Abstract
Purpose
Mutation in isocitrate dehydrogenase 1 (IDH1) at R132 (IDH1R132MUT) is frequent in low-grade diffuse gliomas and, within glioblastoma (GBM), has been proposed as a marker for GBMs that arise by transformation from lower-grade gliomas, regardless of clinical history. To determine how GBMs arising with IDH1R132MUT differ from other GBMs, we undertook a comprehensive comparison of patients presenting clinically with primary GBM as a function of IDH1R132 mutation status.
Patients and Methods
In all, 618 treatment-naive primary GBMs and 235 lower-grade diffuse gliomas were sequenced for IDH1R132 and analyzed for demographic, radiographic, anatomic, histologic, genomic, epigenetic, and transcriptional characteristics.
Results
Investigation revealed a constellation of features that distinguishes IDH1R132MUT GBMs from other GBMs (including frontal location and lesser extent of contrast enhancement and necrosis), relates them to lower-grade IDH1R132MUT gliomas, and supports the concept that IDH1R132MUT gliomas arise from a neural precursor population that is spatially and temporally restricted in the brain. The observed patterns of DNA sequence, methylation, and copy number alterations support a model of ordered molecular evolution of IDH1R132MUT GBM in which the appearance of mutant IDH1 protein is an initial event, followed by production of p53 mutant protein, and finally by copy number alterations of PTEN and EGFR.
Conclusion
Although histologically similar, GBMs arising with and without IDH1R132MUT appear to represent distinct disease entities that arise from separate cell types of origin as the result of largely nonoverlapping sets of molecular events. Optimal clinical management should account for the distinction between these GBM disease subtypes.
INTRODUCTION
Glioblastoma (GBM), also known as grade 4 astrocytoma, is the most aggressive intrinsic brain tumor in adults and continues to be associated with extremely poor outcomes.1 Evidence to date indicates that the cells of origin for GBM may be either neural stem cells or their more differentiated progeny.2–4 Most GBMs arise with no prior clinical history of a precursor lesion and are referred to as primary or de novo GBMs. A minority of GBM cases, known as secondary GBMs, develop from lower-grade astrocytomas or oligodendrogliomas and bear different genomic abnormalities than primary GBM.5–7 Despite recent studies suggesting that molecular subsets of GBM differ in response to current treatments,8,9 standard treatment for all patients with primary GBM10 is a regimen combining radiation and temozolomide. The recent identification of R132 mutations in isocitrate dehydrogenase 1 (IDH1R132MUT) in the majority of low-grade gliomas and secondary GBMs, with relative exclusion from primary GBMs, implicates IDH1R132MUT as a defining marker and key oncogenic event for GBMs that evolve from lower-grade glioma.11–16 Herein, we sought to develop a detailed portrait of untreated GBMs arising with IDH1R132MUT with the aim of gaining insights into the manner in which IDH1R132MUT gliomas develop and to determine whether IDH1R132MUT GBM is a distinct disease entity.
PATIENTS AND METHODS
The focus of our investigation is 618 patients with newly diagnosed untreated primary (de novo) GBMs (cohorts A through F) and 235 patients with newly diagnosed untreated lower-grade diffuse gliomas (cohorts G and H). Patients with GBM included two cohorts compiled for this investigation (cohorts A and B), plus patients associated with previously reported studies17–20 (cohorts C through F). Sequence analysis of IDH1R132 and p53R273 included 105 additional samples of diffuse glioma (cohort I). A summary of patient cohorts is provided in Table 1. Additional details regarding Methods and patient cohorts are provided in the Data Supplement.
Table 1.
Summary of Patient Cohorts
| Cohort | Source Institution | Tumor Type | Sample Type | No. of Patients | Ratio of Males to Females | No. ofIDH1R132MUT Tumors | Mean Age(years) | Range of Ages(years) | Figure | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| A | UCLA, KPLA | Primary GBM | Newly diagnosed* | 389 | 1.5 | 19 | 56.8 | 24–90 | 1A-B, 2A, 3A-C, 4A; Appendix A1A-A1C, A2B,A3D-A3E, A4A-A4C | N/A |
| B | MDACC | Primary GBM | Newly diagnosed | 70 | 1.7 | 8 | 54.6 | 20–80 | 1A, 2A, 3A, 3C, 4A; Appendix A1A, A3E, A4A, A4C | N/A |
| C | UCSF, MDACC | Primary GBM | Newly diagnosed* | 98 | 1.7 | 22 | 46.3 | 19–82 | 2A-B, 3A, 3C, 4A; Appendix A1A, A3E, A4A | Phillips et al17 |
| D | UCSF | Primary GBM | Newly diagnosed | 29 | 1.9 | 0 | 56.0 | 27–76 | 2A, 3A, 3C, 4A; Appendix A3E, A4A | Chen et al18 |
| E | UCSF, MDACC | Primary GBM | Newly diagnosed | 14 | N/D | 0 | 11.7 | 2–17 | 3A, 3C; Appendix A4A | Schiffman et al19† |
| F | UKE | Primary GBM | Newly diagnosed* | 18 | 1.3 | 0 | 65.3 | 32–85 | 2A, 3A, 3C, 4A; Appendix A1A, A3E, A4A | Günther et al20† |
| G | UCSF, MDACC | AA | Newly diagnosed | 77 | 1.3 | 42 | 38.5 | 7–75 | 2A, 3C, 4A; Appendix A2A-A2B, A3E, A4A | Phillips et al17† |
| H | UCLA, KPLA | Grades II and III diffuse glioma | Newly diagnosed | 158 | 1.1 | 103 | 40.1 | 15–79 | 3C, 4A; Appendix A3A-A3C, A3E | N/A |
| I | Various | Grades II to IV diffuse glioma | Newly diagnosed and recurrent | 105 | N/D | 37 | N/D | 4A | N/A |
NOTE. Samples from newly diagnosed patients were obtained before treatment. Tumor type is according to WHO criteria.
Abbreviations: AA, anaplastic astrocytoma; GBM, glioblastoma; IDH1R132MUT, mutation in isocitrate dehydrogenase 1 (IDH1) at R132; KPLA, Kaiser Permanente Los Angeles; MDACC, MD Anderson Cancer Center; N/A, not applicable; N/D, not done; UCLA, University of California at Los Angeles; UCSF, University of California at San Francisco; UKE, Universitätsklinikum Hamburg-Eppendorf.
Includes matched recurrent specimens for a subset of cases.
Cohort includes additional specimens not described in original publication.
