverse Transcription Kit with random hexamer primers (Applied Biosystems, Carlsbad, CA, USA). RNA was extracted from all other samples using the RNeasy kit (Qiagen, Valencia, CA, USA) and transcribed to first-strand cDNA using the SuperScript III kit (Invitrogen, Carlsbad, CA, USA). Primary human B-cells were isolated from eight healthy donors spontaneously donating to the Mayo Clinic Blood Donor Center following approval by the institutional review board. Peripheral blood mononuclear cells were isolated from unused filter cones28 by Ficoll-Hypaque gradient centrifugation followed by immunomagnetic separation of resting B-cells using the MACS untouched B-cell isolation kit (Miltenyi, Auburn, CA, USA). All mRNA quantitation was performed using TAQman pre-designed gene expression assays (Applied Biosystems; UCH-L1 Hs00188233_m1, PHLPP Hs10597868_m1) using the ABI PRISM 7900 Sequence Detection System (Applied Biosystems) according to the manufacturer. All data were normalized to levels of the housekeeping gene tata-binding protein (Applied Biosystems 4333769F).
For other information, please refer to the Supplementary materials and methods.
Results
Generation of a UCH-L1 transgenic mouse
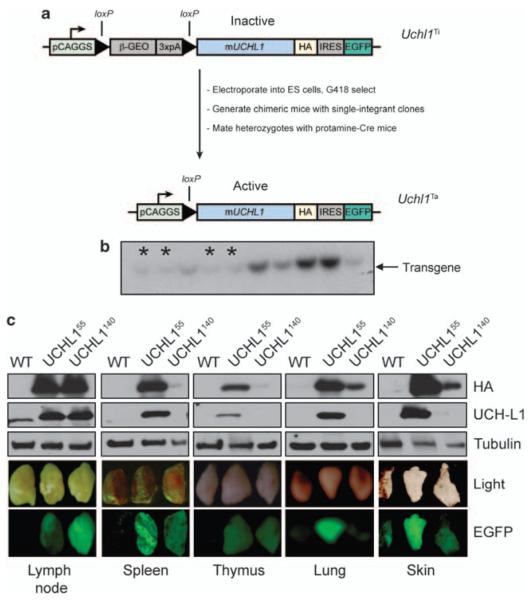
The murine Uchl1 cDNA was cloned from mouse brain cDNA with a C-terminal hemagglutinin tag and was used to generate a conditional, Cre-loxP-regulated transgene construct.29 The transgenic vector contains the CAGGS promoter consisting of the cytomegalovirus immediate enhancer and the chicken β-actin promoter (Figure 1a). The cDNA was inserted downstream of a loxP-flanked ‘stop’ cassette that separates the CAGGS promoter from the UCH-L1 cDNA, thus preventing the expression of the transgene. The stop cassette encodes a fusion protein between the β-galactosidase and neomycin-resistance reading frames (β-Geo) that provides drug resistance. Downstream of the UCH-L1 cDNA is an internal ribosome entry site and an enhanced green fluorescence protein (EGFP) cDNA that allows easy genotyping of offspring. Following electroporation, neomycin-resistant clones were screened by Southern blot and single integrants were selected (Figure 1b). We achieved germline transmission of two independent clones carrying the inactive transgene, Uchl1Ti55 and Uchl1Ti40. We activated the expression of UCH-L1 by crossing these clones with mice expressing the Cre recombinase under the control of the testis-specific promoter protamine (protamine-Cre), which leads to excision of the stop cassette in the male germline.30 Transgene-positive and protamine-Cre-positive males were then mated with C57BL/6 females to produce heterozygous active transgenic (Uchl1Ta, hereafter referred to as Uchl1) mice. These mice are identifiable by green fluorescence in the skin resulting from the expression of EGFP. Expression of the transgene is observed in most tissues (Figure 1c and Supplementary Figure S1) by EGFP, hemagglutinin and UCH-L1 immunoblots, although some clone-specific variation is noted. Although in many tissues we observed a good correlation between the observed EGFP fluorescence and transgene expression, some inconsistencies are seen. These mice have no outwardly apparent phenotype and are fertile.
Figure 1.
Generation of Uchl1 mice. (a) Schematic overview of the generation of Uchl1transgenic mice. (b) Representative southern blot of G418-resistant ES cell clones with indicated single integrant clones marked with an asterisk (*). The other darker bands likely represent clones with multiple integrations. (c) Expression of UCH-L1 in selected tissues as shown by the presence of EGFP and by immunoblotting for HA or UCH-L1. Results are representative of three independent experiments from three pairs of mice. Abbreviations: b-geo, beta-galactosidase/Neomycin resistance fusion protein; EGFP, enhanced green fluorescent protein; HA, hemagglutinin tag; IRES, internal ribosome entry site; PCAGGS, chicken actin promoter; 3XpA, polyadenylation sequence.
Uchl1 mice are susceptible to spontaneous tumors
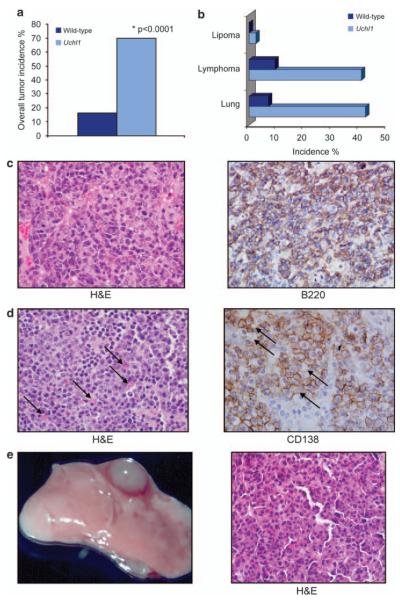
To test whether UCH-L1 acts as an oncogene in vivo, we determined the tumor incidence of a cohort of Uchl1 and wild-type mice at 16–18 months. Mice were humanely killed and dissected for tumors. Although there were some mice with signs of ill health at this age, most mice were overtly healthy, and had no obvious behavioral tendencies. There was a striking increase in the overall tumor incidence in Uchl1 mice, with 70% of mice having macroscopically visible tumors, whereas 16% of wild-type mice had tumors (P<0.0001, Figure 2a). There was no significant difference in the average ages at the time of killing between the cohorts of Uchl1 and wild-type littermates (17.2 vs 17.9 months, respectively). The rate of tumors was nearly identical in each Uchl1 clone (Uchl155=71%, Uchl1140=69%). Consistent with the observed tumor associations in the existing literature, the predominant tumors were evenly split between lymphomas and lung adenomas, with 14% having tumors of both types (Figure 2b). The lymphomas were primarily localized to the spleen, cervical and axillary lymph nodes, although 18% had large abdominal masses. Representative specimens from each tumor type were embedded in paraffin and examined by hematoxylin and eosin staining. The lymphomas were classified according to the Bethesda proposals for classification of lymphoid neoplasms in mice.31 The masses were of B-cell lineage as evidenced by staining with B220, and had a range of cellular morphologies suggestive of immunoblastic large B-cell lymphoma or plasmacytoma (Figure 2c). The lung tumors were classified as bronchiolo-alveolar adenoma/carcinoma—usually with a solid growth pattern (Figure 2d). These data show for the first time that UCH-L1 acts as an oncogene in vivo.
Figure 2.
