Abstract
Mechanical stress plays an essential role in tissue development and remodeling. In this study, we determined the role of microRNA in chondrocyte mechanotransduction. Using microarray, we identified miR-365 as a mechanoresponsive microRNA in parallel to mechanical induction of Indian hedgehog (Ihh) in primary chicken chondrocytes cultured in 3-dimensional collagen scaffoldings under cyclic loading (1 Hz, 5% elongation). Interestingly, expression of miR-365 is elevated in the prehypertrophic zone of the growth plate, coinciding with the Ihh expression region in vivo. MiR-365 significantly stimulates chondrocyte proliferation and differentiation. MiR-365 increases expression of Ihh and the hypertrophic marker type X collagen, whereas anti-miR-365 inhibits the expression of these genes. We identified histone deacetylase 4 (HDAC4), an inhibitor of chondrocyte hypertrophy, as a target of miR-365. MiR-365 inhibits both endogenous HDAC4 protein levels as well as the activity of a reporter gene bearing the 3′-untranslated region of HDAC4 mRNA. Conversely, inhibition of endogenous miR-365 relieves the repression of HDAC4. Mutation of the miR-365 binding site in HDAC4 mRNA abolishes miR-365-mediated repression of the reporter gene activity. Overexpression of HDAC4 reverses miR-365 stimulation of chondrocyte differentiation markers including Ihh, Col X, and Runx2. Moreover, inhibition of miR-365 abolishes mechanical stimulation of chondrocyte differentiation. Taken together, miR-365 is the first identified mechanically responsive microRNA that regulates chondrocyte differentiation via directly targeting HDAC4.—Guan, Y.-J., Yang, X., Wei, L., Chen, Q. MiR-365: a mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4.
Keywords: mechanotransduction, HDAC4, Indian hedgehog
MicroRNAs (miRNAs) are a class of short, noncoding RNA molecules involved in numerous biological processes. Hundreds of miRNAs have been identified in various organisms, and many are evolutionarily conserved. An estimated one-third of all mammalian mRNAs are regulated by miRNAs, demonstrating the essential role of miRNAs in controlling gene expression (1, 2). Functioning at the post-transcriptional level, miRNAs repress translation and/or facilitate mRNA degradation by binding to the 3′-untranslated region (3′-UTR) of mRNA (1, 3–5). MiRNAs play an important role in regulating differentiation and development across a wide range of organisms and tissue types. Recent publications have shown that Dicer, an essential component for biogenesis of miRNAs, plays a significant role in chondrogenic differentiation (6, 7). Tissue-specific deletion of the Dicer gene results in abnormal cartilage development in mice (7). Dicer plays a critical role in maintaining the proliferating pool of chondrocytes through regulation of chondrocyte proliferation and inhibition of premature differentiation to postmitotic hypertrophic chondrocytes. This finding suggests that miRNAs play a critical role in skeletal development. However, the role of miRNA in mechanotransduction has yet to be demonstrated.
Mechanotransduction is the process of translating mechanical stimulation into cellular responses. Mechanical stress plays a fundamental role in regulating cellular activities during tissue morphogenesis. Cyclic loading has been shown to stimulate chondrocyte proliferation and subsequent differentiation (8). However, the precise biochemical and molecular mechanisms by which mechanical stimuli affect chondrocyte differentiation remain unclear. Mechanical forces are thought to be mediated by “mechnosensitive genes” in cells. Expression of these genes is altered in response to mechanical stimulation (9). We identified Indian hedgehog (Ihh), a member of the vertebrate hedgehog morphogen family, as a dynamic mechanoresponsive gene in chondrocytes. Mechanical induction of Ihh is necessary for mechanical stimulation of chondrocyte proliferation (8). Therefore, Ihh is a critical mediator of mechanotransduction in chondrocytes. In addition, Ihh also regulates chondrocyte differentiation via parathyroid hormone-related peptide (PTHrP)-dependent and -independent mechanisms (10, 11).
Chondrocyte differentiation is also regulated by a variety of factors in addition to Ihh and PTHrP. Histone deacetylase 4 (HDAC4) is a major regulator of cartilage development and endochondral ossification by functioning as a potent inhibitor of chondrocyte hypertrophy. HDAC4-deficient mice develop ectopic chondrocyte hypertrophy, whereas overexpression of HDAC4 in proliferating chondrocytes inhibits chondrocyte hypertrophy and differentiation (12). HDAC4 regulates chondrocyte hypertrophy and endochondral bone formation by inhibiting the expression and activity of transcriptional factor Runx2, which is essential to both chondrocyte and osteoblast differentiation (12–14). However, it is not clear how the expression of the HDAC4 gene itself is regulated during chondrocyte differentiation.
To determine the role of miRNAs in mechanotransduction, we screened for mechanosensitive miRNA with the miRNA microarray approach. We identified miR-365 as one of the few miRNAs up-regulated in chondrocytes in response to cyclic loading. We provide the evidence that miR-365 is a critical mediator to stimulate chondrocyte proliferation and differentiation in response to mechanical loading. Specifically, miR-365 promotes chondrocyte hypertrophy and differentiation by targeting the 3′-UTR of HDAC4 mRNA and suppressing the protein levels of HDAC4.
MATERIALS AND METHODS
Expression plasmid, miRNA mimic, inhibitors, and RNA oligos and antibodies
A series of plasmids or oligonucleotides was transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA,USA) following the manufacturer's protocol. miR-365 mimic and negative control mimic were purchased from Dharmacon (Lafayette, CO, USA). These mimics are double-stranded RNA oligonucleotides, chemically modified with the Dharmacon On-Target modifications, which can effectively mimic the function of endogenous mature miRNA. MiR-365 inhibitor and negative control inhibitor were purchased from Dharmacon. These inhibitors are RNA oligonucleotides with novel secondary structures designed to inhibit the function of endogenous miRNA (9). HDAC4 expression plasmids were purchased from ADDgene (Cambridge, MA, USA). pGL3-RunX2-Luc (0.6 kb of Rat Runx2 promoter) was kindly provided by Dr. Gary Stein (University of Massachusetts Medical School, Worcester, MA, USA; ref. 15). pGL-HDAC4 was kindly provided by Dr. Da-Zhi Wang (Children's Hospital Boston and Harvard Medical School, Boston, MA, USA; ref. 16).
