Abstract
Hepatitis C virus (HCV) relies on host lipid metabolic pathways for its replication, assembly, secretion and entry. HCV induces de novo lipogenesis, inhibits β-oxidation and lipoprotein export resulting in a lipid enriched cellular environment critical for its proliferation. We investigated the effects of a hypolipidemic agent, nordihydroguaiaretic acid (NDGA) on host lipid/fatty acid synthesis and HCV life cycle. NDGA negated the HCV induced alteration of host lipid homeostasis. NDGA decreased sterol regulatory element binding protein (SREBP) activation and enhanced expression of genes involved in β-oxidation. NDGA inhibited very low-density lipoprotein (VLDL) secretion by affecting mediators of VLDL biosynthesis. Lipid droplets (LDs), the neutral lipid storage organelles, play a key role in HCV morphogenesis. HCV induces accumulation and perinuclear distribution of LDs whereas NDGA most notably reduced the overall number and increased the average size of LDs. The antiviral effects of NDGA resulted in reduced HCV replication and secretion.
Conclusions
NDGA-mediated alterations of host lipid metabolism, LD morphology and VLDL transport appear to negatively influence HCV proliferation.
Keywords: SREBP, VLDL, β-oxidation, Lipid metabolism, MTP
The Hepatitis C virus (HCV) infection represents a global pandemic with an estimated 3% of world population infected (1). Chronic HCV infection results in severe liver diseases such as steatosis, cirrhosis and hepatocellular carcinoma (1,2). HCV is a single-stranded positive sense RNA virus of Flaviviridae family. HCV is critically dependent on host lipid metabolism for its proliferation (3). Lipidomic analysis during acute HCV infection identified temporal fluctuations in specific lipid species (e.g. phospholipids and sphingomyelins) that play key roles in viral replication, virion morphogenesis/assembly and secretion (4). HCV is shown to deregulate lipid homeostasis by modulating key players such as SREBPs, PPARα, and CPT1a, resulting in increased lipogenesis, lipid uptake and reduced β-oxidation, creating a lipid rich microenvironment conducive for viral proliferation (5-8). Inhibiting SREBP activation, FAS activity or cholesterol biosynthesis abrogates HCV replication (9-11).
HCV replication occurs in close proximity to LDs and this proximity facilitates the downstream viral morphogenetic events (12,13). HCV infection induces accumulation and redistribution of LDs around the perinuclear region (14). Several reports link HCV secretion to cellular very low-density lipoprotein (VLDL) secretion. Silencing apolipoprotein B-100 (ApoB) or pharmacological inhibition of microsomal triglyceride transfer protein (MTP) inhibits HCV secretion (15,16). ApoE is also shown to be involved in HCV entry/infectivity and virion morphogenesis (17,18). HCV infection causes a decline in VLDL secretion (19,20) and correlates with hypoβlipoproteinemia seen in chronic hepatitis C patients (21). These observations strongly support the role of VLDL during HCV secretion and highlight the importance of intracellular lipid enriched microenvironment, and lipoprotein export for HCV proliferation. Reagents/bioactive molecules that can perturb the viral interference on cellular lipid homeostasis can potentially hamper viral infectious process.
NDGA {l,4-bis (3,4-dihydroxyphenyl) 2,3-dimethyl-butane}, is a phenolic antioxidant derived from desert shrub Larrea tridentata (22). It has antioxidant, anti-inflammatory and anti-proliferative properties and has been used as traditional medicine against diabetes, and other health problems (22). NDGA was used to stabilize hepatic lipid deregulation in high fat diet-fed mice by improving lipid breakdown (23,24). The hypolipidemic effects of NDGA prompted this investigation to determine its influence on HCV infection. Here, we report the effect of NDGA on various aspects of HCV lifecycle eventually leading to inhibition of viral proliferation.
Materials and Methods
Reagents and plasmids
NDGA and BSA-conjugated Oleic acid are from Sigma-Aldrich, St Louis, USA. Antibodies against actin, PPARα, and FAS are from Santa Cruz Biotechnology, Santa Cruz, CA. Source of other antibodies used; anti-ApoB (Chemicon International, Temecula, CA), anti-SREBP-1 and anti-LC3 (Abcam, Cambridge, MA), anti-HCV core, (Affinity BioReagents, Golden, CO). Monoclonal anti-HCV-NS5A and JC1-p7-RLuc2A plasmid (25) are gifts from Dr. C. Rice (Rockefeller University, NY). Anti-ApoE was gift of Dr. G. Luo, (University of Kentucky, KY). Reporter plasmids SRE-Luc, FAS-Luc, LDLr-Luc, MTP-Luc and HCV-5’ NCR-Luc plasmids have been described previously (5, 20, 26). JFH1 clone was a gift of Dr. T. Wakita (Tokyo, Japan). The genotype 2a (JFH1) full-length (FL-Feo) and sub-genomic replicon and genotype 1b sub-genomic replicon (BM45-Feo) plasmids containing neomycin-luciferase fusions are gifts of Dr. David Wyles, (University of California, San Diego).
Cell Culture, HCV infection and Foci Forming Unit (FFU) assay
The Huh7 subgenomic or full-length JFH replicon cells were maintained in 500 μg/ml Geneticin. Huh7.5.1 cell line was a gift of Dr. Frank Chisari (Scripps Institute, La Jolla, CA). NDGA was dissolved in DMSO and added to culture medium with DMSO concentration not exceeding 0.1%. The cytotoxicity of NDGA was determined at 48h by using TOX-8 cytotoxicity kit (Sigma-Aldrich, St. Louis, USA), as per manufacturer's instructions. In-vitro synthesized viral genomic RNAs (RiboMax RNA production system, Promega, Madison, WI) were electroporated into Huh7.5.1 cells using cytomix buffer as described (27). The cells were passaged at confluency every 2 to 3 days and culture supernatants collected. The infectivity of the supernatant was determined by foci forming unit (FFU) assays as described (28). Huh7 cells infected with JFH virus (Huh7-JFH) at 0.01 MOI were used for experiments 9-12 days post-infection. Huh7.5.1 cells infected with JCI or JC1p7-RLuc2A virus at 0.01 MOI were used 5 days post-infection. The relative levels of JC1p7-RLuc2A reporter virions in culture supernatants were analyzed by infecting naïve Huh7.5.1 cells and measuring the transduced renilla luciferase activity 72h post-infection as described (25). Intracellular infectivity was determined as described previously (15) and intracellular/extracellular HCV RNA was quantified by real-time PCR as described (29). HCV core ELISA was done using ORTHO HCV antigen ELISA kit (Wako Chemicals, Richmond, VA) as per manufacturer's instructions. HCV pseudoparticle assays for analyzing HCV entry were carried out as described (30).