UCH-L1 acts as an oncogene in mice. (a) Uchl1 (n = 34; mean age 17.3 months) and wild-type littermates (n = 43; mean age = 17.9 months) were killed and dissected for tumors. The overall incidence of all tumor types is indicated. (b) The spectrum and individual incidence of tumor types observed in (a). (c) Representative lymphoma from an Uchl1 mouse, staining as indicated. The mass is comprised of dense sheets of large lymphocytes with abundant cytoplasm, oval nuclei and a vesicular chromatin pattern. There is prominent anisokaryosis and mitotic figures are abundant. The neoplastic lymphocytes stain positively with the B220 antibody. (d) Representative plasmacytoma from an Uchl1 mouse with staining as indicated. The mass consists of sheets of medium-sized plasma cells characterized by moderate amphophilic cytoplasm and an eccentric round nucleus and marginated chromatin. Some cells contain large, homogenous eosinophilic inclusions (Russell bodies; indicated by arrowheads). Admixed with the plasma cells are lymphoblastic cells. (e) Gross and histological views of representative bronchiolo-alveolar carcinoma seen in an Uchl1 mouse. The mass is comprised of large polygonal type II pneumocytes lining a delicate network of papillary connective tissue. Neoplastic pneumocytes often pile up and destroy the alveolar septae, and occasionally invade the bronchiolar walls. There is increased cytoplasmic basophilia and moderate anisokaryosis.
UCH-L1 expression accelerates the development of lymphoma in Eμ-myc mice
UCH-L1 is frequently expressed at an elevated level in human BL, as seen by RNA microarray studies, as well as at the protein, and DUB activity, level.17,18,32 UCH-L1 expression has also been observed in primary lymphocytes infected with the Epstein–Barr virus in vitro.18 Taken together with our observation of a dramatic incidence of lymphoma in Uchl1 mice, we hypothesized that UCH-L1 is in fact a powerful driving force behind the development of BL in vivo. We therefore asked if enforced UCH-L1 expression could hasten the development of lymphoma in the well-studied Eμ-myc model.33,34 This mouse strain carries a transgene encoding c-myc under the control of the immunoglobulin heavy chain enhancer, thereby mimicking the t(8;14) translocation observed in most cases of BL.35,36 To determine the impact of enforced UCH-L1 expression on Eμ-myc-induced lymphoma, we generated cohorts of approximately 30 mice of Uchl1, Uchl1/Eμ-myc and Eμ-myc alone and observed them for tumor formation.
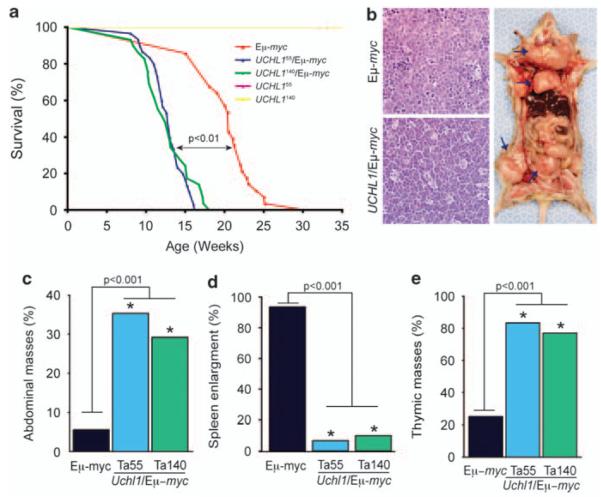
Eμ-myc mice developed lymphoma with 100% penetrance beginning at 13 weeks with a median survival of approximately 21 weeks (Figure 3a). In contrast, Uchl1/Eμ-myc mice developed lymphoma beginning at 8 weeks and had a median survival of only 13 weeks. By 19 weeks, 100% of Uchl1/Eμ-myc mice died of lymphoma. Furthermore, there were some notable differences in the anatomical patterns in tumor development between Eμ-myc and Uchl1/Eμ-myc mice. There were much fewer Uchl1/Eμ-myc mice with splenic enlargement and many more with thymic masses compared with Eμ-myc animals (Figures 3b, d and e). We also noted that significantly more Uchl1/Eμ-myc mice developed abdominal masses compared with Eμ-myc mice (Figure 3c). Abdominal masses are a very common presenting feature in children with BL.37 The tumors from Uchl1/Eμ-myc mice exhibited classic ‘starry sky’ histology, with lymphoblastic cellular morphology. These results confirm that UCH-L1 expression promotes the development of B-cell lymphomas and that these lymphomas closely resemble human BL.
Figure 3.
UCH-L1 accelerates the development of lymphoma in Eμ-myc mice. (a) Survival curves for Uchl1, Eμ-myc and Uchl1/Eμ-myc mice. Please note that the survival curve for clone 55 is not visible, as it is superimposed by that of clone 140, with 100% survival through the course of this experiment. P value represents the results of the log-rank test. (b) Histology of tumors from the indicated genotypes. Representative image of an Uchl1/Eμ-myc mouse with lymphoma. Similar results were seen in four separate masses from separate animals. Arrows indicate (from top to bottom) enlarged lymph nodes, thymic mass and an abdominal mass. (c–e) Graphs depicting the incidence of abdominal masses (c), splenic enlargement (d) and thymic involvement (e). Ta55 and Ta140 indicate Uchl1 mice from clones 55 and 140, respectively. n = 30 mice for all groups. P values represent results from the χ2 test.
UCH-L1 increases lymphoid proliferation and decreases apoptosis in vivo
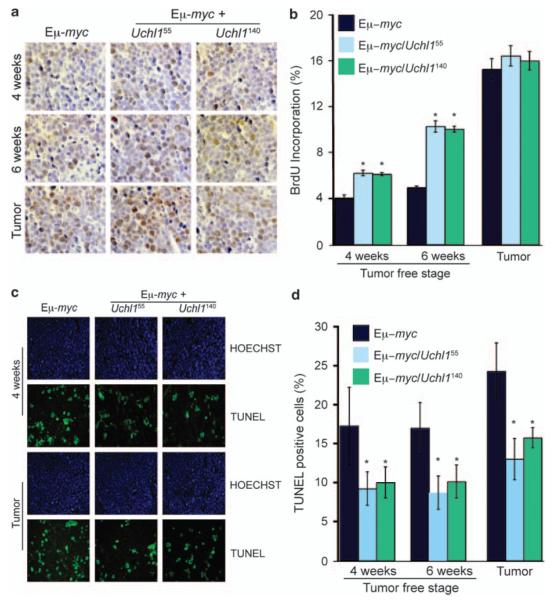
UCH-L1 may accelerate the development of lymphoma by either increasing proliferation or reducing apoptosis in lymphocytes or in the resulting tumors. To determine the effect of UCH-L1 on the rate of proliferation, we performed a BrdU incorporation assay in age-matched Uchl1/Eμ-myc and Eμ-myc mice. We observed an increase in BrdU incorporation in lymph nodes from both 4- and 6-week-old mice. Although the differences were significant at both ages, the increase was most notable at 6 –weeks, with an increase from 5% incorporation in 6-week-old Eμ-myc lymph nodes to approximately 10% in nodes from both clones of Uchl1/Eμ-myc animals (Figures 4a and b). In contrast, mature tumors from both Eμ-myc and Uchl1/Eμ-myc had a much higher proliferation rate that was not further enhanced by UCH-L1. This suggests that UCH-L1 increases the rate of proliferation in lymphocytes before the development of obvious tumors. To determine the rate of apoptosis, we performed a terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay in the same samples. Strikingly, we observed a significant decrease in TUNEL-positive cells in both lymph nodes as well as in established lymphomatous masses in UCH-L1 transgenic mice, with an approximately 50% reduction in TUNEL-positive cells in each condition suggesting reduced apoptosis (Figures 4c and d). Taken together, these results suggest that UCH-L1 enhances myc-driven tumor formation by both increasing the cell proliferation in lymph nodes (presumably pre-malignant lesions) and reducing apoptosis in lymph nodes as well as in lymphomatous tissues.