Cell culture and 3-dimensional (3-D) organotypic chondrocyte culture with mechanical stimulation
Chicken primary proliferative chondrocytes were isolated from the caudal part of 17-d-old embryonic chick sternum (17), while prehypertrophic chondrocytes were isolated from the cephalic part of 17-d-old embryonic chick sternum. Mouse primary chondrocytes were isolated from the rib cage of 6-d-old mice. Cartilage slices were digested enzymatically in 0.2% collagenase type II. Individual cells were grown in DMEM containing 10% fetal calf serum. Chondrocytes were cultured in a 3-D collagen system as described previously (18). The mechanical properties of collagen sponges were described previously (19, 20). Chondrocytes cultured in 3-D collagen scaffoldings form cartilage-like nodules that express specific markers of cartilage, including collagen type II, aggrecan, link protein, and matrilin-1 (18). After incubation of chondrocytes in collagen sponges overnight, the sponges were mechanically loaded to induce 5% elongation at 60 cycles/min, 15 min/h by a computer-controlled biostretcher device (Bio-Stretch; ICCT Technologies, Markham, ON, Canada). Bio-Stretch exerts a uniaxial stretch with square wave patterns, which induces extension of the collagen sponges at the x axis and compression at the y axis. The extent of matrix deformation may be comparable to that experienced in a growth plate in vivo (21). At the indicated mechanical loading duration, chondrocytes were freed from sponges by collagenase digestion (19) and collected for further analysis.
RNA isolation and miRNA microarray
Total RNA including miRNA was extracted from chondrocytes using the miRNeasy Mini Kit (Qiagen, Valencia, CA, USA) by following the manufacturer's instructions. The concentration, purity, and integrity of total RNA were determined by NanoDrop 2000C spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Chicken miRNA microarray assay was performed using LC Science (Houston, TX, USA). We performed our screening using proliferating chondrocytes from embryonic chicken based on the following reasons: the same cells were used for the previous Ihh mechanotransduction study (8, 18, 22); chondrocytes from a specific developmental stage can be isolated; and expression levels of the isolated miRNA can be quantified in different zones of a growth plate. Total RNA from 3 individual samples was pooled in the control and loaded groups, respectively. The control and loaded samples were hybridized to the same microarray for dual-sample analysis. Six repeats were performed from the same sample to ensure the consistency of hybridization. A t test was used for statistical analysis with significance set at P < 0.01. TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org) were utilized to identify target genes for miR-365.
Real-time PCR
Total RNA was reverse transcribed using the miScript reverse transcription kit (Qiagen) and miRNA expression levels were then analyzed using the Taqman miRNA assays (Applied Biosystems Inc., Foster City, CA, USA) per manufacturer's instructions. Quantification of the ubiquitously expressed miRNA, snoRNA U6, was performed as an endogenous control. To determine the expression levels of Col X, Ihh, and Sox 9, total RNA was analyzed using the Reverse Transcription System (Bio-Rad, Hercules, CA, USA) followed by real-time PCR with SYBR Green chemistry. A reaction mixture containing the SYBR Green Master Mix (Qiagen), and the appropriate primers was added to a 96-well plate, together with cDNA template, for a final reaction volume of 20 μl/well, and run for an initial step at 95°C for 10 min, followed by 40 cycles of amplification at 95°C for 15 s and 55°C for 60 s. Absolute quantization of 18s was performed as an endogenous control. All real-time PCRs were carried out in a DNA engine Opticon 2 PCR amplification system (Bio-Rad). HDAC4 Primer assay (chicken) was purchased from Qiagen. The forward and reverse primers for Col X (chicken) were 5′-AGTGCTGTCATTGATCTCATGGA-3′ and 5′-TCAGAGGAATAGAGACCATTGGATT-3′, respectively. The forward and reverse primers for Sox 9 (chicken) were 5′-TCGGTGAAGAACGGGCAGTC-3′ and 5′-TCGCTGATGCTGGAGGATGA-3′, respectively. The forward and reverse primers for Ihh (chicken) were 5′-AAGTCAGAGCACTCGGCTGCC-3′ and 5′-GTTGTCGGCCACGAAGAGCA-3′, respectively. The forward and reverse primers for Runx2 (chicken) were 5′-CTTCGCCGTCCATTCACTCC-3′ and 5′-GTGCATTCGTGGGTTGGAGA-3′, respectively. The forward and reverse primers for COLII (chicken) were 5′-GGACAGCGTGGTGAGAGAGGCTT-3′ and 5′-TCCCTGCCTGGGAGACCGTCA-3′, respectively. The forward and reverse primers for 18s were 5′-CGGCTACCACATCCAAGGAA-3′ and 5′-GCTGGAATTACCGCGGCT-3′, respectively. The same amount of RNA was used for both stretched and nonstretched samples. The 18 S RNA was amplified at the same time and used as an internal control. The cycle threshold values for 18 S RNA and that of samples were measured, and the relative mRNA levels were calculated by computer software. The cycle threshold (Ct) values for 18S RNA and that of samples were measured and calculated by computer software (PE ABI). Relative transcript levels were calculated as x = 2−ΔΔCt, where ΔΔCt =ΔE − ΔC, and ΔE = Ct exp − Ct 18 s; ΔC = Ct ctl − Ct 18 s.
Cell proliferation assay
Chondrocyte proliferation was measured by TACS MTT cell proliferation assays (Trevigen, Gaithersburg, MD, USA) and BrdU cell proliferation assay (EMD Chemicals Inc, Darmstadt, Germany) following the manufacturer's protocol. Briefly, miRNA-transfected chondrocytes (4000 cells/well) were plated on 96-well culture plates and incubated in DMEM/F12 containing 10% FBS. After 15, 24, 48, and 72 h of culture, 10 μl of MTT reagent was added to each well, and the plates were incubated for 4 h. The resulting formazan product was dissolved by detergent, and the absorbance at a wavelength of 590 nm (A590) was read on a SpectraMax 190 absorbance microplate reader (Molecular Devices, Downingtown, PA, USA). For the BrdU proliferation assay, cells were incubated in the presence of BrdU for 24 h. Absorbance was measured using the microplate reader at dual wavelengths of 450 and 540 nm.