Quantitative Real-time PCR
The relative mRNA levels were quantified by comparative CT method by subjecting the cDNA to real-time qPCR using DyNAmo SYBR qRT-PCR mix (Finnzymes, Finland) as per manufacturer's instructions. Gene-specific primers used are detailed in supplementary information.
Immunofluorescence
Fluorescent immunostaining was performed as described (27). For staining lipid droplets 1μg/ml of Bodipy-493/503 was used. The images were analyzed by Olympus FluoView 1000 confocal microscope.
Results
NDGA inhibits HCV replication
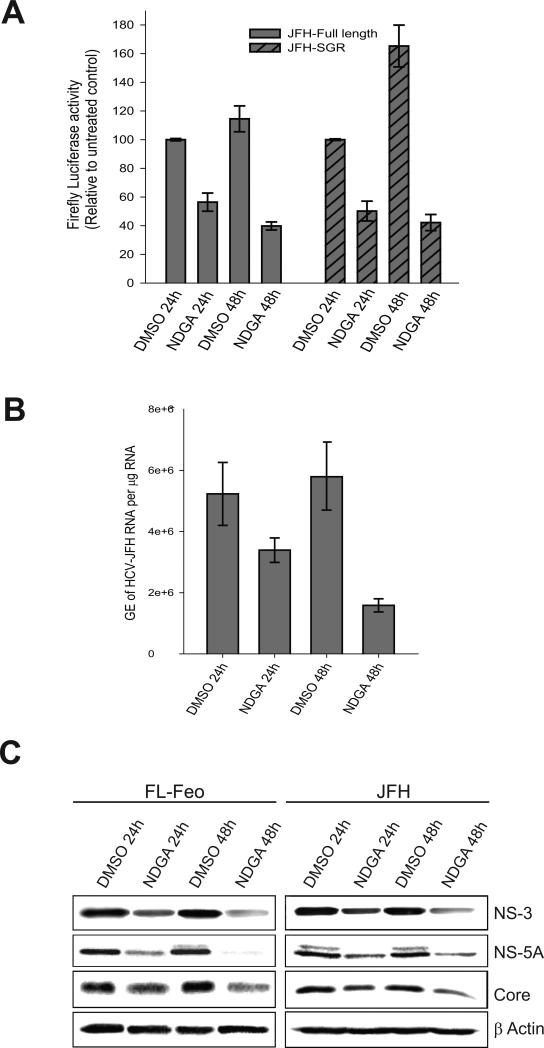
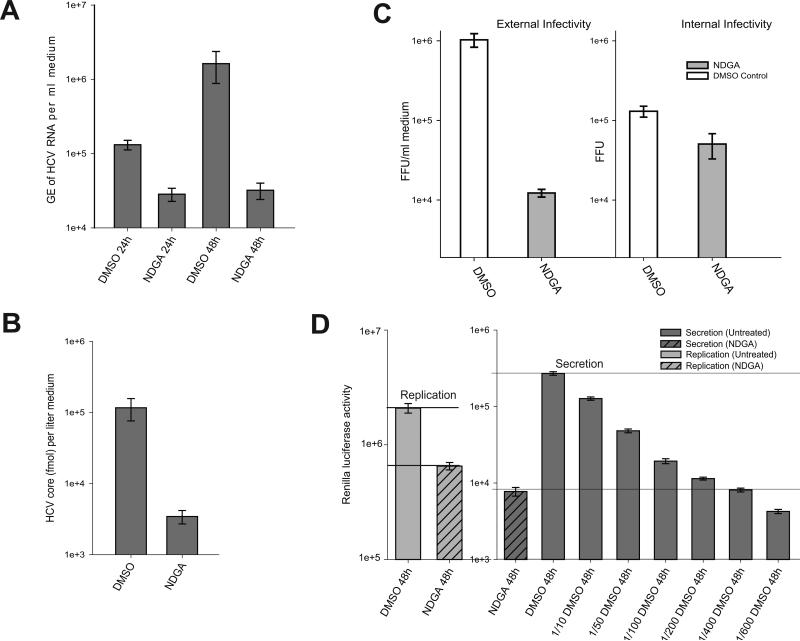
NDGA induces hypolipidemic effects by altering host lipid metabolism (23,24). We investigated the NDGA mediated hypolipidemic actions on HCV-induced lipid homeostasis and its effect on viral infectious process. Initially to evaluate cytotoxicity Huh7 cells were treated with various concentrations (10-100 μM) of NDGA for 48h. NDGA had a dose-dependent effect on Huh7 cell viability with a CC50 value of 70μM (Fig. S1A). Huh7 cells harboring full-length replicon (FL-Feo) of HCV genotype-2a with an in-frame luciferase gene were treated with various concentrations of NDGA for 48h and HCV replication assessed by luciferase activity. NDGA showed a dose-dependent inhibition of HCV replication, with the EC50 value of about 30μM (Fig S1B). For subsequent studies, 35μM NDGA was used. We then analyzed the effect of 35μM NDGA on replication of sub-genomic and full-length HCV-JFH replicons at 24h and 48h post-treatment. In both the sub-genomic and full-length replicon cells the luciferase activity in untreated control cells increased from 24h to 48h but decreased by about 70% after 48h in NDGA treated cells (Fig 1A). The HCV-JFH cell culture based infection system (HCVcc) also yielded similar results. 48h after NDGA treatment the intracellular levels of HCV RNA in infected cells was suppressed by about 70% compared to untreated cells (Fig 1B). In correlation with above results, the immunoblot analysis of HCV proteins after 24h and 48h NDGA treatment in replicon and infected cells also showed similar decline in the HCV protein levels compared to untreated controls (Fig 1C). We also analyzed the effect of NDGA on genotype 1b sub-genomic replicon cells (BM45-Feo). Genotype 1b replication was decreased by about 60% after 48h of NDGA treatment (Fig S2) indicating its genotype-independent effects on viral replication. Together, these results indicate that NDGA inhibited both the HCV RNA and protein expression.
Figure 1. NDGA inhibits HCV replication.