Figure 4.
UCH-L1 enhances proliferation and suppresses apoptosis in pre-malignant tissues. (a, b) Mice at 4 weeks, 6 weeks or those with tumors were killed and analyzed for cell proliferation. Mice of the indicated genotypes3 were injected with BrdU and killed 6 h later. BrdU incorporation was assessed by immunohistochemistry (a). Quantitation (b) reflects the average percentage of BrdU-positive cells in 10 random fields. Images are representative. (c, d) Tissues from the same samples as in (a, b) were analyzed using the TUNEL assay using immunofluorescence (c). Quantitation (d) reflects the average numbers of TUNEL-positive cells in 10 random fields. Data and statistics for all panels reflect the results from tissues from three mice per genotype. *P<0.05 (chi-square).
UCH-L1 is required for the growth and survival of malignant B-cells
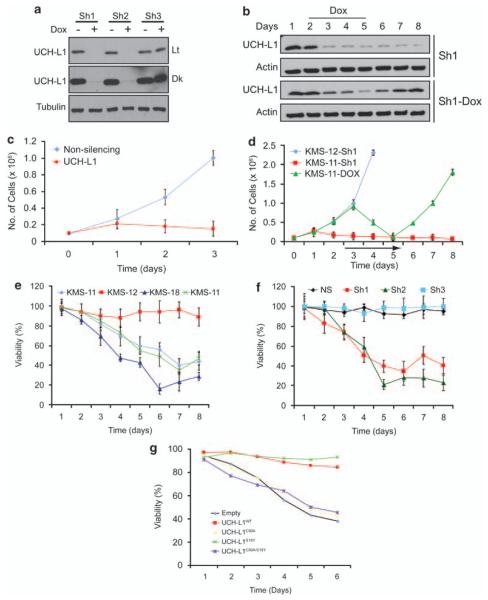
Although our data show that UCH-L1 acts as an oncogene, it is not clear whether malignant B cells are subject to ‘oncogene addiction’—a state in which transformed cells require the continued expression of the inciting oncogene. A recent report using RNA interference showed a modest reduction in proliferation in BL cell lines depleted of UCH-L1, suggesting that it is required for some of the ongoing malignant behaviors in these established lines.19 It is notable, however, that high levels have also been seen in other B-cell cancers, including the lethal plasma-cell malignancy and multiple myeloma.38 This observation led us to question whether UCH-L1 may be involved in the pathogenesis of other B-cell cancers as well. To address these issues, we selected a series of human B-cell lines on the basis of their varying expression of UCH-L1 and transducibility with lentiviral vectors. Of these cell lines, we analyzed three UCH-L1-positive myeloma lines (KMS-11, KMS-18 and KMS-28) and one that lacks UCH-L1 (KMS-12) as visualized by immuno-blotting (Supplementary Figure S2). To determine the role of UCH-L1 in the survival and growth of malignant B cells, we transduced the UCH-L1-expressing line KMS-11 with either non-silencing or the UCH-L1-targeting Sh1-encoding lentivirus. Cells transduced with non-silencing virus continue to proliferate robustly following selection (Figure 5c). In contrast, while growing at a similar rate for the first 1–2 days, we observe a dramatic decline in cell numbers in KMS-11 cells at days 3 and beyond, coincident with the observed decline in UCH-L1 protein levels on those days. To eliminate the possibility of toxicity resulting from viral integration, we repeated these experiments with doxycycline (dox)-inducible small hairpin RNA (shRNA) constructs and included the UCH-L1-negative cell line KMS-12. Whereas KMS-12 cells continue to proliferate after transduction with UCH-L1-targeting shRNA, we again observe a dramatic loss of cell numbers in KMS-11 cells transduced with either the constitutive or doxycycline-inducible knockdown constructs (Figure 5d). The inhibition of proliferation is reversible, as cells resume a normal growth rate following removal of doxycycline. These results were verified in two other UCH-L1-expressing cell lines (KMS-18 and KMS-28; Figure 5e) and with another UCH-L1 targeting shRNA (Figure 5f). These results strongly indicate that UCH-L1-expressing cells require continued enzyme expression for growth and survival.
Figure 5.
UCH-L1 is required for the survival of UCH-L1-expressing malignant B-cells. (a) KMS-11 cells transduced with the indicated Dox-inducible constructs were incubated with Dox for 5 days and then assayed for knockdown by immunoblotting. The blots shown are representative of more than three independent experiments. (b) KMS-11 cells transduced with the indicated shRNA constructs were sampled on the indicated days for expression of UCH-L1 by immunoblotting. Doxycycline was included on the indicated days. Protein loading was controlled by actin. (c) KMS-11 cells were transduced with the indicated shRNA constructs and selected as above. Cell numbers were determined by trypan blue exclusion at the indicated times. (d) KMS-11 or KMS-12 cells were transduced with the indicated shRNA constructs, and cell numbers were determined as in (c). Doxycycline was included on days 3 through 5 as indicated by the black line. (e) The indicated cell lines were transduced with the Sh1-construct and viability was determined by MTS assay. (f) KMS-28 cells were transduced with the indicated shRNA constructs and selected with puromycin. Viability, as determined by MTS assay, was determined in triplicate on the indicated day and results for non-silencing shRNA were set at 100%. (g) KMS-28 cells transduced with DOX-inducible Sh1 were transduced with the indicated shRNA-resistant UCH-L1 constructs, and then incubated in doxycycline to deplete endogenous UCH-L1. Viability was monitored daily by the MTS assay. Time is expressed as days in doxycycline. All growth curves and viability assays were performed in triplicate and were performed on at least three independent occasions. Abbreviations: Dk, dark; Lt, light exposure; NS, non-silencing shRNA; Sh1 = UCH-L1 targeting construct 1; Sh2 = UCH-L1 targeting construct 2.
UCH-L1 requires its de-ubiquitinase activity to support the survival of malignant B-cells
While clearly bearing the sequence signature of a DUB, the functional targets of UCH-L1 have been the subject of debate from some time. Biochemical substrate profiling experiments show UCH-L1 to have a very fast hydrolysis rate towards small Ub-adducts (for example, Ub-ethyl ester) and a very slow rate towards large protein adducts (for example, di-ubiquitin).14 Adding further complexity, UCH-L1 was found to possess an unusual ligase activity that maps to a region distinct from the DUB active site. A mutation known to reduce the risk of Parkinson’s disease (UCH-L1S18Y) was found to reduce the ligase activity, while not affecting the DUB activity.16,39 To gain further mechanistic insight into the ability of UCH-L1 to promote survival and proliferation of malignant B-cells, we determined the ability of various UCH-L1 constructs to rescue cell death that results from UCH-L1 depletion. One of our potent UCH-L1-targeting shRNA constructs (Sh1) anneals with sequences in the 3′ untranslated region of the UCH-L1 mRNA. Therefore, our cDNA constructs, which lack the 3′-untranslated region, are shRNA resistant and can be used to complement the depletion of endogenous UCH-L1. To examine the requirement for UCH-L1’s catalytic activities in mediating cell survival, we generated lentivirus constructs encoding wild-type UCH-L1 (UCH-L1WT), with mutations affecting the de-ubiquitinase activity (UCH-L1C90A, Supplementary Figure S3), or its putative ligase activity (UCH-L1S18Y). After selection of stable integrants, doxycycline was added to deplete endogenous UCH-L1, and cell viability was monitored daily. As expected, cell death was entirely prevented by expressing UCH-L1WT (Figure 5g). In contrast, expression of UCH-L1C90A could not rescue the cells from depletion-induced death. Cells transduced with UCH-L1 encoding a mutation in the putative ligase region of UCH-L1 (UCH-L1S18Y) were fully rescued from depletion-induced cell death. A minor role for ligase activity cannot be excluded, however, as this mutation was not reported to eliminate all ligase activity. These data indicate that UCH-L1 promotes the survival and proliferation of transformed cells in a mechanism requiring de-ubiquitinase activity, but not its putative ligase activity.