Western blot analysis
Total protein was extracted from cells in collagen sponges with or without cyclic loading for indicated incubation periods, as described previously (23). Equal amounts of cell lysates were loaded in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulouse membrane. Immunoblotting was coupled with fluorescent signal detection using an Odyssey fluorescence scanner (LI-COR Biosciences, Lincoln, NE, USA). The following antibodies were used for this study: anti-HDAC4 antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA); anti-actin (Cell Signaling Technology, Danvers, MA, USA); anti-mouse-IRDye800 (Roche; Rockland Immunochemicals, Gilbertsville, PA, USA) and anti-rabbit-Alexa Fluor 680 (Molecular Probes, Eugene, OR, USA). Quantification of Western blot data was performed using the Odyssey software.
Site-directed mutagenesis
pGL-HDAC4-mut was derived from pGL-HDAC4 by mutating miR-365 seed sites within the HDAC4 3′-UTR sequence (see Supplemental Fig. S2C for details) using the QuikChange XL Site-directed Mutagenesis kit (Stratagene, La Jolla, CA, USA). Mutations were generated by replacing 3 consecutive base pairs at the 3′ region of the seed site. The following primer pair was used for pGLHDAC4 mut1: 5′-GGGCCCACGTGTTTTATGGGCCGCGATACATATATAAATATATAG-3′, and 5′-CTATATATTTATATATGTATCGCGGCCCATAAAACACGTGGGCCC-3′.
Luciferase assay
HDAC4 reporter gene construct was assayed by cotransfection of PGL3HDAC4 or its mutant (0.5 μg) with 100 nmol miR-365 mimic or control miRNA in chicken chondrocytes. The effect of miR-365 on Runx2 promoter activity was assayed by transfection of Runx2 luciferase reporter gene. Transfection control was performed with an equal amount of empty vector DNA. Transfection was performed using Lipofectamine 2000 (Invitrogen) in 12-well plates per manufacturer's instructions. Cultures were harvested after 48 h of incubation. Luciferase activities were determined using the Dual-Luciferase Reporter assay system (Promega, Madison, WI, USA) using the DLR-ready luminometer. All firefly luciferase values were normalized for transfection efficiency using the PRL-TK, Renilla luciferase value. The assays were performed in triplicate.
Statistical analysis
Results are expressed as means ± sd. Statistical analysis was carried out using a 2-way ANOVA and Turkey's test analysis with significance set as P < 0.05 or P < 0.01.
RESULTS
Identification of mechanoresponsive miRNA
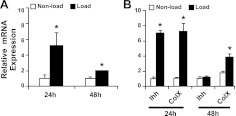
To identify mechanoresponsive miRNAs, we screened the expression profile of miRNAs by microarray according to the time course of mechanical induction of Ihh and Col X. Cyclic loading of these chondrocytes greatly induced the mRNA levels of Ihh and Col X after 24 h incubation, and the extent of mechanical stimulation decreased after 48 h incubation (Fig. 1B). Therefore, we performed miRNA microarray analysis using chondrocytes after 24 h mechanical loading. After exclusion of miRNA expressed below significant signal levels, 24 miRNA were up-regulated and 25 miRNA were down-regulated by mechanical loading out of a total 518 miRNA screened (P<0.01; see Supplemental Fig. S1). MiR-365 had the most significant P value (7.49E-07) of all the miRNAs whose expressions were altered by mechanical loading. To validate this result, we performed real-time PCR of miR-365 with miRNA assay. Consistent with the microarray data, the expression level of miR-365 was increased 5.6-fold on mechanical stimulation for 24 h. This increase was attenuated after 48 h mechanical loading, in parallel to the mechanical stimulation pattern of Ihh and Col X (Fig. 1A). A similar mechanoresponsive pattern also exists for miR-365 in mouse chondrocytes (data not shown). In addition, miR-365 is conserved across species, including chickens, mice, and humans (Supplemental Fig. S2B). We therefore used both primary chicken and mouse chondrocytes for this study.
Figure 1.
Regulation of miR-365 expression levels by mechanical loading. Proliferating chondrocytes isolated from the caudal part of sterna of 17 d chicken embryos were cultured in 3-D collagen sponges. Cyclic loading was applied to induce 5% deformation of the sponge at 1 Hz, 15 min/h for 24 and 48 h. Expression levels of miR-365 (A) and Ihh and ColX mRNA (B) were quantified by real-time RT-PCR. Values are means ± sd (n=3 cell culture experiments). *P < 0.01 vs. nonload.
MiR-365 expression in a growth plate
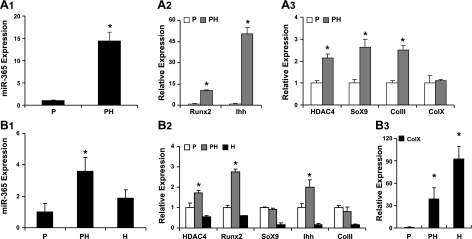
To determine the expression pattern of miR-365 during chondrocyte differentiation, we quantified its expression levels in proliferating and prehypertrophic chondrocytes. The level of miR-365 increased 14.7-fold in the prehypertrophic chondrocytes, in parallel to the expression pattern of Ihh (Fig. 2A1). The mRNA levels of Ihh increased 50-fold and those of Runx2 increased 10.2-fold in the prehypertrophic chondrocytes (Fig. 2A2). The mRNA levels of Col 2A1 and Sox9 increased ∼2.5 fold; the HDAC4 mRNA levels increased 2.1-fold but with little increase of Col X mRNA in the prehypertrophic chondrocytes (Fig. 2A3). We also quantified the expression levels of miR-365 in all three zones (proliferating, prehypertrophic, and hypertrophic) of tibia growth plates (Fig. 2B1). The expression level of miR-365 was elevated in the prehypertrophic zone, coinciding with the expression pattern of Ihh mRNA in vivo (Fig. 2B2). Col X mRNA was used as a marker for the hypertrophic zone (Fig. 2B3).