RESULTS
IDH1R132MUT GBMs Are Phenotypically Distinct
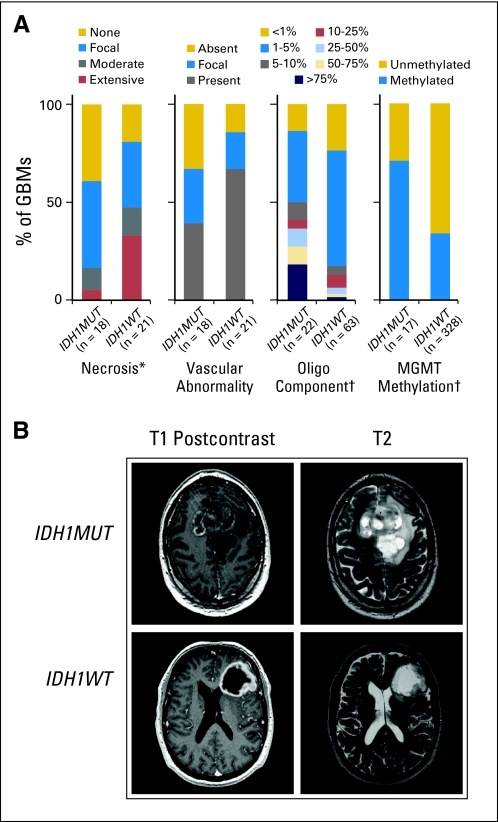
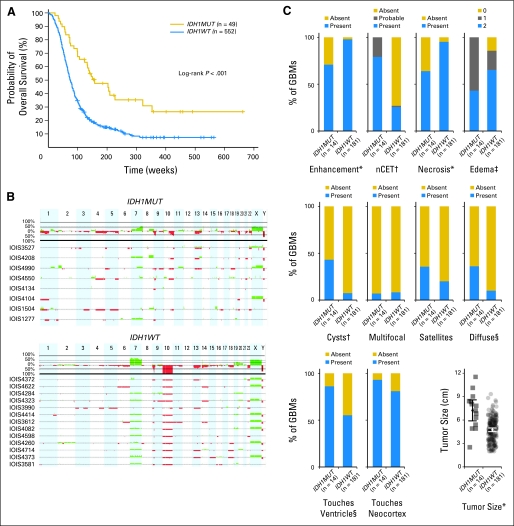
To characterize IDH1R132MUT and IDH1R132-wild type (IDH1R132WT) GBMs in the absence of confounding treatment effects, we pursued a comparison within treatment-naive patients with primary GBM. From 618 de novo GBMs, we identified 49 IDH1R132MUT tumors, all of which occurred in adults. Within adult patients, we confirmed that IDH1R132MUT GBMs manifest longer overall survival12,13,16,21 (Appendix Fig A1A, online only) and showed more frequent promoter methylation of O6-methylguanine–DNA methyltransferase (MGMT)8,21,22 as illustrated in Figure 1A. Histologic characterization of a sampling of IDH1R132MUT and IDH1R132WT GBMs demonstrated similar levels of cell proliferation on the basis of MIB-1 staining (Data Supplement), but revealed a lesser extent of necrosis in IDH1R132MUT GBMs (Fig 1A) and a nonsignificant trend toward less frequent occurrence of vascular abnormalities (Fig 1A). Consistent with a previous report,12 examination of an expanded series of samples revealed a statistically significant, albeit modest, increase in the percentage of cells with oligodendroglial morphology in IDH1R132MUT GBMs (Fig 1A). To examine whether tumors in our series of GBMs harbored the co-deletion of chromosome arms 1p/19q commonly observed in oligodendrogliomas, we assessed DNA copy number alterations by using array comparative genomic hybridization in a series of samples and found that a minority of IDH1R132MUT GBMs (two of eight) and none of the IDH1R132WT GBMs (zero of 13) displayed 1p/19q co-deletion (Appendix Fig A1B). There was a striking difference between the two GBM subsets regarding loss of chromosome 10; this alteration was absent in all IDH1R132MUT GBMs but was present in all but one sample of IDH1R132WT GBM (Appendix Fig A1B).
Fig 1.
Glioblastomas (GBMs) with mutations in isocitrate dehydrogenase 1 (IDH1) at R132 (IDH1R132MUT; IDH1MUT) are phenotypically distinct from IDH1R132-wild type (IDH1R132WT; IDH1WT) GBMs. (A) Histologic assessment of IDH1R132MUT and IDH1R132WT GBMs reveals decreased necrosis (*P < .05) and oligodendroglial content (†P < .005), with a trend toward decreased vascular abnormalities (P < .075) for IDH1R132MUT GBMs (Data Supplement). Frequency of O6-methylguanine–DNA methyltransferase (MGMT) promoter methylation is increased in IDH1R132MUT GBMs (†P < .005; Data Supplement). (B) Representative pretreatment contrast-enhanced T1-weighted images (left) with corresponding T2-weighted images (right) of a patient with IDH1R132MUT (IDH1MUT; upper panel) and a patient with IDH1R132WT (IDH1WT; lower panel).
By using previously defined parameters,23 we examined available preoperative cranial magnetic resonance images. Consistent with our histologic findings, detection of necrosis was less frequent in IDH1R132MUT GBMs; moreover, IDH1R132MUT GBMs exhibited more frequent non-enhancing tumor component, larger size at diagnosis, lesser extent of edema, and increased prevalence of cystic and diffuse components (Fig 1B and Appendix Fig A1C). In addition, the IDH1R132MUT GBMs demonstrated greater frequency of contact with brain ventricles, although interpretation of this finding may be confounded by the larger size of IDH1R132MUT GBMs (Appendix Fig A1C). Overall, the radiographic and histologic features that distinguish IDH1R132MUT GBMs resemble characteristics of lower-grade gliomas and are consistent with a less aggressive clinical course.