Huh7-JFH replicon cells harboring full-length or sub-genomic HCV genome of genotype 2a (JFH) and Huh7 cells persistently infected with culture derived HCV JFH virus were treated with 35μM NDGA for the indicated time. The control cells were treated with DMSO. A) HCV replication in the replicon cells was determined as a function of the firefly luciferase activity. B) HCV replication in JFH infected cells determined by qRT-PCR of intracellular HCV RNA and data shown as genome equivalents (GE) of HCV RNA per μg total RNA. The experiments were carried out in triplicate and data shown are mean ± SEM. C) Western blot analysis showing indicated HCV proteins in the full-length genotype 2a JFH replicon (FL-Feo) and infected (JFH) cell lysates after NDGA treatment. β-actin is used as protein loading control.
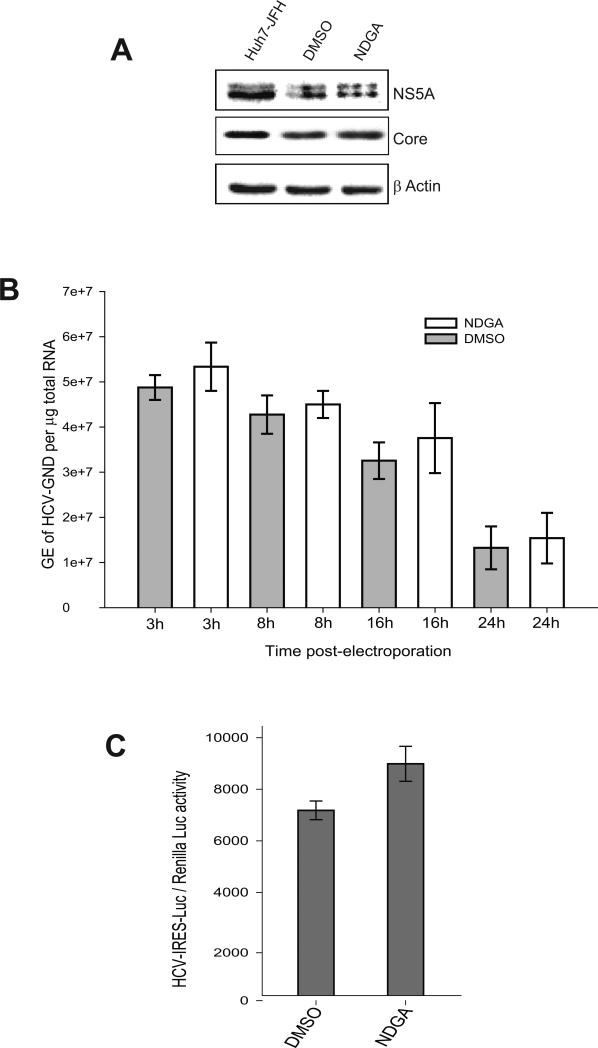
We next investigated if NDGA treatment also affects HCV IRES-dependent translation. Huh7 cells pretreated with NDGA for 24h were electroporated with replication defective GND mutant (JFH-GND) HCV RNA and continued to be treated with NDGA. The GND mutant RNA is defective in replication but is translated efficiently. Cell lysates were analyzed for HCV protein expression by Western blot at 8h post-electroporation. We observed no significant difference in levels of HCV Core and NS5A proteins in NDGA pretreated or untreated cells (Fig 2A). The intracellular levels of GND mutant RNA was determined by qRT-PCR at 3h, 8h, 16h and 24h post-electroporation in both NDGA treated and untreated cells to determine the electroporation efficiency and RNA decay. The results show that NDGA does not affect RNA stability (Fig 2B). Overall these results suggest that NDGA treatment does not hinder HCV RNA translation or mediate its degradation. The effect of NDGA on HCV-IRES translation was also analyzed using luciferase reporter plasmid HCV-5’NCR-Luc (26). No significant variation in the HCV-IRES luciferase reporter activity was observed (Fig 2C). These results suggest that the inhibition in HCV RNA replication and protein expression caused by NDGA is not a consequence of inhibition in HCV IRES-mediated translation.
Figure 2. NDGA does not affect HCV-IRES translation.
Huh7 cells pretreated with NDGA or DMSO treated for 48h were electroporated with replication defective JFH GND RNA and cell lysates analyzed for HCV protein expression 8h post electroporation. A) Western blot analysis showing expression of HCV core and NS5A proteins. Cell lysates of persistently infected cells (Huh7-JFH) were used as positive control and β-actin as protein loading control. B) Intracellular levels of JFH-GND RNA in respective cell lysates at 3h, 8h, 16h and 24h post-electroporation determined by qRT-PCR analysis and data shown as genome equivalents (GE) of HCV RNA per μg total RNA. C) The HCV-IRES activity determined by HCV-IRES luciferase reporter plasmid HCV 5’NCR-Luc. Huh7 cells pretreated with NDGA or DMSO for 24h were transfected with HCV 5’NCR-Luc RNA and treatment with NDGA or DMSO continued for further 24h prior to estimation of firefly luciferase activity in cell lysates. Renilla luciferase plasmid was used as transfection control.