Figure 2.
Expression of miR-365 in embryonic chicken sterna and tibia growth plate. A1–3) Total RNA was isolated from the caudal part (P, proliferating) and the cephalic part (PH, prehypertrophy) of sterna of 17-d chicken embryos. Expression levels of miR-365 (A1); Runx2 and Ihh (A2); and HDAC4, Sox9, Col II, and Col X (A3) were quantified by real time RT-PCR. B1–3) Tibia growth plate was isolated by dissection of 17-d chicken embryos. Three zones (P, proliferating; PH, prehypertrophy; and H, hypertrophy) of the growth plate were separated under dissection microscope as described previously (37). Total RNA was isolated from the cartilage pieces of the 3 zones, respectively. Expression levels of miR-365 (B1); HDAC4, Runx2, Sox9, Ihh, and Col II (B2); and Col X (B3) were quantified by real-time RT-PCR. Values are means ± sd (n=6). *P < 0.05.
MiR-365 stimulates chondrocyte proliferation
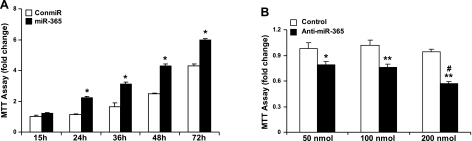
To determine whether miR-365 affects chondrocyte proliferation, a miR-365 mimic was transfected into primary growth plate chondrocytes. MiR-365 significantly enhanced chondrocyte growth at 24, 36, 48, and 72 h post-transfection, as revealed by MTT cell proliferation assay (Fig. 3A) and by BrdU cell proliferation assay (Supplemental Fig. S3). Conversely, when a miR-365 inhibitor was transfected into primary growth plate chondrocytes, it inhibited cell growth in a dose-dependent manner (Fig. 3B).
Figure 3.
MiR-365 stimulates chondrocyte proliferation. A) Expression of miR-365 promotes chondrocyte proliferation. Primary chicken proliferating chondrocytes were transfected with a miR-365 mimic (miR-365) or a control miRNA as negative control (conmiR). After transfection of miRNAs at 100 nmol, cells were incubated in F12 medium containing 10% FBS for 15, 24, 36, 48, and 72 h. Cell growth was measured by an MTT-based cell proliferation assay. Values are means ± sd (n=3). *P < 0.05. B) A miR-365 inhibitor inhibits chondrocyte growth in a dose-dependent manner. Primary chicken proliferating chondrocytes were transfected with a miR-365 inhibitor (anti-miR-365) or a control miRNA inhibitor (control) at indicated concentrations. After incubation for 48 h, cell growth was measured by an MTT-based cell proliferation assay. Values are means ± sd (n=3). *P < 0.05, **P < 0.01 vs. control; #P < 0.05 vs. 50 nmol.
MiR-365 enhances chondrocyte differentiation
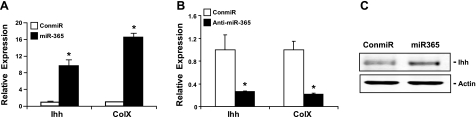
To determine whether miR-365 affects chondrocyte differentiation, we quantified the mRNA expression levels of Ihh, a marker of prehypertrophic chondrocytes, and those of Col X, a marker of hypertrophic chondrocytes, by real-time RT-PCR. Transfection of a miR-365 mimic significantly increased mRNA levels of Ihh and Col X (Fig. 4A), while transfection of a miR-365 inhibitor significantly decreased expression of both genes (Fig. 4B). Furthermore, the Ihh protein level was up-regulated by miR-365 (Fig. 4C).
Figure 4.
MiR-365 enhances chondrocyte differentiation. A) Expression of miR-365 increases chondrocyte differentiation. Primary chicken proliferating chondrocytes were transfected with a miR-365 mimic (miR-365) or control miRNA (conmiR) at 100 nmol, respectively. Total RNA was extracted 48 h post-transfection. The mRNA levels of Ihh and ColX were quantified by real-time RT-PCR. Values are means ± sd (n=3). *P < 0.05. B) Inhibition of miR-365 down-regulates mRNA levels of Ihh and Col X. Primary chicken prehypertrophic chondrocytes were transfected with a miR-365 inhibitor (anti-miR-365) or a control miRNA inhibitor (control) at 100 nmol, respectively. At 48 h post-transfection, mRNA levels of Ihh and Col X were quantified by real-time RT-PCR. Values are means ± sd (n=3). *P < 0.05. C) Expression of miR-365 increases Ihh protein levels. Whole-cell lysates were collected and subjected to Western blot with an antibody against Ihh.
HDAC4 is a target of miR-365
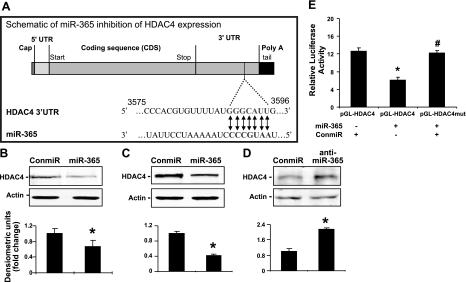
To identify target genes of miR-365, we performed a bioinformatics search. Two different algorithms, TargetScan and miRanda, consistently predicted HDAC4, an established negative regulator of chondrocyte differentiation, as a putative target of miR-365. The predicted seeding site was in the HDAC4 3′-UTR (Fig. 5A), which is evolutionarily conserved across different species. To determine whether miR-365 inhibits HDAC4 protein expression, we quantified HDAC4 protein levels by Western blot analysis. Expression of miR-365 mimic significantly inhibited HDAC4 protein levels in both chicken and mouse chondrocytes (Fig. 5B, C). Conversely, expression of a miR-365 inhibitor stimulated the endogenous levels of HDAC4 protein in chondrocytes (Fig. 5D).