Restricted Gene Expression of IDH1R132MUT GBMs
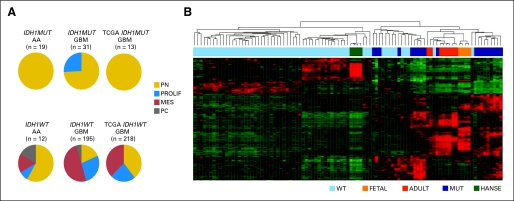
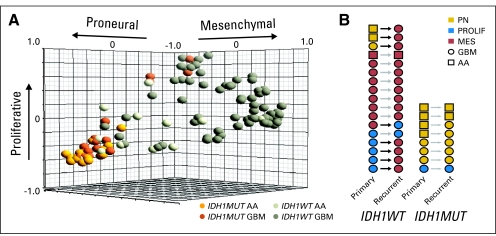
By examining transcriptional signatures of both the newly diagnosed high-grade astrocytomas in this investigation and The Cancer Genome Atlas (TCGA) primary GBM data set, we found that the majority of IDH1R132MUT tumors express the Proneural17 subtype signature (Fig 2A and Appendix Fig A2A, online only). This Proneural signature has previously been reported as a positive prognostic indicator17 and is substantially similar to the TCGA Proneural signature associated with IDH1R132MUT GBMs that resembles the signature of oligodendroglia.9 A minority of IDH1R132MUT GBMs possessed the Proliferative signature, and none possessed the Mesenchymal signature associated with angiogenesis and poor outcome17 (Fig 2A and Appendix Fig A2A). In contrast, IDH1R132WT GBMs displayed all three signatures, with a preponderance of the Mesenchymal subtype. Strikingly, evaluation of gene expression subtype in matched sample pairs from high-grade astrocytomas obtained at initial diagnosis and after recurrence (Appendix Fig A2B) showed that all IDH1R132MUT tumors maintained their original subtype, although several IDH1R132WT GBMs shifted to the Mesenchymal subtype. Thus, IDH1R132MUT tumors differ not only in their presentation but also in their pattern of disease progression and do not share the propensity of IDH1R132WT GBMs to adopt a Mesenchymal phenotype.
Fig 2.
Glioblastomas (GBMs) with mutations in isocitrate dehydrogenase 1 (IDH1) at R132 (IDH1R132MUT; IDH1MUT) are restricted from Mesenchymal gene expression and show greater similarity to transcriptional signatures of brain cells than to those of neural stem cells. (A) Gene expression analysis reveals a robust positive association of IDH1R132MUT high-grade astrocytomas with the Proneural signature and an absolute exclusion of the Mesenchymal gene signature from IDH1R132MUT tumors. Subtypes are labeled as follows: Proneural (PN), Proliferative (PROLIF), and Mesenchymal (MES). PC denotes poorly classified tumors. Pie charts depict tabulated data for all tumors assigned to subtype. AA, anaplastic astrocytoma (grade 3 astrocytoma) cohort G (Data Supplement); GBM, cohorts A-D, and F (Data Supplement); TCGA, newly diagnosed GBM samples from The Cancer Genome Atlas (TCGA) data set (Cancer Genome Atlas Research Network: Nature 455:1061-1068, 2008). (B) Heatmap of hierarchical clustering of GBMs in cohort C based on gene list derived from most variable genes between IDH1R132MUT and IDH1R132-wild type (IDH1R132WT; IDH1WT) GBMs (Data Supplement). IDH1R132WT GBMs (WT) show similarity to human adult neural stem cells for experimentation (HANSE), whereas IDH1R132MUT GBMs (MUT) show similarity to fetal/adult brain tissue (FETAL, ADULT).
By using agreement of differential expression (AGDEX)24 to compare expression profiles of the human tumors with a published embryonic mouse forebrain gene expression data set,25 we found that global expression profiles of IDH1R132WT GBMs resemble mouse neural stem cells, and IDH1R132MUT GBMs resemble lineage-committed neural precursors (AGDEX, +0.147; P < .004; Data Supplement). A separate hierarchical clustering analysis that used the genes most differentially expressed between IDH1R132MUT and IDH1R132WT GBMs reveals similarity of IDH1R132MUT samples to normal fetal or adult brain parenchyma and similarity of IDH1R132WT GBMs to cultured adult neural stem cells26 (Fig 2B). Because both fetal and adult brain samples are enriched for differentiating or mature neural cell types, these findings underscore the greater similarity of IDH1R132MUT GBMs to lineage-committed neural cells than to stem cells.
IDH1R132MUT GBMs Are Spatially and Temporally Restricted
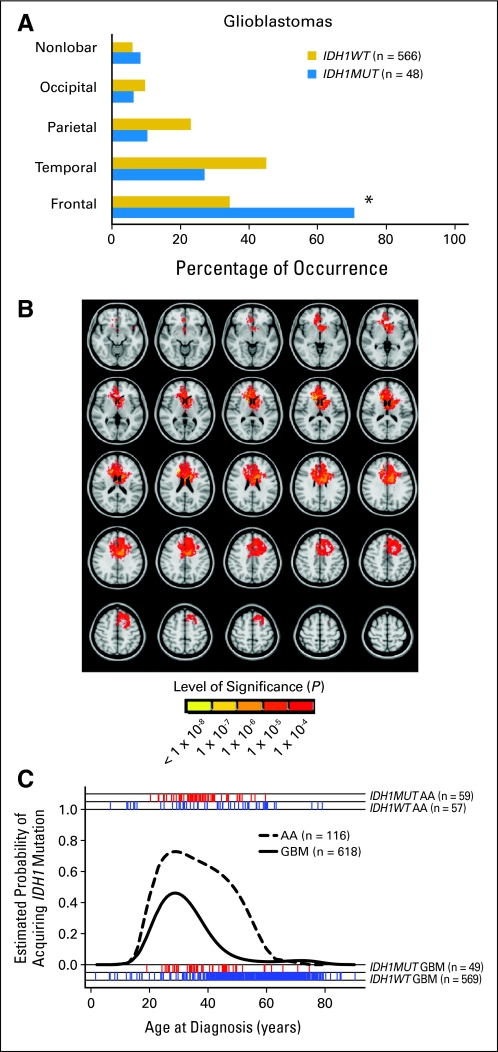
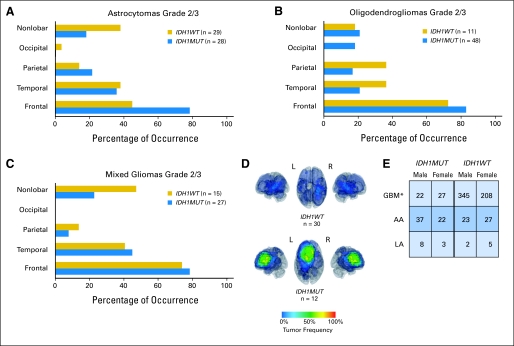
Tabulating the location of IDH1R132MUT and IDH1R132WT GBMs, we found a striking predominance of frontal lobe involvement of IDH1R132MUT GBMs that contrasts with the more widespread distribution of IDH1R132WT GBMs (Fig 3A). Regardless of histologic subtype, IDH1R132MUT gliomas displayed a nearly identical percentage of frontal lobe involvement (Appendix Figs A3A-A3C, online only).
Fig 3.