NDGA subverts HCV induced alterations of lipogenesis
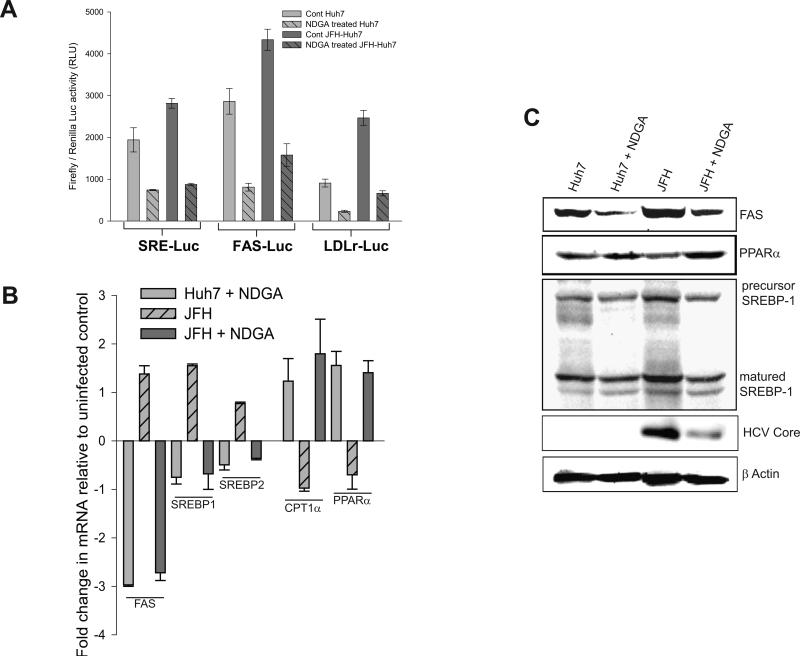
HCV gene expression stimulates lipogenesis (3). HCV activates master regulators of cholesterol/lipid biosynthetic enzymes, the SREBP-1 and -2 and reduces expression of PPARα and CPT1a (5-8). To determine the effect of NDGA on SREBP activation, HCV infected and uninfected Huh7 cells were transfected with SRE-luciferase reporter plasmids and treated with NDGA for 48h post-transfection. Luciferase activity was reduced by about 60% in both the infected and uninfected NDGA treated cells (Fig 3A). Further, we used luciferase reporter plasmids in which promoters of fatty acid synthase (FAS) or low-density lipoprotein receptor (LDLr) genes control transcription. In both cases, 48h NDGA treatment resulted in about a 60% decline in luciferase activity compared to untreated controls (Fig 3A). In contrast HCV infection stimulated the transcription of SREBPs, FAS and LDLr (Fig. 3A, see cont Huh7 vs cont JFH-Huh7). Both FAS and LDLr promoters contain SREBP binding sites and are upregulated by SREBPs in addition to other transcription factors (31,32). We then evaluated variations in the mRNA levels of a few representative genes such as FAS, SREBP1c, SREBP2, PPARα and CPT1a in HCV infected and uninfected Huh7 cells treated with and without NDGA for 48h. HCV infection increased levels of FAS mRNA and reduced PPARα and CPT1a mRNA (Fig 3B), consistent with the previous reports. In contrast, 48h NDGA treatment subverted HCV lipogenic signaling by increasing PPARα and CPT1a mRNA levels and reducing FAS mRNA (Fig 3B). Immunoblot analysis of FAS, PPARα and SREBP-1 proteins in HCV infected and uninfected Huh7 cells treated with or without NDGA further support the data in Fig. 3B (Fig 3C). Overall these results suggest that NDGA mediated inhibition of HCV replication may be a consequence of its effects on lipid metabolism. To evaluate this hypothesis, NDGA treated cells were supplemented with oleic acid (100 μM). Oleic acid supplementation could only partially restore HCV replication. This may be due to multiple effects of NDGA on lipid metabolism (Fig S3A).
Figure 3. NDGA subverts the HCV-altered/induced lipid biosynthetic proteins.
Uninfected (Huh7) or persistently infected (Huh7- JFH) cells were transiently transfected with the respective reporter plasmids and treated with NDGA or DMSO for 48h. The cell lysate were then used to determine firefly luciferase activity. Renilla luciferase plasmid was used as transfection control. A) The firefly luciferase activity of the indicated reporter plasmids normalized against renilla luciferase activity after NDGA or DMSO treatment in Huh7 and HCV infected Huh7 (Huh7-JFH) cells. B) Alterations in mRNA levels of the indicated genes determined by qRT-PCR as described in Material and Methods in NDGA or DMSO treated Huh7 and persistently infected Huh7-JFH cells relative to the DMSO treated Huh7 cells. C) Western blot analysis of the lysate used for experiment described in B for indicated proteins. All experiments were carried out in triplicates and data shown are mean ± SEM.
NDGA reduces the number and enhances size of Lipid droplets
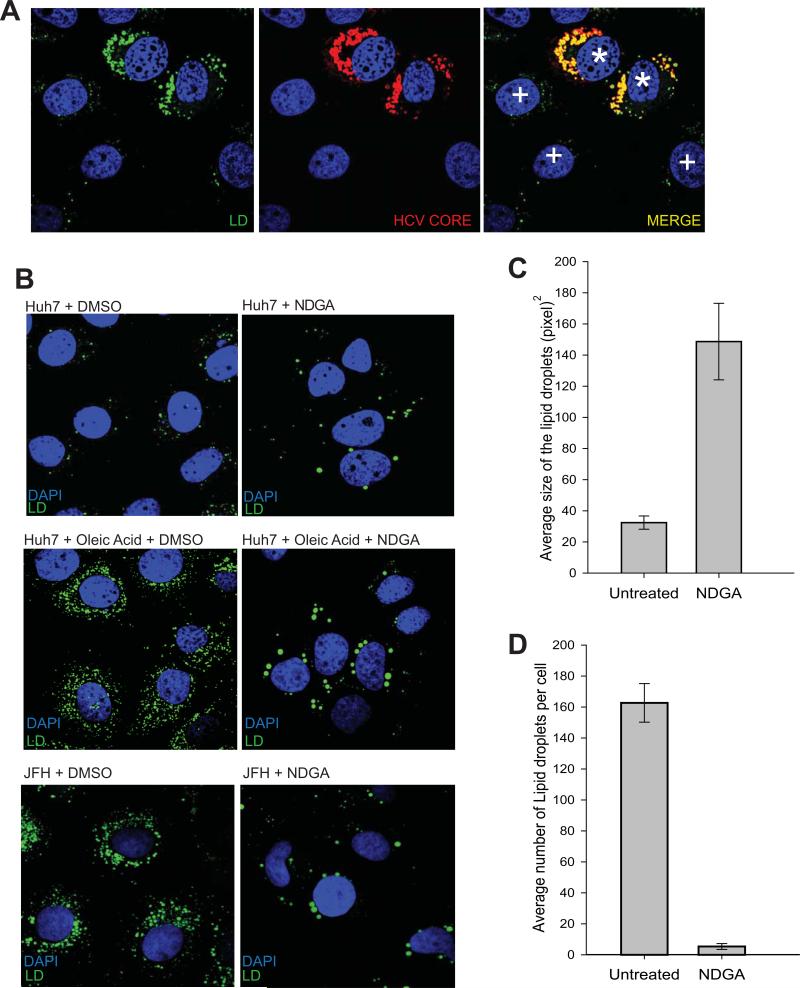
Alteration of host lipid metabolism associated with HCV infection leads to accumulation of lipid droplets (2,3,13,14). Our studies thus far showed that NDGA subverted HCV mediated alterations of lipid metabolism by preventing SREBP activation and inducing genes involved in lipid breakdown. We studied the effect of NDGA on accumulation and arrangements of LDs in HCV infected cells. In correlation with previous reports (14) we observed that HCV infection led to significant induction and rearrangement of LDs around the perinuclear space in contrast to uninfected cells (Fig 4A, see HCV infected cells marked with * sign vs uninfected cells marked with + sign, in the merged panel). Under normal physiologic conditions, Huh7 cells display few LDs, (see Huh7 cells vs HCV infected or oleic acid-stimulated Huh7 cells in Fig 4B). In oleic acid-stimulated or HCV infected Huh7 cells, NDGA treatment led to a decrease in number and increase in average size of the LDs (Fig 4B and 4C). The increase in size and reduction in number of LDs caused by NDGA treatment (Fig 4C and 4B) could be due to fusion activity between LDs and/or delivery of newly synthesized lipids to preexisting LD (33, 34). It has been shown that lipophagy regulates intracellular lipid stores (35). Further investigations on possible induction of lipophagy by NDGA did not reveal any lipophagy being induced by NDGA treatment (Fig S4A), as there was no colocalization of LDs with the lysosomal protein Lamp1 or enhanced lipidation of LC3-I to LC3-II although the overall levels of LC3 protein increased (Fig S4B).