UCH-L1 expression boosts signaling through the Akt pathway
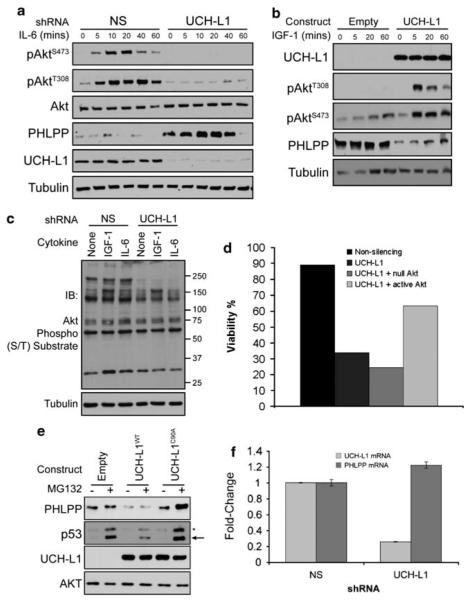
An important pathway that has strong effects on both the survival and proliferative programs is the Akt-signaling network. This pathway is important for the growth and survival in a number of cancers, is known to enhance lymphomagenesis in the Eμ-myc model,40 and has been implicated in altering cell morphology and invasiveness in the UCH-L1-expressing non-small-cell lung cancer line H157.20 To examine the effect of UCH-L1 expression on Akt phosphorylation, we stimulated the UCH-L1-positive cell line KMS-28 with the cytokine interleukin-6, which is known to be an important Akt-activating signal in malignant B-cells.41,42 In cells transduced with a non-silencing shRNA control, we observe low-level baseline phosphorylation with a brisk increase following cytokine exposure (Figure 6a, Supplementary Figure S4). In contrast, in cells expressing a UCH-L1 knockdown construct, phosphorylation was dramatically reduced. These reductions were seen in both pAktT308 and pAktS473, although the latter was usually more dramatic. To exclude global toxicity for the observed decline in Akt phosphorylation, we examined the levels of phospho-STAT5 and found them to be unaltered (data not shown). Confirming these results, introduction of UCH-L1 into HeLa cells (UCH-L1 negative) leads to augmented Akt phosphorylation on both pAktT308 and pAktS473 (Figure 6b). To test whether the increased Akt phosphorylation results in downstream effects, we examined the phosphorylation of Akt substrates using a polyspecific anti-phospho-(RXRXXS/T) antibody. Following UCH-L1 knockdown, we observe a substantial reduction in the phosphorylation of downstream targets, most notably in the higher molecular masses (Figure 6c). To assess whether the increase in Akt signaling is important to the pro-survival activity of UCH-L1, we tested the ability of a constitutive Akt mutant (myr-Akt43) to rescue UCH-L1-depleted cells from death (Figure 6d). Although a null Akt construct (AktT308A/S473A) did not improve cell viability, myr-Akt produced a substantial, but not complete, rescue from cell death (Figure 6d). These data suggest that UCH-L1 promotes malignant B-cell survival by boosting signaling through the Akt pathway.
Figure 6.
UCH-L1 expression leads to enhanced Akt signaling in vitro. (a) KMS-28 cells transduced with either control (NS) or the UCH-L1 targeting Sh1 construct were serum starved and stimulated with 10 ng/ml interleukin-6 (IL-6) for the indicated times. Extracts were immuno-blotted for the indicated proteins. (b) HeLa cells were transduced with empty lentivirus (empty) or virus encoding wild-type UCH-L1 and stimulated with 50 ng/ml insulin-like growth factor-1 (IGF-1). Extracts were immuno-blotted for the indicated proteins. (c) KMS-28 cells were transduced with the indicated shRNA constructs and stimulated with the indicated cytokine. Extracts were blotted for phospho-S/T Akt substrates as indicated (d) KMS-28 cells stably transduced with doxycycline-inducible non-silencing, or UCH-L1 targeting shRNAs were transduced with either null Akt (AktT308A/S473A) or constitutively active Akt (myr-Akt). Following the addition of doxycycline, cells were monitored daily for viability with the MTS assay. The results shown were obtained in triplicate on day 6. (e) HeLa cells transduced with either virus or those encoding wild-type UCH-L1 or catalytic mutant UCH-L1 (C90A) were prepared as in (b) without the addition of cytokine. Empty or Sh1 knockdown constructs were incubated with MG132 for 2 h, followed by immunoblotting for the indicated proteins. Extracts were immuno-blotted with the indicated proteins. p53 accumulation confirms proteasome inhibition. Asterisk denotes a non-specific band. The relative decrease in p53 levels seen with UCH-L1WT is not reproducible. (f) PHLPP mRNA was measured by qRT-PCR. RNA was extracted from cells transduced with the indicated shRNA following overnight serum starvation. All reactions were performed in triplicate and values were normalized to those of tata-binding protein (TBP). All immunoblots are representative of at least three independent experiments. qRT-PCR was performed on two independent occasions with similar results. The P-value comparing the levels in non-silencing to UCH-L1 shRNA is >0.05.
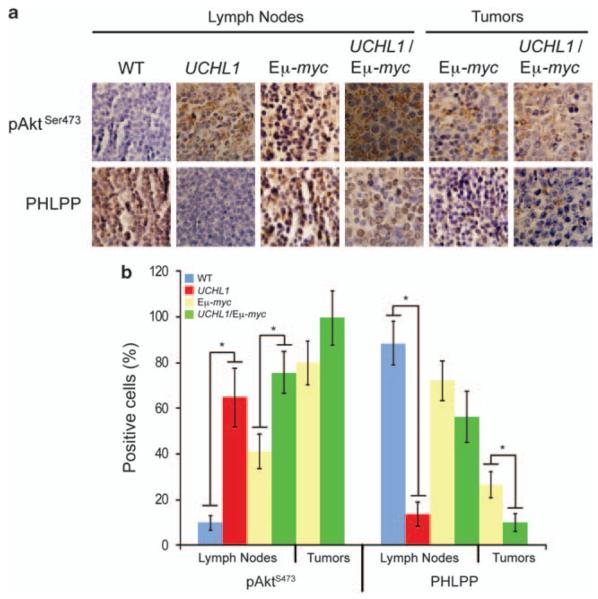
To confirm that these in vitro findings are relevant in vivo, we examined the presence of phospho-Akt in tissues from wild-type and transgenic mice by immunohistochemistry. Wild-type lymph nodes have low levels of pAktS473 (Figure 7). In striking contrast, lymph nodes from age-matched Uchl1 mice have high levels of pAktS473. A similar relationship was seen in tumor tissues, although relative to non-malignant lymph nodes, lymphomatous tissues were found to have a general increase in pAktS473. Together, these data strongly suggest that UCH-L1 expression leads to enhanced Akt phosphorylation in vitro and in vivo.
Figure 7.