Spatial and temporal restriction of gliomas arising with mutations in isocitrate dehydrogenase 1 (IDH1) at R132 (IDH1R132MUT; IDH1MUT). (A) IDH1R132MUT de novo glioblastomas (GBMs) occur more frequently in the frontal lobe compared with IDH1R132-wild type (IDH1R132WT; IDH1WT) GBMs (*P < .001, two-tailed Fisher's exact test, with Bonferroni correction for multiple comparisons; Data Supplement). (B) Area of differential involvement (ADIFFI) analysis of axial magnetic resonance images. Voxels that are more frequently involved in IDH1R132MUT GBMs can be seen to cluster in the frontal lobe near the rostral extension of the lateral ventricle. All voxels with P < .0001 for differential involvement are depicted, as indicated in the color-coded bar. (C). IDH1R132MUT GBMs are temporally restricted. Logistic regression curves show the probability of IDH1R132MUT occuring in anaplastic astrocytomas (AAs; grade 3 astrocytoma) and GBMs as a function of age. The red (IDH1R132MUT) and blue (IDH1R132WT) tick marks show actual ages of patients (Data Supplement).
Overlay of tumor areas from a series of all available IDH1R132MUT GBMs with digitized pretreatment magnetic resonance images confirmed the high frequency of frontal lobe involvement, whereas overlay of a random sampling of IDH1R132WT tumors failed to demonstrate any frequently involved regions (Appendix Fig A3D). By performing a voxel-wise Fisher's exact test to isolate the area of differential involvement, we found that IDH1R132MUT GBMs were distributed at increased frequency in the area of the frontal lobe surrounding the rostral extension of the lateral ventricle (Fig 3B).
By examining tumor genotype as a function of age for both GBMs and grade 3 astrocytoma (anaplastic astrocytoma), we found that the relative frequency of IDH1R132MUT tumors rises sharply in the third decade of life and decreases in the fourth or fifth decade (Fig 3C). Thus, relative to IDH1R132WT tumors, IDH1R132MUT tumors appear to arise at greatest frequency within a more restricted time period. Interestingly, within adult GBMs, a significant difference in sex ratios was seen as a function of IDH1R132 status, consistent with previous reports of trends for a greater fraction of female patients with secondary versus primary GBM27,28 (Appendix Fig A3E).
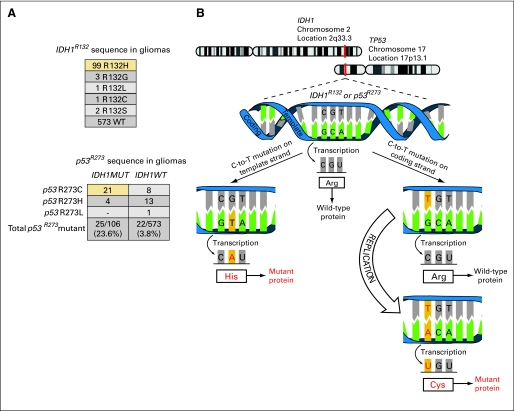
IDH1R132MUT GBMs Show Preponderance of Template Strand Mutation in IDH1 and Coding Strand Mutation in p53
In agreement with previous reports,13,16,29 our data showed a higher frequency of p53 mutation in IDH1R132MUT versus IDH1R132WT high-grade astrocytomas (Appendix Fig A4A, online only). Consistent with the well-documented propensity for C>T mutation at cytosine phosphate guanine (CpG) sites,30 data from our sample set showed that the most common mutations in both p53 and IDH1 are at Arg residues encoded by the codon CGT (IDH1R132 and p53R273; Figs 4A and 4B). For both IDH1R132 and p53R273, C>T mutations on template and coding strands will result in substitutions of His or Cys, respectively. By sequencing an expanded series of grades 2 to 4 gliomas for these codons in IDH1 and p53, we found, within IDH1R132MUT gliomas, a marked and unexpected contrast between the prevalence of Cys and His substitutions in the two proteins (P < .001 with Fisher's exact test; Fig 4A). Given that the probability for mutation is highest for C>T mutations, we deduce that the mutation pattern observed in tumors with mutations in both IDH1R132 and p53R273 is most likely to have occurred by strong selection for IDH1 and p53 mutations on the template versus on the coding strands, respectively (Fig 4B). These findings suggest that if both IDH1R132H and p53R273C mutations occur as C>T mutations in a nonproliferating cell, mutant IDH1 protein will be expressed immediately, whereas mutant p53 protein will not occur until after DNA replication. The predominance of p53R273C in the IDH1R132MUT tumors contrasts with the preference for p53R273H in the IDH1R132WT tumors (P < .005 with Fisher's exact test).
Fig 4.
Evidence for ordered appearance of aberrations in isocitrate dehydrogenase 1 (IDH1) mutant glioblastoma (GBM). (A) Top: Frequencies of sequences found at IDH1R132 show that the predominant substitution is His (yellow box). Bottom: Among gliomas with mutations at IDH1R132 (IDH1MUT) and p53R273, a predominance of Cys substitution at p53R273 is seen (yellow box); for IDH1R132-wild type (IDH1WT) tumors, the preferred substitution at p53R273 is His. (B) Schematic diagram showing that IDH1 mutation on the template strand and p53 mutation on coding strand can select for the expression of IDH1 mutant protein before p53 mutant protein if replication is delayed.
IDH1R132MUT GBMs Demonstrate CpG Island Methylator Phenotype, Focal EGFR Amplification, and Focal PTEN Loss
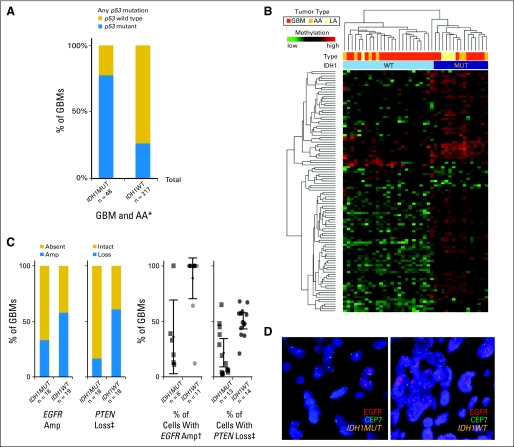
To identify other molecular aberrations that may cooperate with IDH1R132MUT in gliomagenesis, we performed a CpG island methylation profiling analysis on a subset of IDH1R132MUT and IDH1R132WT GBMs and found a distinct pattern of CpG island hypermethylation that was detected in all GBMs and lower-grade gliomas with IDH1R132MUT but was absent from nearly all IDH1R132WT gliomas (Appendix Fig A4B). The methylation pattern in IDH1R132MUT GBMs shows similarity to the recently reported CpG island methylator phenotype (CIMP) found to be closely associated with IDH1R132MUT gliomas31 and adds to the previous suggestion that these coordinated methylation changes occur early in the development of IDH1R132MUT tumors.