Figure 4. NDGA severely affects lipid droplet numbers and morphology.
A) HCV infected Huh7-cells (Huh7-JFH) were stained for LDs using Bodipy and HCV core protein and imaged as described in Material and Methods. The LDs are shown in green and HCV core protein in Red. The * and + signs in the merged panel indicate HCV infected and uninfected Huh7 cells respectively. B) Control Huh7, oleic acid stimulated Huh7 and HCV (JFH) infected Huh7 cells were treated with NDGA or DMSO for 48h and stained for LDs. Upper row represents Huh7 cells treated with DMSO or NDGA. Middle row shows Huh7 cells supplemented with 100 μM BSA-oleic acid complex and treated with DMSO or NDGA. The lower row displays HCV infected (JFH) Huh7 cells treated with DMSO or NDGA. C) Relative quantitation of change in LDs size and number in NDGA or DMSO treated HCV (JFH) infected Huh7 cells. Quantitation was done manually in 20 individual cell images from NDGA treated and untreated conditions using Image J software.
NDGA suppresses HCV virus assembly and secretion
HCV replication occurs in close juxtaposition to LDs and the viral particle morphogenesis requires interaction of core and NS5A proteins with LDs (12, 13). Since NDGA treatment altered distribution, number and morphology of LDs, we investigated if NDGA treatment affects viral assembly and secretion. The intracellular infectivity of JCI infected Huh7.5.1 cells treated with and without NDGA for 48h was evaluated as previously described (15). The results indicate that NDGA treatment causes a reduction in accumulation of matured infectious viral particles compared to untreated cells (Fig 5C). The intracellular infectivity was also determined in NDGA treated cells supplemented with oleic acid, to evaluate if the increase in LD number restores HCV assembly. The results reveal that oleic acid supplementation only partially restored HCV intracellular infectivity not reaching to the levels seen in untreated cells (Fig S3B) probably due to lower availability of viral components compared to untreated cells (Fig S3A).
Figure 5. NDGA inhibits HCV secretion.
A) Persistently infected Huh7.5.1 cells were treated with NDGA or DMSO and the accumulation of HCV viral particles in the culture supernatant evaluated at indicated time points by qRT-PCR analysis of HCV RNA isolated from culture supernatant and data shown as genome equivalents (GE) of HCV RNA per ml culture supernatant. B) The level of HCV core protein in the 48h culture supernatants used in experiment A were analyzed by OrthoHCVcore antigen ELISA kit as described in Material and Methods. C) Infectivity of the 48h culture supernatants used in experiment A (extracellular infectivity) and intracellular infectivity of crude lysates of cells used in experiment A was determined by Foci Forming Unit (FFU) assay as described in Materials and Methods. D) Inhibition in HCV secretion by NDGA assayed using the JCI-p7-Rluc 2A reporter virus. Infectivity of the culture supernatants was determined by estimating the renilla luciferase activity 48h post-infection as described in Materials and Methods. The naïve Huh7.5.1 cells were infected with samples of culture supernatants undiluted or those at various dilutions, collected 48h post-NDGA or DMSO treatment as indicated. Intracellular renilla luciferase activity in lysates of parent cultures was used to determine the effect on HCV replication. All the experiments were carried out in triplicated and the results shown are mean ± SEM.
We then evaluated the secretion of viral particles into culture supernatants in JCI infected Huh7.5.1 cells treated with NDGA for 48h. Analysis of extracellular HCV RNA purified from infectious culture supernatants showed that 48h NDGA treatment led to a 2 log-fold reduction in the HCV secretion compared to untreated control (Fig 5A). Analysis of HCV core protein levels in culture supernatant by ELISA also yielded close to 2 log-fold declines in level of core protein after 48h NDGA treatment (Fig 5B). Foci-forming unit (FFU) assay, a more reliable functional assay for analyzing infectivity of infectious HCV culture supernatants also confirmed that the supernatants of NDGA treated cultures were about 2 log-fold less infectious compared to untreated control (Fig 5C). We also performed a similar functional assay using the JCI-p7Rluc2A reporter virus (25). Huh7.5.1 cell cultures infected with JCI-p7Rluc2A reporter virus were treated with or without NDGA for 48h and naïve Huh7.5.1 cells were infected with log fold dilutions of supernatants of untreated control cultures and with undiluted supernatants from NDGA treated cultures respectively. The infectivity was determined by assaying for transduced renilla luciferase activity in cell lysates 72 h post-infection (Fig 5D). The results indicate that the infectivity of undiluted supernatant from NDGA treated cultures was close to the infectivity of 400-fold diluted untreated culture supernatant (Fig 5D). The level of HCV RNA replication in the NDGA treated and untreated parent cultures was also evaluated by estimating intracellular renilla luciferase activity (Fig 5D). The reference lines drawn on the bar graph depict the log fold difference between HCV replication and secretion levels in NDGA treated and untreated samples. These results indicate that NDGA inhibits HCV replication and assembly by ~60% but more strongly effects HCV secretion by two log-fold (Fig 5D). Next we analyzed the localization of core and NS5A on LDs after NDGA treatment by immunofluorescence (Fig S5). In agreement with earlier results on HCV replication, NDGA treatment resulted in diminution of core and NS5A protein levels and perturbed the typical perinuclear distribution pattern observed in untreated infected cells for both the proteins (Fig S5). However the proteins still localized to the periphery of LDs suggesting that NDGA treatment did not interfere with their interaction to LDs but affects their expression and cytosolic distribution.