Figure 5.
HDAC4 is a direct target of miR-365. A) Diagram of predicted miR-365 sequential pairing with a putative target region (miR-365 binding site) in the 3′-UTR of HDAC4 mRNA. The 5′ end of miR-365 contains a base pair sequence complementary to the putative seed site in HDAC4 mRNA. B, C) MiR-365 inhibits endogenous HDAC4 protein levels in mouse chondrocytes (B) and embryonic chicken chondrocytes (C). Primary mouse or embryonic chicken chondrocytes were transfected with a miR-365 mimic (miR-365) or with a control miRNA (conmiR) at 100 nmol, respectively. At 48 h post-transfection, total protein was extracted from cell lysates. Western blot analysis was performed with an antibody against HDAC4. D) Inhibition of miR-365 enhances the protein levels of HDAC4 in mouse chondrocytes. Primary mouse chondrocytes were transfected with a miR-365 inhibitor (anti-miR-365) or a control miRNA inhibitor (control) at 100 nmol, respectively for 48 h. After total protein was extracted from cell lysates, Western blot analysis was performed with an antibody against HDAC4. Equal amounts of protein were loaded in each lane, and actin was used as a loading control. Values of quantitation of HDAC4 protein levels normalized to actin are presented. E) Inhibition of pGL-HDAC4 reporter gene activity requires the miR-365 seed site. Mouse chondrocytes were transfected with 0.5 μg pGL-HDAC4 reporter gene or pGL-HDAC4 mutant gene (pGL-HDAC4mut) with firefly luciferase and 0.1 μg PRL-TK gene with Renilla luciferase as internal control. At 48 h after transfection, cells were harvested for quantification of dual luciferase activities. Ratio of firefly vs. Renilla luciferase is the relative value of pGL-HDAC4 or pGL-HDAC4mut reporter gene activities. Values are means± sd (n=3). *P < 0.05.
To identify the miR-365 target region in the HDAC4 mRNA, we mutated 3 nucleotides from the putative seed region in the HDAC4 3′-UTR (Supplemental Fig. S2C). While transfection of miR-365 mimic suppressed the luciferase activity of the wild-type HDAC4 3′-UTR reporter, this suppression was abolished in the mutated HDAC4 3′-UTR reporter (Fig. 5E).
MiR-365 stimulates chondrocyte differentiation through HDAC4 inhibition
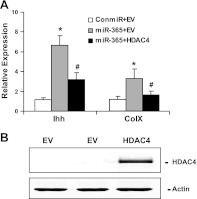
To determine whether HDAC4 is involved in miR-365 stimulation of chondrocyte differentiation, we quantified the mRNA levels of Ihh and Col X by real-time RT-PCR. While expression of miR-365 increased the mRNA levels of Ihh and Col X, cotransfection of HDAC4 abolished this increase (Fig. 6A). Overexpression of HDAC4 could reverse the induction of chondrocyte differentiation by miR-365, thus providing evidence that miR-365 mediates the induction of chondrocyte differentiation via targeting HDAC4 (Fig. 6).
Figure 6.
Overexpression of HDAC4 inhibits miR-365 stimulation of Ihh and Col X mRNA levels. A) Primary mouse chondrocytes were transfected with following 3 groups of constructs. Control: a control miRNA (conmiR) and a cDNA plasmid containing empty vector (EV); miR-365: a miR-365 mimic (miR-365) and a cDNA plasmid containing EV; and miR-365+HDAC4: a miR-365 mimic (miR-365) and a cDNA plasmid containing HDAC4 expression vector (HDAC4). At 48 h post-transfection, total RNA was isolated, and the mRNA levels of Ihh and Col X were quantified by real-time RT-PCR. Values are means± sd (n=3). *P < 0.05 vs. control; #P < 0.05 vs. miR-365. B) HDAC4 overexpression increased HDAC4 protein level. EV, transfection with empty vector. Western blot analysis was performed with cell lysates collected at 48 h post-transfection, with an antibody against HDAC4.
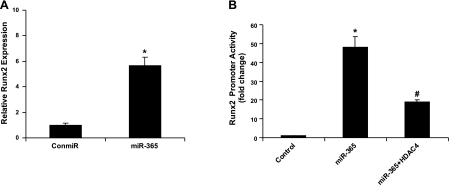
MiR-365 stimulation of Runx2 involves HDAC4
To determine whether miR-365 regulates Runx2, a target gene of HDAC4, we quantified Runx2 mRNA levels and Runx2 promoter activities in chondrocytes. Expression of miR-365 significantly stimulated both Runx2 mRNA levels and its promoter activities (Fig. 7). Cotransfection of HDAC4 significantly inhibited the stimulation of Runx2 promoter activities by miR-365 (Fig. 7B). These data suggest that miR-365, through its inhibition of the HDAC4 pathway, can regulate the expression of downstream genes such as Runx2.
Figure 7.
MiR-365 increases Runx2 gene expression and its luciferase reporter activity. A) Primary chicken chondrocytes were transfected with a miR-365 mimic (miR-365) or a control miRNA (conmiR). At 48 h post-transfection, total RNA was extracted, and the mRNA levels of Runx2 was quantified by real-time PCR. B) Primary chicken chondrocytes were transfected with the Runx2 promoter reporter gene with firefly luciferase and an internal control gene with Renilla luciferase. They were cotransfected with the following 3 groups of constructs. Control: a control miRNA (conmiR) and a cDNA plasmid containing empty vector (EV); miR-365: a miR-365 mimic (miR-365) and a cDNA plasmid containing EV; and miR-365+HDAC4: a miR-365 mimic (miR-365) and a cDNA plasmid containing HDAC4 expression vector (HDAC4). The Runx2 luciferase reporter construct was transfected into primary chicken chondrocytes together with miR-365 or negative control miRNA (conmiR), as well as an internal control plasmid for Renilla luciferase. At 48 h after transfection, cells were harvested for quantification of dual luciferase activities. Ratio of firefly vs. Renilla luciferase is the relative value of Runx2 promoter reporter gene activities. Values are means± sd (n=3). *P < 0.05 vs. control; #P < 0.05 vs. miR-365.
