Abstract
Background
Aberrant methylation of promoter DNA and transcriptional repression of specific tumor suppressor genes play an important role in carcinogenesis. Recently, many studies have investigated the association between cigarette smoking and p16INK4α gene hypermethylation in lung cancer, but could not reach a unanimous conclusion.
Methods and Findings
Nineteen cross-sectional studies on the association between cigarette smoking and p16INK4α methylation in surgically resected tumor tissues from non-small cell lung carcinoma (NSCLC) patients were identified in PubMed database until June 2011. For each study, a 2×2 cross-table was extracted. In total, 2,037 smoker and 765 nonsmoker patients were pooled with a fixed-effects model weighting for the inverse of the variance. Overall, the frequency of p16INK4α hypermethylation was higher in NSCLC patients with smoking habits than that in non-smoking patients (OR = 2.25, 95% CI = 1.81–2.80). The positive association between cigarette smoking and p16INK4α hypermethylation was similar in adenocarcinoma and squamous-cell carcinoma. In the stratified analyses, the association was stronger in Asian patients and in the studies with larger sample sizes.
Conclusion
Cigarette smoking is positively correlated to p16INK4α gene hypermethylation in NSCLC patients.
Introduction
The incidence of lung cancer is increasing worldwide, particularly in developing countries. In China the death rate of lung cancer has been increasing from 7.1 to 30.8 per 100,000 during 1975–2005. People dying due to lung cancer accounted for 23% of total amount of cancer death in 2005 [1]. 80% of primary lung cancers are non-small cell lung carcinoma (NSCLC) which is characterized by a long asymptomatic latency and poor prognosis. Without an early diagnostic approach, over 40% of lung cancer patients develop metastasis at the time of diagnosis and survive for a short time period under a conventional chemotherapy [2]. Only 15% of NSCLC patients can survive over 5 years [3]. Thus, it is essential to identify biomarkers for early prediction of lung cancer.
Cigarette smoking is a well known driving force for lung cancer development. The lifetime risk of developing lung cancer is 17.2% in male smokers and 11.6% in female smokers, which is much higher than that in nonsmokers with 1.3% in male and 1.4% in female [4]. Although most lung cancers are associated with cigarette smoking, it is statistically estimated that 15% of them in males and 53% in females, accounting for about 25% of all lung cancers, are not attributable to cigarette smoking [3]. Lung cancers arising in nonsmokers are more frequently adenocarcinomas, affect females disproportionately more than males, and have regional differences ranging from 10–15% in Europe and North America to 30–40% in Asian countries [5]–[7]. Moreover, nonsmoker lung cancers have improved survival and are more sensitive to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor therapy. It might be due to that the activation of EGFR by gene mutations appears more often in nonsmoker lung cancers [5], [6]. Taken together, lung cancer in nonsmokers would probably be considered a separate cancer category. If so, it would rank as the seventh most common cause of cancer death worldwide [3]. However, whether these clinical-pathological and molecular differences between lung cancer in nonsmokers and smokers are related to cigarette smoking is still unknown.
The interest in cancer-associated changes in gene methylation has grown enormously in recent years with the speculation that the promoter methylation status may provide an early biomarker for tumorigenesis [8]. Silencing of genes by aberrant promoter hypermethylation has been recognized as a key event in cancer initiation and progression [9], [10]. Highly sensitive assays such as methylation-specific PCR (MSP), which could detect one methylated allele in the presence of 103–104 unmethylated alleles [11], [12], have been used to assess gene-promoter methylation in primary tumors, serum, plasma, sputum or specimens from the aerodigestive tract epithelium [13]. Numerous studies have investigated the methylation statuses of specific genes in body fluids and tumor tissues of lung cancer patients, and identified more than 60 genes as being epigenetically silenced in lung tumors [8]. The proof-of-concept studies suggested that gene-specific promoter methylation occurs as an early event in lung cancer. For example, hypermethylation of p16INK4α (also known as cyclin-dependent kinase inhibitor 2A, CDKN2A) or O6-methylguanine-DNA methyltransferase (MGMT) was found in sputum samples 5 to 36 months prior to clinical diagnosis [14]. Moreover, the frequency of p16INK4α hypermethylation increased progressively from 17% in basal-cell hyperplasia to 24% in squamous metaplasia, and to 60% in squamous cell carcinomas [15]. The correlation between gene methylation and recurrence of lung cancer has also been reported [16], [17]. The promoter hypermethylation of several genes including p16INK4α, cadherin 13 (CDH13), Ras association (RalGDS/AF-6) domain family member 1 (RASSF1A) and adenomatous polyposis coli (APC) in NSCLC specimens is associated with early recurrence after surgery [17]. Moreover, the reversibility of DNA methylation modification makes it possible that the expression of genes that have undergone epigenetic silencing becomes reactivated by the inhibitors of DNA methylation. Many agents such as 5-azacytidine and zebularine are currently being tested in clinical trials [18]. Despite the significance of gene promoter methylation in predicting incidence or prognosis and in epigenetic therapy of lung cancer, the manner in which these epigenetic lesions accumulate during carcinogenesis is not completely understood. The missing links among environmental factors, DNA methylation changes, and lung cancer limit the applications of methylated genes as a biomarker for early detection of lung cancers.