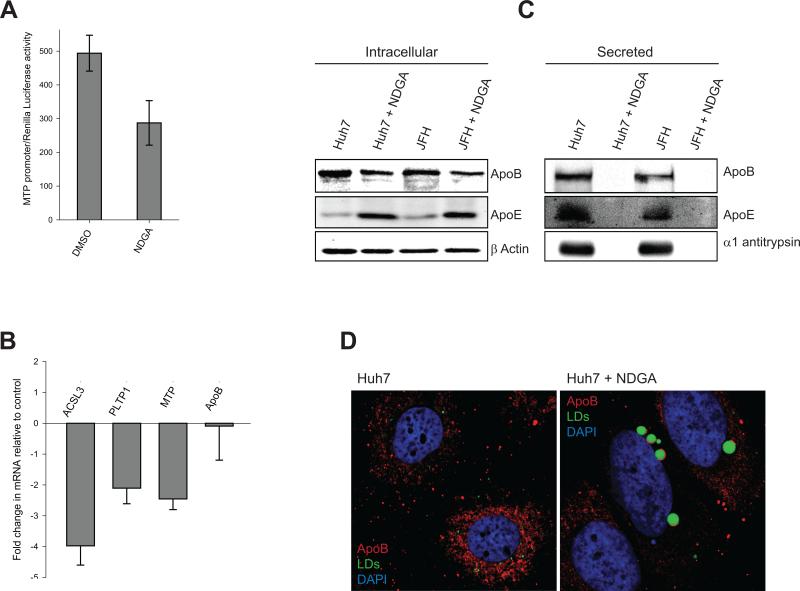
NDGA inhibits VLDL secretion
It is widely acknowledged that HCV co-opts the VLDL secretory pathway for its egress (15,16,36,37). MTP protein initiates VLDL biosynthesis in endoplasmic reticulum (ER) by co-tranlsational lipidation of nascent ApoB in the ER (38). Since NDGA treatment reduced the secretion of virus (Fig. 5), we investigated if NDGA inhibited MTP expression. Huh7 cells transfected with MTP-promoter reporter plasmid were treated with NDGA for 48h. NDGA treatment led to 40% reduction of MTP promoter activity (Fig 6A) indicating that NDGA inhibits MTP gene transcription. Analysis of MTP mRNA levels also yielded similar results (Fig 6B). Unlipidated ApoB is subjected to proteasomal degradation (39), in correlation to this we observed a decline in the intracellular levels of ApoB in NDGA treated cells (Fig 6C) although the ApoB mRNA levels did not significantly alter (Fig 6B), indicating an inhibition of MTP activity by NDGA. Inhibition in VLDL synthesis after MTP mediated ApoB lipidation results in accumulation of lipidated ApoB as crescents on the periphery of the LDs (40). Analysis of cytoplasmic distribution of ApoB in NDGA treated and untreated Huh7 cells by immunofluorescence revealed a characteristic reticular distribution pattern of ApoB in control cells but enhanced number of ApoB crescents on periphery of LDs in NDGA treated cells (Fig 6D). This suggests that NDGA treatment results in the inhibition of VLDL biosynthesis downstream of MTP mediated lipidation of ApoB. Recently, proteins, like acyl-CoA synthetase 3 (ACSL3) and phospholipid transfer protein 1 (PLTP1) have been shown to play crucial role in VLDL biosynthesis and secretion downstream of MTP (37,41). Analysis of mRNA levels of these proteins after NDGA treatment resulted in about 4-fold reduction in ACSL3 mRNA and nearly 2-fold reduction in PLTP1 mRNA (Fig 6B). These observations suggest that NDGA inhibits VLDL secretion by inhibiting MTP expression and activity and also events downstream of MTP. We examined the secretion of VLDL particles by directly monitoring the levels of ApoB and ApoE proteins in the culture supernatants as has been described previously (16). NDGA treatment completely abolished VLDL secretion in both uninfected and HCV infected Huh7 cells (Fig 6C). Interestingly, NDGA also completely abolished secretion of α1anti-trypsin suggesting that it inhibited general intracellular secretory transport (Fig 6C). In contrast to ApoB intracellular levels of ApoE increased with NDGA treatment (Fig 6C) probably due to the accumulation of unsecreted ApoE and not enhanced expression because we observed that ApoE mRNA levels remain constant in NDGA treated and untreated cells (Fig S6). Overall the results suggest that NDGA inhibits VLDL secretion by inhibition of MTP activity and down-regulation of genes essential for VLDL secretion pathway.
Figure 6. NDGA inhibits VLDL secretion.
A) Effect of NDGA on MTP promoter activity. To determine the MTP promoter activity Huh7 cells were transiently transfected with MTP-promoter-Luc reporter plasmid followed by treatment with NDGA or DMSO for 48h. The firefly luciferase activity in cell lysates was then determined to estimate the relative MTP promoter activity. The values were normalized against the renilla luciferase activity, which was used as transfection control. B) Changes in the mRNA levels of indicated genes in NDGA treated Huh7 cells relative to DMSO treated cells were determined by qRT-PCR as described in material and methods. All the experiment were carried out in triplicates and the results shown here are mean ± SEM. C) Intracellular levels of ApoB and ApoE were determined by Western analysis of uninfected Huh7 and infected (JFH) cells treated with NDGA or DMSO for 48h. VLDL secretion in the culture supernatants of above experiment was determined by Western blot analysis of ApoB and ApoE levels in culture supernatants. D) Immunofluorescence of ApoB in NDGA treated or untreated Huh7 cells. ApoB is shown in red and LDs in green and the nuclei counterstained with DAPI in blue.