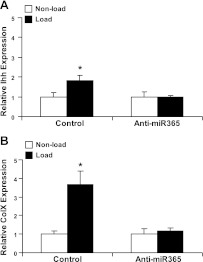
Inhibition of miR-365 impairs mechanical induction of chondrocyte differentiation
To determine whether miR-365 is required for mechanical stimulation of chondrocyte differentiation, we quantified the mRNA levels of differentiation markers Ihh and Col X in 3-D chondrocyte culture after cyclic loading. While the mRNA levels of Ihh and Col X were stimulated significantly by cyclic loading, the presence of miR-365 inhibitor completely abolished this stimulation (Fig. 8).
Figure 8.
Inhibition of miR-365 abolishes mechanical stimulation of Ihh (A) and Col X (B) mRNA levels. Primary chicken chondrocytes were transfected with a miR-365 inhibitor (anti-miR-365) or a control miRNA inhibitor (control) for 48 h followed by cyclic loading for 24 h. Total RNA was extracted, and expression of ColX and Ihh mRNA was quantified by real-time RT-PCR. Ihh and Col X mRNA levels were normalized against the endogenous control, 18S RNA. Values are means ± sd (n=3). *P < 0.05 vs. nonload.
DISCUSSION
In this study, we identified miR-365 as a mechanoresponsive miRNA that regulates chondrocyte proliferation and differentiation. MiR-365 was up-regulated in proliferating cells in patients with breast cancer or endometriosis (24–26), while down-regulated in the growth arrest states of lung fibroblasts (27). It was also up-regulated under stress conditions such as UVB irradiation in NIH3T3 cells (25). Only two targets of miR-365, IL-6 and Bcl-2, have been identified so far (28, 29). Our study is the first to demonstrate the functions of miRNA-365 in chondrocytes, which is involved in mechanotransduction and regulation of proliferation and differentiation. Since miR-365 is conserved across species and expressed in many cell types, it will be interesting to examine whether miR-365 has similar functions in other types of cells in addition to chondrocytes.
MiR-365 has several special features. First, it is mechanoresponsive. With microarray, we screened for miRNAs that are regulated by cyclic loading according to the mechanical activation pattern of Ihh. We hypothesized that a miRNA that is coregulated by mechanical loading with Ihh may play a role in mechanical activation of chondrocytes mediated by Ihh. It was previously shown that Ihh is a critical mediator of mechanotransduction in chondrocytes (8, 30). Mechanical loading induces expression of Ihh, which in turn regulates chondrocyte proliferation and differentiation. The expression of 49 of 518 miRNAs is altered by cyclic loading of chondrocytes. Among them, miR-365 is the most significantly regulated. As validated by real-time RT-PCR, the mechanoresponsiveness of miR-365 is conserved across species including chicken and mouse, the two species tested in this study.
The role of miRNA in mechanotransduction needs to be studied more extensively. Only a few miRNAs have been identified as mechanoresponsive. These include miR-26a, which is up-regulated by mechanical stretch and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3 (31), and miR-23b, which is involved in flow-regulation of Rb phosphorylation and endothelial cell growth (32). In addition, miR-222 was postulated as a potential regulator of articular cartilage mechanotransduction pathway, since its expression patterns in articular cartilage are higher in the weight-bearing anterior medial condyle as compared with the posterior nonweight-bearing medial condyle (33).
Another special feature of miR-365 is that its expression pattern in a growth plate mimics that of Ihh with the characteristically elevated expression in the prehypertrophic zone. The prehypertrophic zone is located in the middle of the growth plate, which is an important region to control the transition from cell proliferation to hypertrophy. Several important regulatory molecules including Ihh and PTHrP receptors are expressed specifically in this region. Similar to Ihh expressed in this region, miR-365 affects both chondrocyte proliferation and differentiation. MiR-365 is both necessary and sufficient to stimulate chondrocyte proliferation. Since miR-365 is predominantly synthesized by the postmitotic prehypertrophic chondrocytes, its stimulatory effect on proliferation is more likely to be paracrine rather than autocrine.
The expression of miR-365 not only overlaps with Ihh but also is both necessary and sufficient for inducing Ihh and the subsequent hypertrophic marker Col X. This underscores the potentially important role of miR-365 in regulating chondrocyte differentiation in the growth plate. Recently, Kobayashi et al. (7) have shown that, in a growth plate, deficiency of Dicer, an essential component of biogenesis of miRNAs, leads to progressive reduction in the proliferating pool of chondrocytes and results in severe growth defects of mice. The researchers propose that these defects are caused by two distinct mechanisms—decreased chondrocyte proliferation and accelerated differentiation into postmitotic hypertrophic chondrocytes—and that these defects appear to be caused by mechanisms downstream or independent of the Ihh-PTHrP signaling pathway (7). Our data indicate that inhibition of miR-365 decreases chondrocyte proliferation, which is consistent with the conclusion of Kobayashi et al. (7). However, inhibition of miR-365 also leads to suppression of Ihh and Col X, suggesting that it regulates chondrocyte differentiation upstream of the Ihh pathway. While the Kobayashi study clearly demonstrates the requirement of miRNA in growth plate development, it represents the end result of inhibiting all miRNAs through knocking out Dicer in cartilage. Different miRNAs may play different roles in regulating cartilage development. Indeed, in addition to miR-365, recent studies have identified specific miRNAs that fulfill differential functions in growth plate development. They include miR-199a, a BMP20-responsive miRNA that adversely regulates early chondrocyte differentiation via targeting of the Smad1 (34); and miR-140, which regulates both cartilage development and homeostasis by targeting ADAMTS-5 (35).