The correlation between cigarette smoking and aberrant gene methylation has been extensively studied, but the results are inconsistent and inconclusive. The present study mainly focused on p16INK4α gene as it is the first gene identified in lung cancer, and is transcriptionally silenced predominantly through aberrant promoter hypermethylation [19]. Here, we performed a literature-based systematic review and meta-analysis to quantitatively analyze the correlation between cigarette smoking and p16INK4α gene methylation in NSCLC patients.
Methods
Ethics Statement
An ethics statement was not required in this study.
Literature search
We systematically searched for all published articles indexed in PubMed database from 1966 to June 18, 2011 with the Medical Subject Headings (MeSH) and corresponding free text: “smok* AND (p16* OR CDKN2A OR INK4A) AND (methylation OR epigene*)”. We also manually searched the references of these publications in order to retrieve additional studies. Only those published as full-text articles and in English or in Chinese were included as candidates.
Inclusion and exclusion criteria
Studies were selected for analysis if they met the following criteria: 1) they were original epidemiological studies on the correlation between cigarette smoking and p16INK4α methylation; 2) they were conducted in lung cancer patients; 3) the specimens used for methylation analysis must include surgically resected primary tumor samples, while other specimens such as sputum, serum, bronchial lavage samples and normal or non-malignant lung tissue may be used, but not essential; 4) p16INK4α methylation status was examined using methylation-specific PCR (MSP) or quantitative MSP (QMSP); 5) the subjects in every study comprised nonsmokers and smokers (former smokers and/or current smokers), irrespective of minor discrepancies of the definition of nonsmokers over all studies. To avoid duplication of data, we carefully checked the author names, research institutions and procedures for enrolling participants. Where several publications reported from the same population data, only the most rounded study with more information was included.
Data collection
For each eligible study, we collected information regarding authors, year and source of publication, country of origin, inclusion criteria, exclusion criteria, histology of lung cancer, types of biological specimen, number of participants, participants' age and gender, smoking behaviour, p16INK4α methylation frequencies in nonsmokers and smokers and the method for methylation detection. All included studies used nonsmokers as a control group, though some of them did not provide the definition of nonsmoker. In studies defining nonsmoker, there were three different definitions of nonsmoker: (1) daily cigarette consumption×years of smoking = 0; (2) less than 100 cigarettes in entire lifetime; (3) less than 20 pack-years. Since it is impossible to redefine nonsmoker based on a unified standard, we combined nonsmokers in our meta-analysis according to their original group in each individual study. Correspondingly, subjects except nonsmokers were smokers comprising former smokers and current smokers, and light, moderate or heavy smokers. Then data was integrated in 2×2 tables demonstrating the methylation/unmethylation of p16INK4α gene according to the cigarette smoking status (smoker/nonsmoker). All data were extracted independently by two reviewers using a standard form. Minor discrepancies were resolved by the authors' discussion.
Meta-analysis and statistical analysis
The foremost analysis examined the differences in the frequency of p16INK4α methylation in lung cancer tissues between smoker and nonsmoker patients. Summary OR was obtained across all studies. Heterogeneity was examined using the I2 statistic, which represents the proportion of variation in the effect sizes that is attributable to true differences across studies rather than to a random error. Results without heterogeneity were pooled using the fixed-effect model, following the Mantel-Haenszel method. Otherwise, the random effect analysis with the method of DerSimonian and Larid was used.