Akt phosphorylation is enhanced in vivo in response to UCH-L1 expression. (a) The indicated tissue sections were prepared from either wild-type or Uchl1 mice. The images are representative of fields. Immunohistochemistry was performed with antibodies targeting pS473 or PHLPP as indicated. (b) Quantitation of immunohistochemistry performed in a. Immunohistochemistry was performed on tissues from three independent mice of each genotype. Values reflect the average percentage of positive cells in 10 random fields from each mouse. Error bars represent the standard deviation. Asterisk denotes P<0.05 by the chi-square test.
UCH-L1 activity leads to reduced levels of the phosphatase PHLPP1
Phosphorylation of Akt is the result of the activity of the kinases PDK1 and the mammalian target of rapamycin (mTORC2).44 PDK1 phosphorylates Akt on T308, whereas mTORC2 acts on S473.45,46 Antagonizing these activities are phosphatases including PP2A,47 PHLPP148 and CTMP.49 To understand the mechanism behind the effect of UCH-L1 on Akt phosphorylation, we determined the levels of these proteins in UCH-L1-positive cells transduced either with non-silencing or with UCH-L1 targeting shRNA. Regardless of the level of UCH-L1, we observed uniform amounts of the kinases PDK1, the TORC2 components mTOR and rictor, as well as the phosphatases PP2A and CTMP, suggesting that UCH-L1 does not function by modulating the levels of these proteins (Supplementary Figure S4). In contrast, we observed a dramatic increase in the phosphatase PHLPP1 levels following depletion of UCH-L1 (Figure 6a and Supplementary Figure S4). The apparent molecular mass of the affected isoform is 170 kDa, consistent with the PHLPP-β isoform.48 The smaller α isoform was also reduced when it was detected, but the inconsistency of its detection prevents us from drawing firm conclusions regarding its regulation. In some experiments (as in Figure 6a) we observed a decline in PHLPP1 levels following 1 h of cytokine stimulation, although this decline was not consistently seen. To confirm the relationship between UCH-L1 and PHLPP1, we transduced HeLa cells with either an empty vector or UCH-L1 and examined the level of PHLPP1 by immunoblotting. We observed an inverse relationship between the expression of UCH-L1 and the levels of PHLPP1 (Figure 6b). To determine whether UCH-L1 similarly affects PHLPP1 levels in vivo, we examined the levels of PHLPP1 in tissues from wild-type and transgenic mice by immunohistochemistry. Whereas PHLPP1 was readily seen in lymph nodes of wild-type mice, we observed a near total lack of the enzyme in sections from Uchl1 mice (Figure 7). UCH-L1 expression led to a reduction in PHLPP levels in the lymph nodes and tumors of Eμ-myc mice as well. These results strongly suggest that UCH-L1 has a suppressive effect on the protein level of the phosphatase PHLPP1 in vitro and in vivo.
Recent reports suggest that PHLPP1 is degraded through activity of the β-TrCP skip, F-box and cullin (SCF) Ub ligase in a phospho-dependent manner.50 As UCH-L1 is a DUB, it is tempting to speculate that its effect on PHLPP1 involves the UPS, although it is clear that UCH-L1 does not salvage PHLPP1 from proteasomal degradation. To examine the effect of proteasomal proteolysis on PHLPP1, and to examine the requirement for UCH-L1 catalytic activity, we incubated HeLa cells, transduced with either empty virus or those encoding UCH-L1WT or UCH-L1C90A, with the proteasome inhibitor MG132. Surprisingly, incubation with MG132 for 2, 6 or 12 h did not change the levels of PHLPP1 in these experiments (Figure 6E and Supplementary Figure S5). We observed that the effect of UCH-L1 on PHLPP1 levels is dependent on catalytic activity, as expression of a catalytic mutant UCH-L1 construct had no effect on PHLPP1 levels compared with the empty vector. These data suggest that UCH-L1 leads to reduced PHLPP1 levels in a non-proteasome-dependent manner.
To refine our understanding of the mechanism behind PHLPP1 modulation, we analyzed the mRNA level of PHLPP1 using quantitative real-time PCR in KMS-28 cells following the knockdown of UCH-L1. If UCH-L1 suppressed the transcription of PHLPP1, we would expect to see an increase in PHLPP1 mRNA levels upon UCH-L1 depletion. Instead, we observed essentially unchanged levels of PHLPP1 mRNA in knockdown cells (Figure 6e) despite an increase at the protein level in immunoblots (Figure 6d). We therefore conclude that UCH-L1 does not alter the level of the PHLPP1 transcript. Taken together, these data suggest that UCH-L1 has an indirect effect on PHLPP1 levels that is independent of transcription and proteasomal degradation but is dependent on UCH-L1 catalytic activity.
UCH-L1 is highly expressed in human lymphoid malignancies
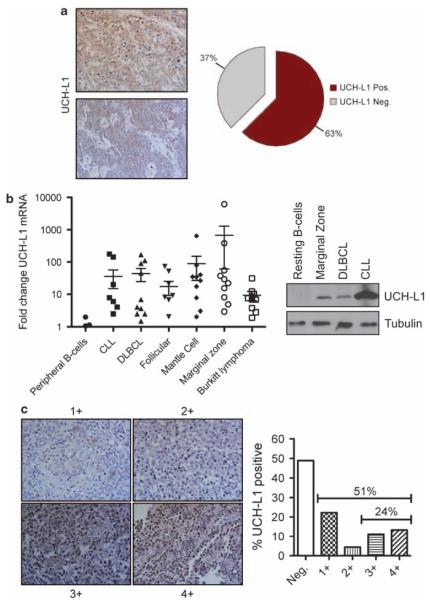
UCH-L1 expression has been observed at the protein, and activity level, in a sampling of cell lines derived from human B-cell malignancies, but the true incidence of expression in primary lesions is unknown. To more accurately determine the incidence of UCH-L1 expression in B-cell malignancies, we examined the frequency of UCH-L1 expression in 24 cases of human BL by immunohistochemistry. We found that 15/24 (62.5%) of cases had widespread expression, confirming that UCH-L1 expression is common in this disease (Figure 8a). To determine whether UCH-L1 expression is common in other forms of B-cell malignancy, we analyzed UCH-L1 mRNA levels in a panel of 59 human primary samples of B-cell lymphoma and leukemia using qRT-PCR. The samples consisted of 10 cases each of chronic lymphocytic leukemia, diffuse large B-cell lymphoma (DLBCL), follicular lymphoma, mantle cell lymphoma, marginal zone lymphoma and nine cases of BL. For comparison, we isolated resting primary human B-cells from 14 normal donors. Consistent with our previous observations, we observed a marked increase in the level of UCH-L1 mRNA in primary samples of B-cell malignancy, with 48% having at least 10-fold increased levels compared with the average seen in resting primary B-cells (Figure 8b). There was variability in the incidence of elevated UCH-L1 expression across disease types, such that 30% of chronic lymphocytic leukemia samples had >10-fold expression and a median 3.96-fold increase whereas 70% of marginal zone lymphoma samples had a greater than 10-fold increase with a median of 25.63-fold increased mRNA abundance. Three out of nine samples (33%) of BL had a greater than 10-fold increase with an overall median of 6.8-fold. To confirm that increased levels of mRNA lead to increased UCH-L1 protein, we prepared extracts from selected samples and compared their expression levels by immunoblotting. Samples with increased levels of UCH-L1 mRNA indeed had high levels of UCH-L1 protein (Figure 8b). We further examined the expression of UCH-L1 in 45 primary B-cell lymphoma specimens by immunohistochemistry. To facilitate quantitation, all samples were classified as having negative or 1–4+ staining based on the numbers of cells positive and the intensity of staining. In this series of mixed B-cell lymphoma subtypes, approximately 50% showed at least 1+ staining and 24% had abundant (3–4+) staining (Figure 8c). In aggregate, these data show that a substantial subset of B-cell lymphoma specimens express UCH-L1 at the mRNA and protein level.