Array comparative genomic hybridization analysis and/or fluorescent in situ hybridization revealed lower frequencies of PTEN loss and fewer instances of EGFR amplification in IDH1R132MUT GBMs versus IDH1R132WT GBMs (Appendix Fig A4C; Data Supplement). A more robust finding from fluorescent in situ hybridization, however, was that, within cases showing any evidence of cells with PTEN loss or EGFR amplification, IDH1R132MUT cases showed a significantly lower percentage of affected cells (Appendix Figs A4C and A4D). The focality of copy number alterations for EGFR and PTEN in the IDH1R132MUT GBMs points to the likelihood that these are late events in the evolution of these tumors.
DISCUSSION
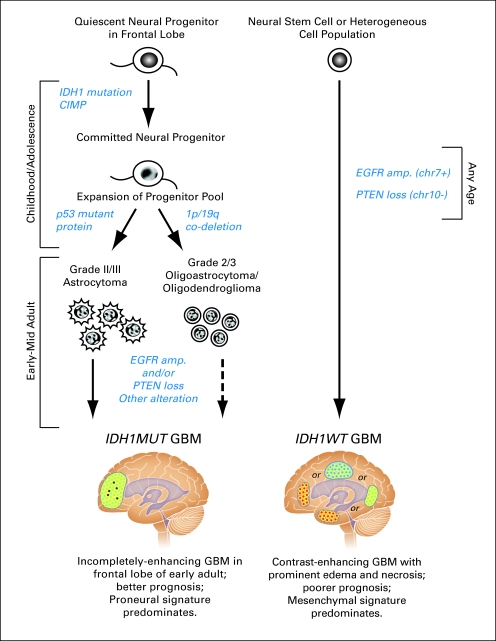
The development of novel therapeutic regimens for human malignancies, particularly those involving targeted therapy, is greatly facilitated by methods for identifying clinically meaningful disease subsets. To gain greater understanding of the utility of IDH1R132MUT as a marker in diffuse glioma, we have conducted a comprehensive analysis of the features of newly diagnosed cases of primary GBMs arising with and without IDH1R132MUT. Our results indicate that, although histologically similar, IDH1R132MUT and IDH1R132WT GBMs differ in their demographic, anatomic, phenotypic, epigenetic, and genomic presentation and follow a different clinical course, supporting a model of two disease entities that most likely arise from separate cell types of origin as the result of largely nonoverlapping sets of molecular events (Fig 5). Increased understanding of the cell of origin and molecular evolution of IDH1R132MUT GBM may aid in development of therapeutic strategies for this tumor type by yielding insights into the biology of these lesions and facilitating the development of animal models.
Fig 5.
Model comparing glioblastomas (GBMs) arising with and without mutations in isocitrate dehydrogenase 1 (IDH1) at R132. We propose that the IDH1R132MUT GBM pathway (left) is initiated by the occurrence of IDH1 mutation and resultant CpG island methylator phenotype (CIMP) in a quiescent neural progenitor residing in the frontal lobe. Although p53 mutation can be present during this time, expression of p53 mutant protein ensues only after expansion of this progenitor pool during late adolescence or early adulthood. According to the proposed model, glioma formation along the IDH1R132MUT pathway requires the ordered appearance of IDH1 mutant protein and CIMP, followed by p53 mutant protein (or 1p/19q co-deletion11). Tumors along this pathway arise from a spatially and temporally restricted neural progenitor population and most frequently maintain a Proneural gene expression signature. Transformation to IDH1R132MUT GBM requires EGFR amplification (amp), PTEN deletion, or other genomic alterations. In contrast, in the IDH1R132WT GBM pathway (right), EGFR amplification, and PTEN loss frequently act in concert to drive GBM formation from a cell population that maintains the ability to adopt a Mesenchymal gene expression signature. chr7+, chromosome 7 gain; chr10−, chromosome 10 loss. IDH1MUT, IDH1R132MUT; IDH1WT, IDH1R132WT.
Our analysis demonstrates that the phenotypic features that distinguish IDH1R132MUT from IDH1R132WT GBMs include better outcome, predominance of frontal lobe location, presentation and maintenance of Proneural expression signature, lesser extent of necrosis and edema, presence of non–contrast-enhancing component, and greater oligodendroglial content. These phenotypic findings, along with our genomic and epigenetic observations, confirm and extend earlier reports that IDH1R132MUT GBMs are distinguished by features associated with lower-grade diffuse glioma and support the contention that all IDH1R132MUT GBMs arise by evolution from lower-grade IDH1R132MUT gliomas, regardless of clinical history.12,14,15,29,31 Differences in demographics (age and sex) and tumor location in patients with IDH1R132MUT and IDH1R132WT GBM add to the features that suggest different etiologies of the two disease entities.
A key finding of this investigation is the discovery of a constellation of restricted phenotypic, spatial, and temporal features of IDH1R132MUT GBMs that is consistent with origin from a non–stem-cell neural precursor pool. By revealing the exclusion of Mesenchymal signature as an absolute feature of IDH1R132H gliomas, our results extend earlier reports of an association between IDH1 mutation and Proneural gene expression.9,32 This observation, and the closer resemblance of IDH1R132MUT GBM transcriptional signatures to those of brain tissue rather than of stem cells suggests that IDH1R132MUT GBM cells do not retain the capability to generate progeny with a broad range of transcriptional profiles.
Although the restriction of IDH1R132MUT GBM gene expression signatures may be a consequence of the actions of IDH1R132MUT, taken with the spatial and temporal homogeneity of IDH1R132MUT GBM presentation, this finding suggests that the cell of origin for IDH1R132MUT GBMs is a neural precursor population with limited differentiation potential that is most abundant during a specific stage and location in forebrain development. Our area of differential involvement analysis demonstrates a strong propensity for IDH1R132MUT gliomas to occur in the frontal lobe, specifically in the area surrounding the rostral extension of the lateral ventricles, indicating this region as a likely location of the cell of origin for many IDH1R132MUT gliomas. Consistent with our findings, predominance of frontal lobe location has been reported in oligodendrogliomas with 1p/19q co-deletion,33,34 a tumor type now known to carry IDH1 mutation at high frequency.13,14,29 Previous studies13,15,16,21,35,36 indicate that although the median age of patients with IDH1R132MUT GBM is younger than that for IDH1R132WT GBM, IDH1R132MUT is rare in the pediatric population. By using logistic regression to overcome the limitations of patient sampling, we were able to determine that the relative probability of a tumor harboring IDH1R132MUT abruptly increases around age 20 and begins to decrease a decade later. This raises the possibility that the cell type of origin for IDH1R132MUT gliomas is most abundant and permissive during a limited developmental time window, possibly coinciding with remodeling of prefrontal cortex in adolescence.37,38 Our findings that Proneural gene expression and increased oligodendroglial histology are associated with IDH1R132MUT GBMs are consistent with an oligodendroglial progenitor cell type of origin, and several studies lend support for oligodendroglial progenitor cells as a cell type of origin for glioma.39–44
Given the strong correlation we confirmed between CIMP and IDH1 mutation, we propose that CIMP is also an early and critical event in the development of IDH1R132MUT gliomas. Recent studies45,46 indicating that IDH1 mutation acts to inhibit a class of alpha-ketoglutarate–dependent enzymes, including proteins that catalyze histone demethylation and hydroxylation of methylated DNA, support the possibility that IDH1 mutation can initiate oncogenesis by inducing an epigenetic block to differentiation in a specific population of CNS cells poised at a particular developmental state. This hypothesis explains the homogeneity in presentation of IDH1R132MUT gliomas and suggests the possibility that these lesions might show sensitivity to therapeutic regimens with differentiating agents.