The meta-analyses were performed using Stata statistical software (version 10.0, Stata Corporation, College Station, Texas, USA). Results were shown in forest plots, where the sizes of the boxes for individual studies were inversely proportional to the variances of the log relative risks, and the horizontal lines represent 95% confidence interval (CI).
The frequencies of p16INK4α methylation in smokers and nonsmokers were compared by Wilcoxon signed rank test. The coefficients of Spearman's rank correlation were calculated between frequency of p16INK4α methylation and sample size. These analyses were performed using SPSS for Windows (version 11.5, SPSS Inc., Chicago, IL, USA).
Results
Study characteristics
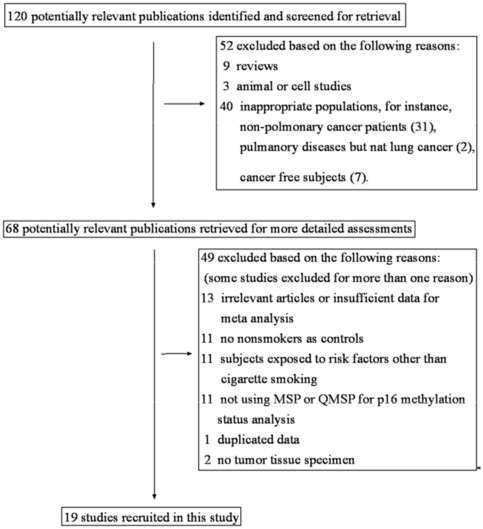
Following the inclusion and exclusion criteria described above, 19 studies [12], [20]–[37] were included in the analysis (Figure 1). The characteristics of these studies are summarized in Table 1. Of these 19 studies, eight defined the subtypes (adenocarcinoma or squamous cell carcinoma) of NSCLCs. Ten studies were conducted in Asia (2 in China, 6 in Japan and 2 in Korea), five were in USA, and the remaining four were in Australia, Greece, Chile, and multi-areas in Asia-Pacific regions (USA, Australia, Japan and Taiwan) respectively (Table 1). In two studies [12], [25], lung adenocarcinoma (AC) patients and squamous cell carcinoma (SCC) patients were analyzed separately, therefore they were treated as separate items in the meta-analysis. As for the primer sequences of MSP, 15 studies used the same primers designed by Herman et al. in 1996 [11]. The primer sequences for detecting methylated p16INK4α gene were 5′-TTA TTA GAG GGT GGG GCG GAT CGC-3′ (sense) and 5′-GAC CCC GAA CCG CGA CCG TAA-3′ (antisense). The size of the PCR product for the methylated reaction was 150 bp. The primer sequences used for the unmethylated promoter were 5′-TTA TTA GAG GGT GGG GTG GAT TGT-3′ (sense) and 5′-CAA CCC CAA ACC ACA ACC ATA A-3′ (antisense). The size of the PCR product for the unmethylated reaction was 151 bp.
Figure 1. Flow diagram of the stepwise selection from associated studies.
Table 1. Characteristics of studies on the overall relationship between cigarette smoking and p16 methylation in lung cancer patients.