Figure 8.
Increased UCH-L1 expression is highly prevalent in human B-cell cancers. (a) UCH-L1 was detected in a panel of 24 primary Burkitt’s lymphoma samples by immunohistochemistry. Two UCH-L1-positive samples are shown along with a graph showing the prevalence of UCH-L1 expression. Only those samples with widespread immunoreactivity were considered positive. (b) UCH-L1 mRNA was measured from a panel of 59 primary human samples, of the indicated histological subtypes, by quantitative real-time PCR and compared with values obtained from a panel of peripheral B-cells purified from eight healthy donors. All values were obtained in triplicate and were normalized to levels of the housekeeping gene tata-binding protein (TBP). (b) Immunoblot of UCH-L1 expression at the protein level in selected primary samples from (a). Results are representative of two independent experiments. (c) Immunohistochemical staining for UCH-L1 in a B-cell lymphoma tissue microarray (n = 45 cases, unrelated to those in (a) or (b)). All cases were quantified based on a scale ranging from negative (no staining) to 4 + (widespread staining).
Discussion
Using a novel transgenic mouse model, we have established that UCH-L1 is a potent oncogene that drives the development of cancer in vivo. Uchl1 transgenic mice develop a high incidence of spontaneous tumors with a tumor spectrum consisting primarily of lymphomas and lung adenomas. This provides direct evidence for an oncogenic role of a DUB. Supporting an important role for UCH-L1 in driving lymphoid malignancies, we find that aberrant UCH-L1 expression dramatically accelerated the development of lymphoma in the Eμ-myc model, and led to the emergence of malignant features that closely resemble human BL. We find that UCH-L1 promotes proliferation and survival of lymphocytes in vitro and in vivo, at least in part, through its ability to boost Akt signaling. We additionally report a novel mechanism by which UCH-L1 affects the Akt pathway by suppressing levels of the antagonistic phosphatase PHLPP1, an event that requires UCH-L1 de-ubiquitinase activity. Taken together with the observed high rate of UCH-L1 expression in primary human B-cell malignancies, these data suggest that UCH-L1 is an important driving force behind the development of these common human diseases.
UCH-L1 is an oncogene that promotes lung tumors and lymphomagenesis
Although the Uchl1 transgene is active in many tissues, we were struck by the relatively limited tumor spectrum in our mice. Previous studies have found high levels of UCH-L1 in a set of human carcinomas, including those of the lung, colon and pancreas. UCH-L1 over-expression has been observed at the transcript and protein level in non-small-cell lung cancer (NSCLC). While not detectable in normal adjacent lung tissue, the presence of UCH-L1 mRNA in tumor samples correlated with more advanced tumor stage, suggesting a possible relationship with pathogenesis.51,52 Results from in vitro studies have provided conflicting results. Incubation of NSCLC cell line H129 with either UCH-L1 siRNA or a specific UCH-L1 inhibitor LDN-57444 resulted in increased proliferation.25 The conclusion from these data was that UCH-L1 is an inhibitor of tumor cell growth. More recently, UCH-L1 expression was found to correlate with more invasive behavior in NSCLC cell lines.20 Our data establish that UCH-L1 acts as an oncogene that initiates the development of lung adenomas and adenocarcinomas. It is possible that the exact subtype of NSCLC may influence the response to unregulated UCH-L1 expression.
Previous studies have observed high levels of UCH-L1 transcript,32 protein38 and enzymatic activity18 in B-cell cancers. Whether UCH-L1 expression in these studies was a cause, or effect, of malignant transformation has been unknown. Our Uchl1 transgenic mice develop B-cell tumors with features resembling human DLBCL and plasma cell malignancies—two human cancers in which increased UCH-L1 expression has been observed by us (DLBCL, this study) and others (DLBCL32 and multiple myeloma38). Our data showing that Uchl1 mice develop spontaneous lymphomas firmly establish that UCH-L1 acts as an oncogene and drives the malignant transformation in these conditions. In our report, we have examined the prevalence of high levels of UCH-L1 in primary lymphoma specimens by qRT-PCR and immunohistochemistry. First, we find that a high proportion of BL specimens have high levels of UCH-L1 mRNA and widespread protein immunoreactivity. High levels of UCH-L1 mRNA have been found in BL, leading to its inclusion in a specific molecular signature of this disease based on gene expression profiling.32 We also observed high levels of UCH-L1 mRNA and protein in a variety of other B-cell cancers, suggesting that UCH-L1 may be an important oncogene in these related but distinct forms of malignancy. It is noteworthy that there was a considerable range in the level of mRNA in the cases analyzed, ranging from some with less than peripheral B-cells to others with several hundred-fold increased levels. Based on our data, it is not possible determine whether UCH-L1 is more important in the pathogenesis of any one lymphoma subtype, and future study will be geared towards determining the potential oncogenic role of UCH-L1 in these varied diseases. It is possible that there is a threshold level beyond which UCH-L1 acts oncogenically, whereas in the other cases UCH-L1 expression has little impact on the malignant phenotype. Unfortunately, both clones of Uchl1 mice have very similar levels of UCH-L1 expression in lymph node samples, making it impossible for us to experimentally determine whether such a threshold exists.
To further examine the impact of UCH-L1 expression on the development of lymphoma in vivo, we chose to combine our novel Uchl1 transgenic model with the well-characterized model Eμ-myc.33,34 Surprisingly, tumor sites in Uchl1/Eμ-myc mice were dramatically different from those in the Eμ-myc parental line, with a great reduction in splenic enlargement and the frequent appearance of large abdominal masses. According to the American Burkitt Lymphoma Registry, while only 20% of patients have involvement of the spleen, 56% present with abdominal masses.53 These observations may suggest that enforced UCH-L1 expression leads to clinical features that more closely approximate those seen in humans. We hypothesize that these differences may reflect alterations in the expression of adhesion molecules that regulate homing to these anatomic locations. Underscoring this possibility are observations, in separate reports, from the groups of Masucci and Lee.19,20 These labs independently observed a strong impact of the expression of UCH-L1 on the adhesion properties as read out as culture morphology19 or cell migration in culture.20 Further demonstrating the importance of UCH-L1 in B-cell malignancy is our observation that UCH-L1 knockdown leads to a dramatic loss of viability and loss in proliferation in our set of B-cell lines. In agreement with our observations, the Masucci group also observed that depletion of UCH-L1 in BL and EBV-transformed lymphoblastoid cell lines leads to decreased proliferation.19 In that report, expression of UCH-L1, but not a catalytic mutant version, boosted the proliferation of the UCH-L1-negative lymphoblastoid line CBM. Our observations are that catalytic activity is required to suppress levels of PHLPP1 in malignant B-cells—perhaps providing an explanation for these data.