We demonstrate that miR-365 modulates chondrocyte differentiation by directly targeting HDAC4 through a series of experiments. First, abundance of miR-365 regulates the protein levels of HDAC4 in chondrocytes. However, it does not appear to inhibit HDAC4 mRNA levels (Supplemental Fig S2A). The HDAC4 mRNA levels are increased in the prehypertrophic zone in the growth plate, where miR-365 is expressed. This finding is consistent with the notion that chondrocytic miRNAs do not directly regulate target RNA abundance (7). Second, we identified the miR-365 target region in the 3′-UTR of the HDAC4 mRNA. After different computer algorithms consistently predicted the putative target region of miR-365 in the 3′UTR of the HDAC4 mRNA, we mutated 3 base pairs in that region. While miR-365 inhibits the activity of the reporter gene harboring the wild-type 3′-UTR of the HDAC4 mRNA, it fails to do so to the reporter gene harboring mutations in the target region. This indicates that this region in the 3′-UTR of the HDAC4 mRNA is required for miR-365 inhibition of HDAC4 translation. Third, inhibition of HDAC4 is required for miR-365 up-regulation of Ihh, Col X, and Runx2.
HDACs modulate cell growth by governing chromatin structure and repressing the activity of specific transcription factors. HDAC4 is a class II HDAC expressed in prehypertrophic chondrocytes; it inhibits chondrocyte hypertrophy and endochondral bone formation by interacting with and suppressing the activity of Runx2, a transcription factor necessary for chondrocyte hypertrophy (12). However, one important question is how chondrocyte hypertrophy proceeds despite HDAC4, a potent inhibitor of chondrocyte differentiation. Our data suggest that miR-365 stimulates chondrocyte differentiation by reducing the protein levels of HDAC4, a negative regulator of chondrocyte hypertrophy. It may also explain how up-regulation of miR-365 by mechanical loading enhances chondrocyte differentiation.
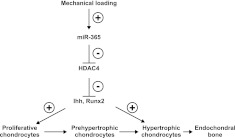
Our data also strongly suggest that miR-365 stimulation of chondrocyte differentiation through inhibiting HDAC4 also involves inducing its downstream molecules, including Ihh and Runx2 mRNA. Consistent with this hypothesis, the mRNA levels of Ihh and Runx2 are greatly increased in HDAC4−/− mice (12). It has also been shown previously that Runx2 regulates Ihh-promoter activities (36). Taken together, our data suggest a model in which miR-365, activated by mechanical loading, stimulates chondrocyte differentiation by inhibiting HDAC4 and inducing Runx2 and Ihh expression (Fig. 9). Currently, we are testing this model in vivo.
Figure 9.
Model for mechanical activation of chondrocyte proliferation and differentiation in a growth plate. A proposed mechanotransduction pathway activating chondrocyte proliferation and differentiation is shown. Cyclic loading induces proliferation and differentiation through regulating several key genes expressed in the prehypertrophic zone of the growth plate, including miR-365, HDAC4, Ihh, and Runx2.
In summary, the data here show for the first time that mechanical regulation of chondrocyte proliferation and differentiation is mediated by a miRNA, miR-365. MiR-365 functions by inhibiting its direct target HDAC4 at the post-transcription level and inducing expression of Ihh and Runx2, important regulators of endochondral bone formation. These findings not only provide new insights into mechanotransduction signaling pathways, but also raise intriguing possibilities of using miRNA for modulating cartilage tissue engineering in regenerative medicine.
Supplementary Material
Acknowledgments
The authors thank Dr. Gary Stein (University of Massachusetts Medical School, Worcester, MA, USA) and Dr. Da-Zhi Wang (Children's Hospital Boston and Harvard Medical School, Boston, MA, USA) for providing plasmid constructs, and Paul Shultz for critical reading of the manuscript.
This study is supported by U.S. National Institutes of Health grants AG 017021, AG 014399, AR 059142 and RR024484.
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 2. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 3. Harris T. A., Yamakuchi M., Ferlito M., Mendell J. T., Lowenstein C. J. (2008) MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. U. S. A. 105, 1516–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan S. P., Slack F. J. (2006) microRNA-mediated silencing inside P-bodies. RNA Biol. 3, 97–100 [DOI] [PubMed] [Google Scholar]
- 5. Bushati N., Cohen S. M. (2007) microRNA functions. Annu. Rev. Cell Dev. Biol. 23, 175–205 [DOI] [PubMed] [Google Scholar]
- 6. Harfe B. D., McManus M. T., Mansfield J. H., Hornstein E., Tabin C. J. (2005) The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. U. S. A. 102, 10898–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kobayashi T., Lu J., Cobb B. S., Rodda S. J., McMahon A. P., Schipani E., Merkenschlager M., Kronenberg H. M. (2008) Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc. Natl. Acad. Sci. U. S. A. 105, 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu Q., Zhang Y., Chen Q. (2001) Indian hedgehog is an essential component of mechanotransduction complex to stimulate chondrocyte proliferation. J. Biol. Chem. 276, 35290–35296 [DOI] [PubMed] [Google Scholar]
- 9. Rubin J., Rubin C., Jacobs C. R. (2006) Molecular pathways mediating mechanical signaling in bone. Gene 367, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kobayashi T., Soegiarto D. W., Yang Y., Lanske B., Schipani E., McMahon A. P., Kronenberg H. M. (2005) Indian hedgehog stimulates periarticular chondrocyte differentiation to regulate growth plate length independently of PTHrP. J. Clin. Invest. 115, 1734–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mak K. K., Kronenberg H. M., Chuang P. T., Mackem S., Yang Y. (2008) Indian hedgehog signals independently of PTHrP to promote chondrocyte hypertrophy. Development 135, 1947–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vega R. B., Matsuda K., Oh J., Barbosa A. C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J. M., Richardson J. A., Karsenty G., Olson E. N. (2004) Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119, 555–566 [DOI] [PubMed] [Google Scholar]