| First author | Year | Location | Histology | Age(y) | Sample size(n) | p16 methylation in smoker | p16 methylation in nonsmoker |
| Sanchez-Cespedes [19] | 2001 | USA | NSCLC | 66±3 | 47 | 7/33 | 5/14 |
| Zochbauer-Muller [20] | 2001 | Australia | NSCLC | 28–81 | 107 | 27/98 | 0/9 |
| Kim [21] | 2001 | USA | NSCLC | 67±11 | 185 | 49/172 | 2/13 |
| Yanagawa [22] | 2002 | Japan | NSCLC | 67±2 | 51 | 13/37 | 1/14 |
| Toyooka [12] | 2003 | Asia-Pacific | AC | 26–87 | 295 | 47/183 | 6/112 |
| Toyooka [12] | 2003 | Asia-Pacific | SCC | 26–87 | 189 | 58/172 | 5/17 |
| Yanagawa [23] | 2003 | Japan | NSCLC | 39–86 | 75 | 21/55 | 2/20 |
| Toyooka [24] | 2004 | Japan | AC | No data | 217 | 29/120 | 10/97 |
| Toyooka [24] | 2004 | Japan | SCC | No data | 138 | 46/130 | 1/8 |
| Kim [25] | 2004 | Korea | SCC | No data | 125 | 37/117 | 3/8 |
| Divine [26] | 2005 | USA | AC | 33–86 | 203 | 81/157 | 18/46 |
| Liu [27] | 2006 | USA | NSCLC | 65±10 | 122 | 51/81 | 13/41 |
| Nakata [28] | 2006 | Japan | NSCLC | 40–85 | 202 | 38/139 | 9/63 |
| Toyooka [29] | 2006 | Japan | AC | No data | 164 | 24/86 | 7/78 |
| Georgiou [30] | 2007 | Greece | NSCLC | 45–75 | 27 | 20/24 | 2/3 |
| Guzman [31] | 2007 | Chile | NSCLC | 66±9 | 65 | 39/54 | 10/11 |
| Kim [32] | 2007 | Korea | NSCLC | 41–82 | 99 | 18/79 | 4/20 |
| Tessema [33] | 2009 | USA | AC | 66 | 175 | 67/100 | 48/75 |
| Wang [34] | 2010 | China | AC | 46–84 | 56 | 9/20 | 10/36 |
| Yanagawa [35] | 2011 | Japan | AC | 39–86 | 62 | 7/36 | 2/26 |
| Zhang [36] | 2011 | China | NSCLC | 32–79 | 198 | 82/144 | 18/54 |
Combined results and subgroup analyses
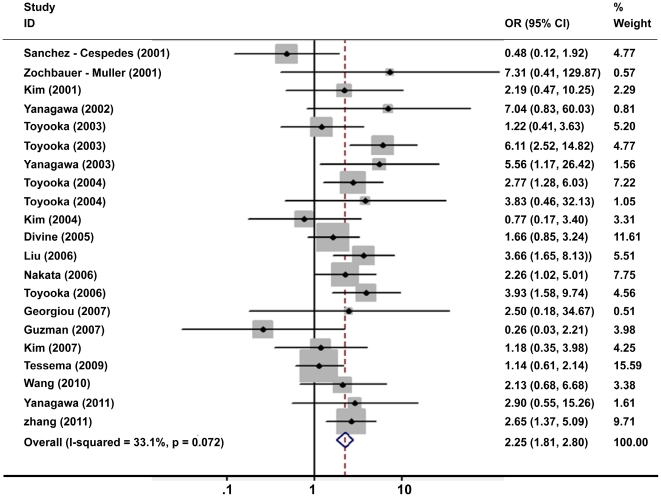
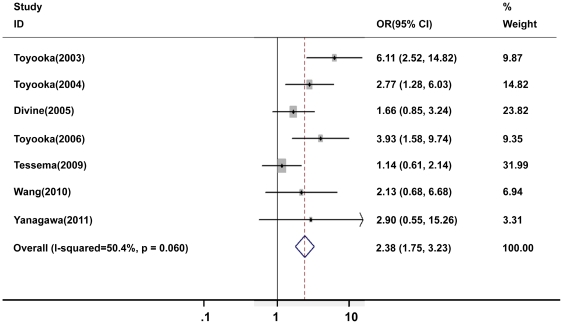
In general, the frequencies of p16INK4α methylation ranged from 19 to 83% (median 34%) in smoker patients, which were much higher than those in nonsmoker patients with range from 0 to 91% (median 20%). In the meta-analysis, 2802 NSCLC patients including 2037 smokers and 765 nonsmokers were included in pooling the overall correlation estimation. Under the fixed-effects model, the pooled odds ratio (OR) of p16INK4α methylation in smoker patients, compared to nonsmoker patients, was 2.25 with 95% CI = 1.81–2.80 (Figure 2). Of six studies only including AC patients, the subgroup analysis showed no significant difference in the association between cigarette smoking and p16INK4α methylation when comparing AC patients (OR = 2.38, 95%CI = 1.75–3.23) (Figure 3) and all NSCLC patients (OR = 2.25, 95% CI = 1.81–2.80).
Figure 2. Meta-analysis of cigarette smoking and p16INK4α methylation in all NSCLC patients.
Figure 3. Meta-analysis of cigarette smoking and p16INK4α methylation in lung adenocarcinoma patients.