Several investigations13,16,18 demonstrate that most IDH1R132MUT astrocytomas harbor mutations of p53, and one study12 revealed instances in which IDH1 mutation was apparent before the appearance of p53 mutation. Our findings reveal the existence of a mechanism that is capable of ensuring sequential appearance of IDH1 mutant protein before p53 mutant protein, regardless of the order in which the mutations occur. Specifically, our sequencing data on IDH1 and p53 provides evidence for strand asymmetry of CpG mutations,47 demonstrating a strong preponderance of presumed C>T mutations on template versus coding strand for IDH1R132 and p53R273, respectively. Although mutation of IDH1 on the template strand permits immediate translation of IDH1 mutant protein, mutation of p53R273 on the coding strand allows the appearance of p53 mutant protein only after DNA replication. Thus, in a quiescent cell, the pattern of mutations we observed at high frequency in IDH1 mutant glioma will result in appearance of IDH1 mutant protein to be followed by p53 mutant protein only after a cycle of DNA replication. The preferred substitutions seen in gliomas harboring mutations at both IDH1R132 and p53R273 stand in stark contrast to the substitution pattern in cancers with mutations in only one of these genes. Specifically, for IDH1R132, the preferred substitution in acute myeloid leukemia is Cys,48–50 and for p53R273, the predominant substitution is His51 (including the IDH1R132WT gliomas in this study). Our finding of the frequent co-occurrence of IDH1R132H and p53R273C suggests that sequential appearance of IDH1 mutant protein before p53 mutant protein may be critical for formation of IDH1 mutant gliomas.
Our analysis suggests a model in which IDH1R132MUT GBMs arise in a stepwise fashion as the result of a series of sequenced molecular alterations that cooperate with normal developmental events (Fig 5 and Data Supplement). We propose that although IDH1R132MUT, induction of CIMP, and p53R273MUT often occur in a quiescent neural stem cell, tumors arise only from lineage-committed progeny following a wave of proliferation related to forebrain maturation that triggers appearance of mutant p53 protein and loss of cell cycle control. This oncogenic process results in a low-grade glioma that subsequently acquires additional genomic alterations that promote malignant transformation to GBM. Previous studies11,14,52 provide strong evidence that PTEN loss via loss of chromosome arm 10q is an event that occurs during transition to secondary GBM. Our cytogenetic observations of focal EGFR amplification and PTEN loss in IDH1R132MUT GBMs are consistent with the proposal that these events occur during the evolution of lower-grade IDH1R132MUT glioma to GBM.
In our model, the production of IDH1 mutant protein is the initial event in an orchestrated process that leads to the stepwise emergence of a distinct GBM entity that has a less aggressive clinical course than other GBMs. In contrast to the heterogeneous presentation of most GBMs, IDH1R132MUT GBMs arise at high frequency in early adult life as frontal lobe lesions with a constellation of radiographic, histologic, and transcriptional features that relates these lesions to the lower-grade diffuse gliomas from which we contend they arise. This investigation adds to a growing body of data that suggests that histologically similar brain tumors may represent distinct disease entities arising as a result of vulnerability of different stem-cell and progenitor populations to particular oncogenic alterations.24,53,54 IDH1R132MUT glioma may be an especially interesting example of the dependence of oncogenesis on normal developmental processes, because the cell of origin we propose may be uniquely abundant in human brain.
Supplementary Material
Acknowledgment
We thank SiliconMed for providing clinical data aggregation and search support, the University of California at Los Angeles Brain Tumor Translational Resource for providing logistic and technical support, and Shadi Lalezari for providing assistance with final manuscript preparation and submission.
Appendix
Fig A1.
Glioblastomas (GBMs) with mutations in isocitrate dehydrogenase 1 (IDH1) at R132 (IDH1R132MUT; IDH1MUT) are phenotypically distinct from IDH1R132-wild type (IDH1R132WT; IDH1WT) GBMs. (A) Kaplan-Meier survival curves of adult patients with de novo GBMs depict increased overall survival in patients with IDH1R132MUT versus IDH1R132WT tumors (log-rank test P < .001). Tick marks denote censored patients. All newly diagnosed adult (≥ 18 years) patients with available survival data are included in this analysis (Data Supplement). (B) Relative DNA copy number estimates of a sampling of IDH1R132MUT GBMs (upper panel) and IDH1R132WT GBMs (lower panel) demonstrates a minority of instances of co-deletion of 1p/19q in IDH1R132H GBMs (Data Supplement). Green and red deflections from the x axis indicate copy number gains and losses, respectively. The top portion of each panel depicts the percentage of tumors surveyed that exhibited copy number change events. The bottom traces in each panel show, for each sample (identified by case ID), a ratio of the fold change in intensity signals between the test and reference sample, transformed by using log2. (C) Scoring of a panel of radiologic variables in a sampling of GBMs reveals decreased occurrence of enhancement (*P < .005), increased frequency of non–contrast-enhancing tumor (nCET; †P < .001), decreased incidence of necrosis (*P < .005), decreased extent of edema (‡P < .01), increased presence of cysts (†P < .001), increased incidence of diffuse component (§P < .05), increased frequency of touches ventricle (§P < .05), and increased tumor size (*P < .005) for IDH1R132MUT compared with IDH1R132WT; Data Supplement). Occurrences of multifocal lesions, satellites, and touches neocortex were not different.
Fig A2.