Discussion
Targeting host factors that facilitate viral proliferation can be an effective strategy in designing specific antiviral agents. The various stages of HCV viral life cycle such as entry, replication and viral assembly and/or secretion are dependent on cellular lipid metabolism (3,9,15). Here we explored the hypolipidemic effects of NDGA on HCV life cycle. HCV infection generates a lipid rich microenvironment conducive for its proliferation by deregulating lipid homeostasis (5-8, 19,20). Here, we show that NDGA subverts HCV induced alterations of lipid homeostasis and thwarts cellular lipid enrichment induced by HCV. NDGA reduced the activation of SREBPs and their target genes FAS and LDLr (Fig 3A and B). Unlike HCV infection, NDGA enhanced the expression of PPARα and its downstream target CPT1a (Fig 3B), implying that NDGA decreases hepatic lipogenesis and enhances catabolic breakdown of lipids. NDGA prevented fatty liver in high-fat fed mice by upregulating the AMPK kinase, a central player in lipid and glucose homeostasis (24). Interestingly, HCV infection has been shown to inhibit AMPK activation via protein kinase B and activation of AMPK abrogated lipid accumulation and inhibited HCV replication (42). HCV infection induces modified membranous structures, on which it assembles its replication complexes that engage in RNA replication activities (43). Such membrane alterations require enrichment and reorganization of specific lipids and lipid-protein interactions (10,44). NDGA through its hypolipidemic actions can alter such effects and inhibit HCV replication. NDGA treatment of persistently infected or replicon cells which possess well established replication complexes resulted in a modest decline in replication whereas the inhibition was more profound in cells in which infection was yet to be established (data not shown). The most notable effect of NDGA was on the LDs. NDGA-treated HCV infected cells displayed reduced number and enlarged LDs (Fig 4B). The LDs are enclosed by phospholipid monolayers and when phospholipids are limiting, fusion of LDs is induced to overcome the reduced surface-to-volume ratio of droplets (34). However, the mechanism behind the observed increase in the size of LDs after NDGA treatment is not clearly understood. HCV infection results in induction and relocation of LDs which play a key role in virion morphogenesis (12-14). Carboxyl-esterase 1 (CES1), an enzyme that regulates LDs abundance and size has recently been shown to be activated during HCV infection and positively influence HCV secretion (45). NDGA treatment reduced intracellular infectivity of infected cells (Fig 5C) although NDGA did not influence the interaction of HCV core or NS5A to LDs (Fig S4) suggesting that the negative influence of NDGA on virion assembly is a consequence of alteration of numbers of LDs. Numerous studies support the notion that HCV exploits VLDL secretion for egress (15,16,36). NDGA treatment abrogated ApoB/VLDL secretion (Fig 6) via reduction in MTP transcript levels (Fig 6A and 6B). In addition NDGA also affects MTP activity per se which correlates with the observed decline in intracellular levels of ApoB although the mRNA levels are not affected (Fig 6B and 6C) because unlipidated ApoB has been shown to be subjected to proteasomal degradation (39). We also observed increased instances of ApoB crescents on LD surfaces after NDGA treatment (Fig 6D). ApoB crescents comprise lipidated ApoB destined for degradation and result from inhibition of VLDL pathway downstream of ApoB lipidation (40). These studies suggest that NDGA downregulates VLDL secretion by affecting MTP expression/activity and by inhibiting subsequent steps downstream of ApoB lipidation probably by downregulating PLTP1 and ACSL3 (Fig 6B), two important genes involved in VLDL assembly. Correlating to these results although oleic acid supplementation to NDGA treated cells increased intracellular infectivity it did not significantly enhance the secretion of virus (Fig S6C). Apart from inhibiting VLDL secretion, NDGA also affected the secretion of hepatic secretory proteins such as α1-antitrypsin indicating a global inhibition of secretory pathway. Indeed, NDGA has been shown to inhibit secretory protein transport by affecting the ER to Golgi transport (46).
In summary, our studies show that NDGA due to its overall effects on host lipid metabolism can potentially serve to abrogate HCV infection. Developing NDGA derivatives with enhanced specificity, potency and less cytotoxicity holds promise as an antiviral agent against not only HCV but also other RNA viruses, which rely on host lipid metabolism.
Supplementary Material
Acknowledgments
Financial Support: This work is supported by NIH grants DK077704, DK083479 and AI085087.
Abbreviation
- HCV
hepatitis c virus
- NDGA
nordihydroguaiaretic acid
- SREBP
sterol regulatory element binding protein
- PPARα
peroxisome proliferator-activated receptor alpha
- CPT1a
carnitine palmitoyl acyl-CoA transferase 1a
- VLDL
very low-density lipoprotein
- LD
lipid droplets
- apoB
apolipoprotein B
- ER
endoplasmic reticulum
- MTP
microsomal triglyceride transfer protein
- FFU
foci forming unit
- FAS
fatty acid synthase
References
- 1.Pawlotsky JM. Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol. 2004;12:96–102. doi: 10.1016/j.tim.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Negro F, Sanyal AJ. Hepatitis C virus, steatosis and lipid abnormalities: clinical and pathogenic data. Liver Int. 2009;29(Suppl 2):26–37. doi: 10.1111/j.1478-3231.2008.01950.x. [DOI] [PubMed] [Google Scholar]
- 3.Syed GH, Amako Y, Siddiqui A. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol Metab. 21:33–40. doi: 10.1016/j.tem.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, et al. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waris G, Felmlee DJ, Negro F, Siddiqui A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol. 2007;81:8122–8130. doi: 10.1128/JVI.00125-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Lerat H, Kammoun HL, Hainault I, Merour E, Higgs MR, Callens C, Lemon SM, et al. Hepatitis C virus proteins induce lipogenesis and defective triglyceride secretion in transgenic mice. J Biol Chem. 2009;284:33466–33474. doi: 10.1074/jbc.M109.019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dharancy S, Malapel M, Perlemuter G, Roskams T, Cheng Y, Dubuquoy L, Podevin P, et al. Impaired expression of the peroxisome proliferator-activated receptor alpha during hepatitis C virus infection. Gastroenterology. 2005;128:334–342. doi: 10.1053/j.gastro.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 8.Cheng Y, Dharancy S, Malapel M, Desreumaux P. Hepatitis C virus infection down-regulates the expression of peroxisome proliferator-activated receptor alpha and carnitine palmitoyl acyl-CoA transferase 1A. World J Gastroenterol. 2005;11:7591–7596. doi: 10.3748/wjg.v11.i48.7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, et al. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sagan SM, Rouleau Y, Leggiadro C, Supekova L, Schultz PG, Su AI, Pezacki JP. The influence of cholesterol and lipid metabolism on host cell structure and hepatitis C virus replication. Biochem Cell Biol. 2006;84:67–79. doi: 10.1139/o05-149. [DOI] [PubMed] [Google Scholar]