- 13. Kang J. S., Alliston T., Delston R., Derynck R. (2005) Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 24, 2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hug B. A. (2004) HDAC4: a corepressor controlling bone development. Cell 119, 448–449 [DOI] [PubMed] [Google Scholar]
- 15. Drissi H., Luc Q., Shakoori R., Chuva De Sousa Lopes S., Choi J. Y., Terry A., Hu M., Jones S., Neil J. C., Lian J. B., Stein J. L., Van Wijnen A. J., Stein G. S. (2000) Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J. Cell. Physiol. 184, 341–350 [DOI] [PubMed] [Google Scholar]
- 16. Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen Q., Johnson D. M., Haudenschild D. R., Goetinck P. F. (1995) Progression and recapitulation of the chondrocyte differentiation program: cartilage matrix protein is a marker for cartilage maturation. Dev. Biol. 172, 293–306 [DOI] [PubMed] [Google Scholar]
- 18. Wu Q. Q., Chen Q. (2000) Mechanoregulation of chondrocyte proliferation, maturation, and hypertrophy: ion-channel dependent transduction of matrix deformation signals. Exp. Cell. Res. 256, 383–391 [DOI] [PubMed] [Google Scholar]
- 19. Liu M., Skinner S. J., Xu J., Han R. N., Tanswell A. K., Post M. (1992) Stimulation of fetal rat lung cell proliferation in vitro by mechanical stretch. Am. J. Physiol. 263, L376–383 [DOI] [PubMed] [Google Scholar]
- 20. Makihira S., Yan W., Ohno S., Kawamoto T., Fujimoto K., Okimura A., Yoshida E., Noshiro M., Hamada T., Kato Y. (1999) Enhancement of cell adhesion and spreading by a cartilage-specific noncollagenous protein, cartilage matrix protein (CMP/Matrilin-1), via integrin alpha1beta1. J. Biol. Chem. 274, 11417–11423 [DOI] [PubMed] [Google Scholar]
- 21. Cohen B., Chorney G. S., Phillips D. P., Dick H. M., Buckwalter J. A., Ratcliffe A., Mow V. C. (1992) The microstructural tensile properties and biochemical composition of the bovine distal femoral growth plate. J. Orthop. Res. 10, 263–275 [DOI] [PubMed] [Google Scholar]
- 22. Kanbe K., Yang X., Wei L., Sun C., Chen Q. (2007) Pericellular matrilins regulate activation of chondrocytes by cyclic load-induced matrix deformation. J. Bone Miner. Res. 22, 318–328 [DOI] [PubMed] [Google Scholar]
- 23. Yuan Z. L., Guan Y. J., Chatterjee D., Chin Y. E. (2005) Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 307, 269–273 [DOI] [PubMed] [Google Scholar]
- 24. Ohlsson Teague E. M., Van der Hoek K. H., Van der Hoek M. B., Perry N., Wagaarachchi P., Robertson S. A., Print C. G., Hull L. M. (2009) MicroRNA-regulated pathways associated with endometriosis. Mol. Endocrinol. 23, 265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo L., Huang Z. X., Chen X. W., Deng Q. K., Yan W., Zhou M. J., Ou C. S., Ding Z. H. (2009) Differential expression profiles of microRNAs in NIH3T3 cells in response to UVB irradiation. Photochem. Photobiol. 85, 765–773 [DOI] [PubMed] [Google Scholar]
- 26. Wang X. Y., Wu M. H., Liu F., Li Y., Li N., Li G. Y., Shen S. R. Differential miRNA expression and their target genes between NGX6-positive and negative colon cancer cells. Mol. Cell Biochem. 345, 283–290 [DOI] [PubMed] [Google Scholar]
- 27. Maes O. C., Sarojini H., Wang E. (2009) Stepwise up-regulation of microRNA expression levels from replicating to reversible and irreversible growth arrest states in WI-38 human fibroblasts. J. Cell. Physiol. 221, 109–119 [DOI] [PubMed] [Google Scholar]
- 28. Xu Z., Xiao S. B., Xu P., Xie Q., Cao L., Wang D., Luo R., Zhong Y., Chen H. C., Fang L. R. (2011) miR-365, a novel negative regulator of interleukin-6 gene expression, is cooperatively regulated by Sp1 and NF-{kappa}B. J. Biol. Chem. 286, 21401–21412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qin B., Xiao B., Liang D., Xia J., Li Y., Yang H. (2011) MicroRNAs expression in ox-LDL treated HUVECs: MiR-365 modulates apoptosis and Bcl-2 expression. Biochem. Biophys. Res. Commun. 410, 127–133 [DOI] [PubMed] [Google Scholar]
- 30. Tang G. H., Rabie A. B., Hagg U. (2004) Indian hedgehog: a mechanotransduction mediator in condylar cartilage. J. Dent. Res. 83, 434–438 [DOI] [PubMed] [Google Scholar]
- 31. Mohamed J. S., Lopez M. A., Boriek A. M. Mechanical stretch up-regulates microRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3beta. J. Biol. Chem. 285, 29336–29347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang K. C., Garmire L. X., Young A., Nguyen P., Trinh A., Subramaniam S., Wang N., Shyy J. Y., Li Y. S., Chien S. Role of microRNA-23b in flow-regulation of Rb phosphorylation and endothelial cell growth. Proc. Natl. Acad. Sci. U. S. A. 107, 3234–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dunn W., DuRaine G., Reddi A. H. (2009) Profiling microRNA expression in bovine articular cartilage and implications for mechanotransduction. Arthritis Rheum. 60, 2333–2339 [DOI] [PubMed] [Google Scholar]
- 34. Lin E. A., Kong L., Bai X. H., Luan Y., Liu C. J. (2009) miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J. Biol. Chem. 284, 11326–11335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miyaki S., Sato T., Inoue A., Otsuki S., Ito Y., Yokoyama S., Kato Y., Takemoto F., Nakasa T., Yamashita S., Takada S., Lotz M. K., Ueno-Kudo H., Asahara H. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 24, 1173–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoshida C. A., Yamamoto H., Fujita T., Furuichi T., Ito K., Inoue K., Yamana K., Zanma A., Takada K., Ito Y., Komori T. (2004) Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 18, 952–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y., Chen Q. (2000) Changes of matrilin forms during endochondral ossification. Molecular basis of oligomeric assembly. J. Biol. Chem. 275, 32628–32634 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.