Glioblastomas (GBMs) with mutations in isocitrate dehydrogenase 1 (IDH1) at R132 (IDH1R132MUT; IDH1MUT) are restricted from Mesenchymal gene expression. Gene expression analysis reveals a robust positive association of IDH1R132MUT high-grade astrocytomas with the Proneural signature and an absolute exclusion of the Mesenchymal gene signature from IDH1R132MUT tumors. Subtypes and their respective centroids are labeled as follows: Proneural (PN), Proliferative (PROLIF), and Mesenchymal (MES). (A) The three-dimensional graphical plot illustrates the similarity between individual tumor samples in cohorts C and G to each of the three centroids created by K means clustering to define tumor subtypes.19 (B) Matched primary and recurrent pairs of tissue show that IDH1R132MUT GBMs and anaplastic astrocytomas (AAs; grade 3 astrocytoma) fail to acquire a Mesenchymal gene expression signature on recurrence (Data Supplement). IDH1WT, IDH1R132WT.
Fig A3.
Spatial restriction of gliomas arising with mutations in isocitrate dehydrogenase 1 (IDH1) at R132 (IDH1R132MUT; IDH1MUT). Location of grade 2 and 3 astrocytomas (A), oligodendrogliomas (B), and mixed gliomas (C) show that, as for GBMs, the majority of IDH1R132MUT gliomas of these histologic subtypes involve the frontal lobe (Data Supplement). (D) Three-dimensional representations of frequency maps generated by overlaying tumor regions determined from digital magnetic resonance images of patient tissue with IDH1R132-wild type (IDH1R132WT; IDH1WT) and IDH1R132MUT GBMs, depicting frequent frontal lobe involvement in patients with IDH1R132MUT tumors. In contrast to IDH1R132MUT GBMs, IDH1R132WT GBMs are more broadly distributed throughout the forebrain (Data supplement). (E) Gender distribution of adult patients with astrocytoma as a function of tumor grade and IDH1R132 mutation status. *P < .05 for IDH1R132MUT versus IDH1R132WT GBM. AA, anaplastic astrocytoma; LA, low-grade astrocytoma.
Fig A4.
Evidence for ordered appearance of aberrations in isocitrate dehydrogenase 1 (IDH1) mutant glioblastoma (GBM). (A) Tumors with a mutation in IDH1 at R132 (IDH1R132MUT; IDH1MUT) show increased mutation frequency of p53 at any site within exons 4 to 10 in a series of GBMs and anaplastic astrocytomas (AAs; grade 3 astrocytoma) compared with IDH1R132-wild type (IDH1R132WT; IDH1WT) tumors (*P < .001; Data Supplement). (B) Unsupervised hierarchical clustering of DNA methylation data profiling of a series of glioma samples reveals the presence of a CpG island methylator phenotype strongly correlated with IDH1R132MUT (Data Supplement). (C) Quantitative evaluation of fluorescent in situ hybridization for EGFR and PTEN in a series of newly diagnosed GBMs (Data Supplement). Bar graphs (left) depict a nonsignificant trend toward less frequent EGFR amplification (Amp; P < .191) and reduced frequency of cases with IDH1R132MUT GBM showing PTEN loss (+P < .05). Scatter plots (right) show the frequency of cells within individual cases that show these copy number alterations, indicating decreased percentage of cells with EGFR amplification (†P < .001) and PTEN loss (‡P < .05) in IDH1R132MUT GBMs. Cases in which no cells showed detectable alterations are not included in the scatter plots or statistical analysis of percentage of cells. (D) Representative example of fluorescent in situ hybridization using probes to EGFR (red) and to the centromere of chromosome 7 (CEP7, green), showing an example of focal amplification in an IDH1R132MUT GBM and widespread amplification in an IDH1R132WT GBM. LA, low-grade astrocytoma; MUT, mutant; WT, wild type.
Footnotes
See accompanying editorial on page 4473
Supported in part by Grants No. K08 CA124479 (A.L.) from the National Cancer Institute, National Institutes of Health; and No. LA1300/4-1 (K.L. and M.W.) from the Deutsche Forschungsgemeinschaft and the Deutsche Krebshilfe.
Presented at the 15th Annual Meeting of the Society for Neuro-Oncology, Montreal, Quebec, Canada, November 18-21, 2010, and at the 102nd Annual Meeting of the American Association for Cancer Research, Orlando, FL, April 2-6, 2011.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Samir Kharbanda, Genentech (C); Franklin Peale, Genentech (C); William F. Forrest, Genentech (C); Kanan Pujara, Genentech (C); Ajay Pandita, Genentech (C); Robert H. Soriano, Genentech (C); Sankar Mohan, Genentech (C); Somasekar Seshagiri, Genentech (C); Zora Modrusan, Genentech (C); Zhaoshi Jiang, Genentech (C); Heidi S. Phillips, Genentech (C), Merck (C) Consultant or Advisory Role: Albert Lai, Genentech Advisory Board (C); Whitney B. Pope, Genentech/Roche (C); Phioanh Leia Nghiemphu, Genentech (C); Michael D. Prados, Genentech (U); Manfred Westphal, Genentech (U); Katrin Lamszus, Genentech (U) Stock Ownership: Samir Kharbanda, Roche; Franklin Peale, Roche; William F. Forrest, Roche; Kanan Pujara, Roche; Ajay Pandita, Roche; Robert H. Soriano, Roche; Sankar Mohan, Roche; Somasekar Seshagiri, Roche; Zora Modrusan, Roche; Zhaoshi Jiang, Roche; Heidi S Phillips, Roche, Merck Honoraria: Whitney B. Pope, Genentech/Roche; Timothy Cloughesy, Genentech, Roche, Agios Pharmaceuticals, Eli Lilly, Novartis Research Funding: Albert Lai, Genentech; Kenneth D. Aldape, Brain Tumor Founder's Collaborative, Doctor Marnie Rose Foundation; Linda M. Liau, Agios Pharmaceuticals, Northwest Biotherapeutics; Michael D. Prados, Genentech; Timothy Cloughesy, Genentech Expert Testimony: Michael D. Prados, Genentech (U) Other Remuneration: Ajay Pandita, Genentech; Sankar Mohan, Genentech
AUTHOR CONTRIBUTIONS
Conception and design: Albert Lai, Samir Kharbanda, Whitney B. Pope, Timothy Cloughesy, Heidi S. Phillips
Financial support: Albert Lai, Timothy Cloughesy
Administrative support: Timothy Cloughesy
Provision of study materials or patients: Albert Lai, Nils O. Schmidt, Kenneth D. Aldape, Paul S. Mischel, Linda M. Liau, Phioanh Leia Nghiemphu, C. David James, Michael D. Prados, Manfred Westphal, Katrin Lamszus, Timothy Cloughesy, Heidi S. Phillips
Collection and assembly of data: Albert Lai, Samir Kharbanda, Whitney B. Pope, Anh Tran, Orestes E. Solis, Franklin Peale, Kanan Pujara, Jose A. Carrillo, Ajay Pandita, Robert H. Soriano, Nils O. Schmidt, Sankar Mohan, William H. Yong, Somasekar Seshagiri, Zora Modrusan, Kenneth D. Aldape, Cameron J. Escovedo, Weidong Chen, Phioanh Leia Nghiemphu, C. David James, Michael D. Prados, Manfred Westphal, Katrin Lamszus, Timothy Cloughesy, Heidi S. Phillips
Data analysis and interpretation: Albert Lai, Samir Kharbanda, Whitney B. Pope, Anh Tran, William F. Forrest, Franklin Peale, Jose A. Carrillo, Ajay Pandita, Benjamin M. Ellingson, Chauncey W. Bowers, Sankar Mohan, Somasekar Seshagiri, Zhaoshi Jiang, Cameron J. Escovedo, Timothy Cloughesy, Heidi S. Phillips