- 11.Yang W, Hood BL, Chadwick SL, Liu S, Watkins SC, Luo G, Conrads TP, et al. Fatty acid synthase is up-regulated during hepatitis C virus infection and regulates hepatitis C virus entry and production. Hepatology. 2008;48:1396–1403. doi: 10.1002/hep.22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 13.McLauchlan J. Hepatitis C virus: viral proteins on the move. Biochem Soc Trans. 2009;37:986–990. doi: 10.1042/BST0370986. [DOI] [PubMed] [Google Scholar]
- 14.Boulant S, Douglas MW, Moody L, Budkowska A, Targett-Adams P, McLauchlan J. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic. 2008;9:1268–1282. doi: 10.1111/j.1600-0854.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 15.Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82:2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Jr., Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci U S A. 2007;104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Owen DM, Huang H, Ye J, Gale M., Jr Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99–108. doi: 10.1016/j.virol.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirandola S, Realdon S, Iqbal J, Gerotto M, Dal Pero F, Bortoletto G, Marcolongo M, et al. Liver microsomal triglyceride transfer protein is involved in hepatitis C liver steatosis. Gastroenterology. 2006;130:1661–1669. doi: 10.1053/j.gastro.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 20.Domitrovich AM, Felmlee DJ, Siddiqui A. Hepatitis C virus nonstructural proteins inhibit apolipoprotein B100 secretion. J Biol Chem. 2005;280:39802–39808. doi: 10.1074/jbc.M510391200. [DOI] [PubMed] [Google Scholar]
- 21.Petit JM, Benichou M, Duvillard L, Jooste V, Bour JB, Minello A, Verges B, et al. Hepatitis C virus-associated hypobetalipoproteinemia is correlated with plasma viral load, steatosis, and liver fibrosis. Am J Gastroenterol. 2003;98:1150–1154. doi: 10.1111/j.1572-0241.2003.07402.x. [DOI] [PubMed] [Google Scholar]
- 22.Lu JM, Nurko J, Weakley SM, Jiang J, Kougias P, Lin PH, Yao Q, et al. Molecular mechanisms and clinical applications of nordihydroguaiaretic acid (NDGA) and its derivatives: an update. Med Sci Monit. 16:RA93–100. [PMC free article] [PubMed] [Google Scholar]
- 23.Kelley GL, Allan G, Azhar S. High dietary fructose induces a hepatic stress response resulting in cholesterol and lipid dysregulation. Endocrinology. 2004;145:548–555. doi: 10.1210/en.2003-1167. [DOI] [PubMed] [Google Scholar]
- 24.Lee MS, Kim D, Jo K, Hwang JK. Nordihydroguaiaretic acid protects against high-fat diet-induced fatty liver by activating AMP-activated protein kinase in obese mice. Biochem Biophys Res Commun. 401:92–97. doi: 10.1016/j.bbrc.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 25.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amako Y, Sarkeshik A, Hotta H, Yates J., 3rd Siddiqui A. Role of Oxysterol Binding Protein in Hepatitis C Virus infection. J Virol. 2009 doi: 10.1128/JVI.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi T, Katsume A, Tanaka T, Abe A, Inoue K, Tsukiyama-Kohara K, Kawaguchi R, et al. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology. 1999;116:636–642. doi: 10.1016/s0016-5085(99)70185-x. [DOI] [PubMed] [Google Scholar]
- 30.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osborne TF. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J Biol Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- 32.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welte MA. Fat on the move: intracellular motion of lipid droplets. Biochem Soc Trans. 2009;37:991–996. doi: 10.1042/BST0370991. [DOI] [PubMed] [Google Scholar]
- 34.Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, et al. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nahmias Y, Goldwasser J, Casali M, van Poll D, Wakita T, Chung RT, Yarmush ML. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology. 2008;47:1437–1445. doi: 10.1002/hep.22197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao H, Ye J. Long chain acyl-CoA synthetase 3-mediated phosphatidylcholine synthesis is required for assembly of very low density lipoproteins in human hepatoma Huh7 cells. J Biol Chem. 2008;283:849–854. doi: 10.1074/jbc.M706160200. [DOI] [PubMed] [Google Scholar]
- 38.Hussain MM, Shi J, Dreizen P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res. 2003;44:22–32. doi: 10.1194/jlr.r200014-jlr200. [DOI] [PubMed] [Google Scholar]
- 39.Gibbons GF, Wiggins D, Brown AM, Hebbachi AM. Synthesis and function of hepatic very-low-density lipoprotein. Biochem Soc Trans. 2004;32:59–64. doi: 10.1042/bst0320059. [DOI] [PubMed] [Google Scholar]
- 40.Ohsaki Y, Cheng J, Suzuki M, Fujita A, Fujimoto T. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J Cell Sci. 2008;121:2415–2422. doi: 10.1242/jcs.025452. [DOI] [PubMed] [Google Scholar]
- 41.Okazaki H, Goldstein JL, Brown MS, Liang G. LXR-SREBP-1c-phospholipid transfer protein axis controls very low density lipoprotein (VLDL) particle size. J Biol Chem. 285:6801–6810. doi: 10.1074/jbc.M109.079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mankouri J, Tedbury PR, Gretton S, Hughes ME, Griffin SD, Dallas ML, Green KA, et al. Enhanced hepatitis C virus genome replication and lipid accumulation mediated by inhibition of AMP-activated protein kinase. Proc Natl Acad Sci U S A. 107:11549–11554. doi: 10.1073/pnas.0912426107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 44.Deng Y, Almsherqi ZA, Ng MM, Kohlwein SD. Do viruses subvert cholesterol homeostasis to induce host cubic membranes? Trends Cell Biol. 20:371–379. doi: 10.1016/j.tcb.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blais DR, Lyn RK, Joyce MA, Rouleau Y, Steenbergen R, Barsby N, Zhu LF, et al. Activity-based protein profiling identifies a host enzyme, carboxylesterase 1, which is differentially active during hepatitis C virus replication. J Biol Chem. 285:25602–25612. doi: 10.1074/jbc.M110.135483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujiwara T, Takami N, Misumi Y, Ikehara Y. Nordihydroguaiaretic acid blocks protein transport in the secretory pathway causing redistribution of Golgi proteins into the endoplasmic reticulum. J Biol Chem. 1998;273:3068–3075. doi: 10.1074/jbc.273.5.3068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.