UCH-L1 leads to reduced levels of PHLPP1, and boosts Akt signaling
While exploring the mechanism by which UCH-L1 promotes cell growth and survival in B-cell malignancies, we observed a striking effect of UCH-L1 on the Akt signaling pathway. The Akt signaling pathway provides critical survival and proliferative signals to many types of cancers, including lymphoma.40,54,55 This is underscored by the development of small-molecule inhibitors by pharmaceutical companies currently in phase II trials for cancer.56,57 In a recent report exploring the differential invasive tendencies of non-small-cell lung cancer cell lines, UCH-L1 was found to be an important mediator with invasiveness and cell migration.20 In agreement with our data, this study also showed a modest effect of UCH-L1 on Akt in H157 cells. There have been several reports of the efficacy of Akt inhibitors, such as perifosine, in producing cell death in numerous cancer cell types including myeloma cells.58 The impact of enhanced Akt signaling on lymphomagenesis in the Eμ-myc model has been previously shown. Using a retroviral transduction stem-cell transplantation approach, Wendel et al.40 showed that a mutant Akt with constitutive activity strongly accelerated the development of lymphoma when combined with the Eμ-myc transgene. These data underscore the importance of the Akt pathway in malignant B-cells and provide precedence for the suggestion that UCH-L1 promotes the survival and proliferation of malignant B-cells through its effect on Akt signaling.
Our data suggest that UCH-L1 expression leads to a profound downregulation of the phosphatase PHLPP1. PHLPP1 specifically removes phosphate groups from AktS473 and as such counteracts the activity of the mTORC2 complex.48 Activation of Akt kinase activity requires phosphorylation both on T308 and S473, making PHLPP1 capable of inactivating the Akt pathway. Conversely, reduced levels of PHLPP1 lead to increased activity of the Akt pathway. Suggesting a direct link between reduced PHLPP1 and cancer, PHLPP1 expression is reduced in some cancers, including colon cancer, glioblastoma cell lines, and in primary colon cancer specimens, in a manner that correlates with the levels of Akt phosphorylation.48,59 Our data reveal that UCH-L1 has a strong negative effect on the levels of PHLPP1 and we observe a strong inverse correlation between PHLPP1 and pAktS473 in vivo. Given the stable levels of other kinases and phosphatases that function on Akt, it seems likely that the observed effect of increased Akt phosphorylation is due to reduced levels of PHLPP1.
The mechanism by which UCH-L1 affects the levels of PHLPP1 is currently not clear. We have found that UCH-L1 does not affect the level of PHLPP1 transcript and does not direct PHLPP1 for proteasomal degradation. These results suggest that UCH-L1 may affect the level of PHLPP1 in other ways, such as by altering the rate of translation. How might this occur? There is a strong relationship between the Akt signaling pathway and the mTOR kinase.60 In addition to its activity as an Akt-activating kinase, mTOR can alternately assemble into mTORC1 complexes, where it regulates cap-dependent translation, in part through phosphorylation of 4E-BP1 and the S6 kinase. Both of these events have a positive effect on the translation of capped mRNAs. In addition, mTORC1 signaling negatively regulates signaling from PI3-kinase and thereby blunts the activation of Akt. Although we have measured levels of mTOR, as well as the mTORC1 component raptor and mTORC2 component rictor and found no changes, it is possible that UCH-L1 affects the assembly of these complexes in such a way that mTORC2 is favored, leaving fewer mTOR molecules to assemble into mTORC1—leading to a decline in translation. Currently, little is known regarding the regulation of mTOR complex assembly. It is also possible that UCH-L1 enhances non-proteasomal proteolytic pathways that lead to a degradation of PHLPP1. UCH-L1 is commonly found in aggresomes seen in the brains of patients with Parkinson’s disease, and a causative role in aggresome formation has been postulated for UCH-L1.61 It is possible in cells expressing high levels of UCH-L1 that PHLPP1 becomes sequestered in aggresomes, although whether this would reduce the total amount of PHLPP1 detected in SDS lysates is doubtful. Lastly, it is also possible that PHLPP is degraded through a lysosomal pathway, such as through autophagy; however, ubiquitination is thought to serve as a degradative signal in autophagy (as well as for the proteasome), requiring an indirect mechanism that connects high levels of a DUB and increased autophagy.62 Further studies are needed to clarify these potential mechanisms and to understand how UCH-L1 may impact these pathways.
Supplementary Material
Acknowledgements
We thank Ying Zhang and members of the van Deursen lab for helpful discussions. We are grateful to Rick Bram, Mitchell Cairo, Andre Catic and Victor Quesada for critically reading the manuscript, as well as Drs Fang Jin and Darren Baker for their help with histopathology. PJG is a Harriet H Samuelsson Foundation Pediatric Research Cancer Scientist, is a Basic Science Scholar of the American Society of Hematology, is supported by the Howard Hughes Medical Institute Early-Career Physician Scientist Award, and is a GAFCR research fellow. JvD is supported by the NIH.
Footnotes
Conflict of interest The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
References
- 1.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 2.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 3.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Schmitz R, Hansmann ML, Bohle V, Martin-Subero JI, Hartmann S, Mechtersheimer G, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206:981–989. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novak U, Rinaldi A, Kwee I, Nandula SV, Rancoita PM, Compagno M, et al. The NF-\{kappa\}B negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood. 2009;113:4918–4921. doi: 10.1182/blood-2008-08-174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honma K, Tsuzuki S, Nakagawa M, Tagawa H, Nakamura S, Morishima Y, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009;114:2467–2475. doi: 10.1182/blood-2008-12-194852. [DOI] [PubMed] [Google Scholar]
- 7.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliveira AM, Perez-Atayde AR, Inwards CY, Medeiros F, Derr V, Hsi BL, et al. USP6 and CDH11 oncogenes identify the neoplastic cell in primary aneurysmal bone cysts and are absent in so-called secondary aneurysmal bone cysts. Am J Pathol. 2004;165:1773–1780. doi: 10.1016/S0002-9440(10)63432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- 10.Hussain S, Zhang Y, Galardy PJ. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle. 2009;8:1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 11.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 12.Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 13.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen CN, Krantz BA, Wilkinson KD. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry. 1998;37:3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson KD, Lee KM, Deshpande S, Duerksen-Hughes P, Boss JM, Pohl J. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science. 1989;246:670–673. doi: 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 17.Gavioli R, Frisan T, Vertuani S, Bornkamm GW, Masucci MG. c-myc overexpression activates alternative pathways for intra-cellular proteolysis in lymphoma cells. Nat Cell Biol. 2001;3:283–288. doi: 10.1038/35060076. [DOI] [PubMed] [Google Scholar]
- 18.Ovaa H, Kessler BM, Rolen U, Galardy PJ, Ploegh HL, Masucci MG. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc Natl Acad Sci USA. 2004;101:2253–2258. doi: 10.1073/pnas.0308411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolen U, Freda E, Xie J, Pfirrmann T, Frisan T, Masucci MG. The ubiquitin C-terminal hydrolase UCH-L1 regulates B-cell proliferation and integrin activation. J Cell Mol Med. 2009;13:1666–1678. doi: 10.1111/j.1582-4934.2008.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HJ, Kim YM, Lim S, Nam YK, Jeong J, Kim HJ, et al. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2009;28:117–127. doi: 10.1038/onc.2008.364. [DOI] [PubMed] [Google Scholar]