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Clarke J, Butowski N, Chang S. Recent advances in therapy for glioblastoma. Arch Neurol. 2010;67:279–283. doi: 10.1001/archneurol.2010.5. [DOI] [PubMed] [Google Scholar]
- 2.Hambardzumyan D, Squatrito M, Carbajal E, et al. Glioma formation, cancer stem cells, and akt signaling. Stem Cell Rev. 2008;4:203–210. doi: 10.1007/s12015-008-9021-5. [DOI] [PubMed] [Google Scholar]
- 3.Alcantara Llaguno S, Chen J, Kwon CH, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alcantara Llaguno SR, Chen J, Parada LF. Signaling in malignant astrocytomas: Role of neural stem cells and its therapeutic implications. Clin Cancer Res. 2009;15:7124–7129. doi: 10.1158/1078-0432.CCR-09-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: A population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 6.Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci. 2009;100:2235–2241. doi: 10.1111/j.1349-7006.2009.01308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 9.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe T, Nobusawa S, Kleihues P, et al. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nobusawa S, Watanabe T, Kleihues P, et al. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15:6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 13.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balss J, Meyer J, Mueller W, et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 15.Ichimura K, Pearson DM, Kocialkowski S, et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol. 2009;11:341–347. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Chen R, Nishimura MC, Bumbaca SM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 19.Schiffman JD, Hodgson JG, VandenBerg SR, et al. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010;70:512–519. doi: 10.1158/0008-5472.CAN-09-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Günther HS, Schmidt NO, Phillips HS, et al. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008;27:2897–2909. doi: 10.1038/sj.onc.1210949. [DOI] [PubMed] [Google Scholar]
- 21.Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: A prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27:5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 22.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 23.Pope WB, Sayre J, Perlina A, et al. MR imaging correlates of survival in patients with high-grade gliomas. AJNR Am J Neuroradiol. 2005;26:2466–2474. [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson RA, Wright KD, Poppleton H, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaguchi A, Ikawa T, Kasukawa T, et al. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- 26.Müller FJ, Laurent LC, Kostka D, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Deimling A, von Ammon K, Schoenfeld D, et al. Subsets of glioblastoma multiforme defined by molecular genetic analysis. Brain Pathol. 1993;3:19–26. doi: 10.1111/j.1750-3639.1993.tb00721.x. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe K, Tachibana O, Sata K, et al. Overexpression of the EGF receptor and p53 mutations are mutually exclusive in the evolution of primary and secondary glioblastomas. Brain Pathol. 1996;6:217–223. doi: 10.1111/j.1750-3639.1996.tb00848.x. discussion 23–24. [DOI] [PubMed] [Google Scholar]
- 29.Yan H, Bigner DD, Velculescu V, et al. Mutant metabolic enzymes are at the origin of gliomas. Cancer Res. 2009;69:9157–9159. doi: 10.1158/0008-5472.CAN-09-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- 31.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ducray F, Marie Y, Sanson M. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:2248–2249. author reply 2249. [PubMed] [Google Scholar]
- 33.Zlatescu MC, TehraniYazdi A, Sasaki H, et al. Tumor location and growth pattern correlate with genetic signature in oligodendroglial neoplasms. Cancer Res. 2001;61:6713–6715. [PubMed] [Google Scholar]
- 34.Laigle-Donadey F, Martin-Duverneuil N, Lejeune J, et al. Correlations between molecular profile and radiologic pattern in oligodendroglial tumors. Neurology. 2004;63:2360–2362. doi: 10.1212/01.wnl.0000148642.26985.68. [DOI] [PubMed] [Google Scholar]
- 35.Paugh BS, Qu C, Jones C, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 37.Barnea-Goraly N, Menon V, Eckert M, et al. White matter development during childhood and adolescence: A cross-sectional diffusion tensor imaging study. Cereb Cortex. 2005;15:1848–1854. doi: 10.1093/cercor/bhi062. [DOI] [PubMed] [Google Scholar]
- 38.Harris LW, Lockstone HE, Khaitovich P, et al. Gene expression in the prefrontal cortex during adolescence: Implications for the onset of schizophrenia. BMC Med Genomics. 2009;2:28. doi: 10.1186/1755-8794-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Persson AI, Petritsch C, Swartling FJ, et al. Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18:669–682. doi: 10.1016/j.ccr.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindberg N, Kastemar M, Olofsson T, et al. Oligodendrocyte progenitor cells can act as cell of origin for experimental glioma. Oncogene. 2009;28:2266–2275. doi: 10.1038/onc.2009.76. [DOI] [PubMed] [Google Scholar]
- 41.Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, et al. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51:187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 42.Levison SW, Young GM, Goldman JE. Cycling cells in the adult rat neocortex preferentially generate oligodendroglia. J Neurosci Res. 1999;57:435–446. [PubMed] [Google Scholar]
- 43.Aguirre A, Dupree JL, Mangin JM, et al. A functional role for EGFR signaling in myelination and remyelination. Nat Neurosci. 2007;10:990–1002. doi: 10.1038/nn1938. [DOI] [PubMed] [Google Scholar]
- 44.Menn B, Garcia-Verdugo JM, Yaschine C, et al. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodin SN, Rodin AS. Strand asymmetry of CpG transitions as indicator of G1 phase-dependent origin of multiple tumorigenic p53 mutations in stem cells. Proc Natl Acad Sci U S A. 1998;95:11927–11932. doi: 10.1073/pnas.95.20.11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Green A, Beer P. Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med. 2010;362:369–370. doi: 10.1056/NEJMc0910063. [DOI] [PubMed] [Google Scholar]
- 49.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 52.Fujisawa H, Kurrer M, Reis RM, et al. Acquisition of the glioblastoma phenotype during astrocytoma progression is associated with loss of heterozygosity on 10q25-qter. Am J Pathol. 1999;155:387–394. doi: 10.1016/S0002-9440(10)65135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor MD, Poppleton H, Fuller C, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Gibson P, Tong Y, Robinson G, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.