- 21.Rosas SL Bittencourt, Caballero OL, Dong SM, da Costa Carvalho Mda G, Sidransky D, Jen J. Methylation status in the promoter region of the human PGP9.5 gene in cancer and normal tissues. Cancer Lett. 2001;170:73–79. doi: 10.1016/s0304-3835(01)00449-9. [DOI] [PubMed] [Google Scholar]
- 22.Mandelker DL, Yamashita K, Tokumaru Y, Mimori K, Howard DL, Tanaka Y, et al. PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer Res. 2005;65:4963–4968. doi: 10.1158/0008-5472.CAN-04-3923. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita K, Park HL, Kim MS, Osada M, Tokumaru Y, Inoue H, et al. PGP9.5 methylation in diffuse-type gastric cancer. Cancer Res. 2006;66:3921–3927. doi: 10.1158/0008-5472.CAN-05-1511. [DOI] [PubMed] [Google Scholar]
- 24.Mizukami H, Shirahata A, Goto T, Sakata M, Saito M, Ishibashi K, et al. PGP9.5 methylation as a marker for metastatic colorectal cancer. Anticancer Res. 2008;28:2697–2700. [PubMed] [Google Scholar]
- 25.Liu Y, Lashuel HA, Choi S, Xing X, Case A, Ni J, et al. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem Biol. 2003;10:837–846. doi: 10.1016/j.chembiol.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 26.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker DJ, Perez-Terzic C, Jin F, Pitel K, Niederlander NJ, Jeganathan K, et al. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat Cell Biol. 2008;10:825–836. doi: 10.1038/ncb1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietz AB, Bulur PA, Emery RL, Winters JL, Epps DE, Zubair AC, et al. A novel source of viable peripheral blood mononuclear cells from leukoreduction system chambers. Transfusion. 2006;46:2083–2089. doi: 10.1111/j.1537-2995.2006.01033.x. [DOI] [PubMed] [Google Scholar]
- 29.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 30.O’Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morse HC, III, Anver MR, Fredrickson TN, Haines DC, Harris AW, Harris NL, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–258. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- 32.Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 33.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 34.Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167:353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalla-Favera R, Martinotti S, Gallo RC, Erikson J, Croce CM. Translocation and rearrangements of the c-myc oncogene locus in human undifferentiated B-cell lymphomas. Science. 1983;219:963–967. doi: 10.1126/science.6401867. [DOI] [PubMed] [Google Scholar]
- 36.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Link MP, Weinstein HJ. Chapter 24. Malignant non-Hodgkin lymphomas in children. In: Pizzo PA, Poplack DG, editors. Principles and Practice of Pediatric Oncology. 5th edn Lippincott Williams & Wilkins; Philadelphia, PA: 2006. pp. 722–747. [Google Scholar]
- 38.Otsuki T, Yata K, Takata-Tomokuni A, Hyodoh F, Miura Y, Sakaguchi H, et al. Expression of protein gene product 9.5 (PGP9.5)/ubiquitin-C-terminal hydrolase 1 (UCHL-1) in human myeloma cells. Br J Haematol. 2004;127:292–298. doi: 10.1111/j.1365-2141.2004.05205.x. [DOI] [PubMed] [Google Scholar]
- 39.Ragland M, Hutter C, Zabetian C, Edwards K. Association between the ubiquitin carboxyl-terminal esterase L1 gene (UCHL1) S18Y variant and Parkinson’s disease: a HuGE review and meta-analysis. Am J Epidemiol. 2009;170:1344–1357. doi: 10.1093/aje/kwp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 41.Tu Y, Gardner A, Lichtenstein A. The phosphatidylinositol 3-kinase/AKT kinase pathway in multiple myeloma plasma cells: roles in cytokine-dependent survival and proliferative responses. Cancer Res. 2000;60:6763–6770. [PubMed] [Google Scholar]
- 42.Hideshima T, Nakamura N, Chauhan D, Anderson KC. Biologic sequelae of interleukin-6 induced PI3-K/Akt signaling in multiple myeloma. Oncogene. 2001;20:5991–6000. doi: 10.1038/sj.onc.1204833. [DOI] [PubMed] [Google Scholar]
- 43.Andjelkovic M, Alessi DR, Meier R, Fernandez A, Lamb NJ, Frech M, et al. Role of translocation in the activation and function of protein kinase B. J Biol Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 44.Franke TF. PI3K/Akt: getting it right matters. Oncogene. 2008;27:6473–6488. doi: 10.1038/onc.2008.313. [DOI] [PubMed] [Google Scholar]
- 45.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 46.Franke TF, Yang SI, Chan TO, Datta K, Kazlauskas A, Morrison DK, et al. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 47.Beaulieu JM, Sotnikova TD, Marion S, Lefkowitz RJ, Gainetdinov RR, Caron MG. An Akt/beta-arrestin 2/PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell. 2005;122:261–273. doi: 10.1016/j.cell.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 48.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 49.Maira SM, Galetic I, Brazil DP, Kaech S, Ingley E, Thelen M, et al. Carboxyl-terminal modulator protein (CTMP), a negative regulator of PKB/Akt and v-Akt at the plasma membrane. Science. 2001;294:374–380. doi: 10.1126/science.1062030. [DOI] [PubMed] [Google Scholar]
- 50.Li X, Liu J, Gao T. beta-TrCP-mediated ubiquitination and degradation of PHLPP1 are negatively regulated by Akt. Mol Cell Biol. 2009;29:6192–6205. doi: 10.1128/MCB.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hibi K, Westra WH, Borges M, Goodman S, Sidransky D, Jen J. PGP9.5 as a candidate tumor marker for non-small-cell lung cancer. Am J Pathol. 1999;155:711–715. doi: 10.1016/S0002-9440(10)65169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasaki H, Yukiue H, Moriyama S, Kobayashi Y, Nakashima Y, Kaji M, et al. Expression of the protein gene product 9.5, PGP9.5, is correlated with T-status in non-small cell lung cancer. Jpn J Clin Oncol. 2001;31:532–535. doi: 10.1093/jjco/hye113. [DOI] [PubMed] [Google Scholar]
- 53.Levine PH, Kamaraju LS, Connelly RR, Berard CW, Dorfman RF, Magrath I, et al. The American Burkitt’s Lymphoma Registry: eight years’ experience. Cancer. 1982;49:1016–1022. doi: 10.1002/1097-0142(19820301)49:5<1016::aid-cncr2820490527>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 54.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 55.Yoeli-Lerner M, Toker A. Akt/PKB signaling in cancer: a function in cell motility and invasion. Cell Cycle. 2006;5:603–605. doi: 10.4161/cc.5.6.2561. [DOI] [PubMed] [Google Scholar]
- 56.Marsh Rde W, Lima CM Rocha, Levy DE, Mitchell EP, Rowland KM, Jr, Benson AB., III A phase II trial of perifosine in locally advanced, unresectable, or metastatic pancreatic adenocarcinoma. Am J Clin Oncol. 2007;30:26–31. doi: 10.1097/01.coc.0000251235.46149.43. [DOI] [PubMed] [Google Scholar]
- 57.Van Ummersen L, Binger K, Volkman J, Marnocha R, Tutsch K, Kolesar J, et al. A phase I trial of perifosine (NSC 639966) on a loading dose/maintenance dose schedule in patients with advanced cancer. Clin Cancer Res. 2004;10:7450–7456. doi: 10.1158/1078-0432.CCR-03-0406. [DOI] [PubMed] [Google Scholar]
- 58.Hideshima T, Catley L, Yasui H, Ishitsuka K, Raje N, Mitsiades C, et al. Perifosine, an oral bioactive novel alkylphospholipid, inhibits Akt and induces in vitro and in vivo cytotoxicity in human multiple myeloma cells. Blood. 2006;107:4053–4062. doi: 10.1182/blood-2005-08-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene. 2009;28:994–1004. doi: 10.1038/onc.2008.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005;8:179–183. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 61.Ardley HC, Scott GB, Rose SA, Tan NG, Robinson PA. UCH-L1 aggresome formation in response to proteasome impairment indicates a role in inclusion formation in Parkinson’s disease. J Neurochem. 2004;90:379–391. doi: 10.1111/j.1471-4159.2004.02485.x. [DOI] [PubMed] [Google Scholar]
- 62.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.