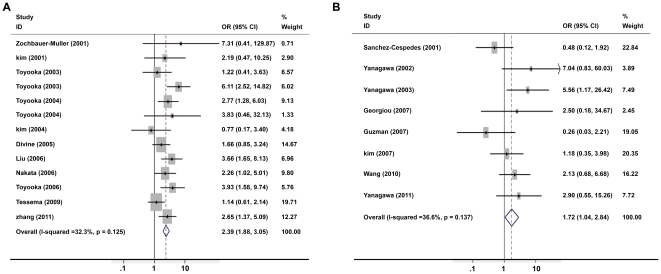
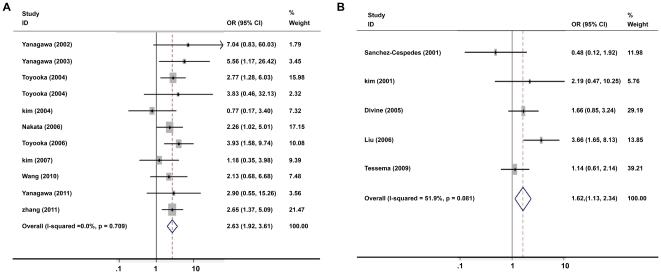
Because there was a borderline significant but moderate degree of heterogeneity among the 19 studies (I2 = 33.1%, P = 0.072), we performed sensitivity analyses to identify potential sources of heterogeneity. Stratification by sample size showed a stronger association in larger-size (>100 patients per study) studies (OR = 2.39, 95% CI = 1.88–3.05, Figure 4A) than that in smaller-size studies (OR = 1.72, 95% CI = 1.04–2.84, Figure 4B). Stratified analysis also revealed that the associations varied among the subjects from different regions. The association between cigarette smoking and p16INK4α methylation tended to be stronger in 9 Asian studies (OR = 2.63, 95%CI = 1.92–3.61) (Figure 5A) compared to the 5 North American studies (OR = 1.62, 95%CI = 1.13–2.34) (Figure 5B). In addition, there was no inter-study heterogeneity in Asian studies (I2 = 0, P = 0.709).
Figure 4. Meta-analysis of cigarette smoking and p16INK4α methylation in NSCLC patients stratified by sample size.
A: sample size >100. B: sample size <100. Stratification by sample size showed a stronger association in studies with a relatively larger sample size.
Figure 5. Meta-analysis of cigarette smoking and p16INK4α methylation in NSCLC patient stratified by study region.
A: Asian studies. B: North American studies. The association between cigarette smoking and p16INK4α methylation tended to be stronger in Asian studies compared to the North American studies.
Four of the 19 included studies [12], [21], [23], [31] and another two excluded studies [38], [39] had compared the frequencies of p16INK4α methylation in adjacent noncancerous tissues or sputum specimens from NSCLC patients with or without smoking habits and no significant differences were found (Table S1). In another 7 excluded studies [40]–[46], p16INK4α methylation was examined in sputum, bronchial lavage samples or blood specimens from cancer-free subjects. We failed to find a link between cigarette smoking and the frequency of p16INK4α methylation (Table S2, Figure S1). These results suggested that cigarette smoking had no impact on p16INK4α hypermethylation in the surrogate samples from NSCLC patients, and that the positive association between cigarette smoking and p16INK4α hypermethylation was not present in health conditions.
Publication bias
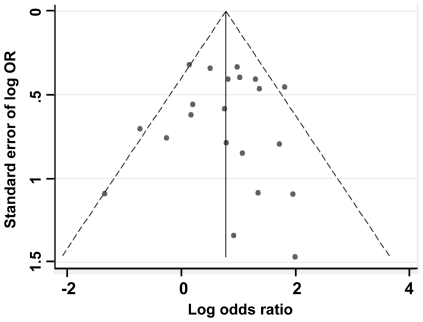
To ensure the quality of this study, we performed a Begger's funnel plot and Egger's tests to eliminate the publication bias of included studies. As shown in Figure 6, the shapes of the funnel plots showed a little asymmetry at the bottom. However, Egger's test, which provided statistical data of funnel plot symmetry, did not show any evidence of publication bias (t = 0, P = 0.998).
Figure 6. Begg's funnel plot for visual assessment of the presence of publication bias for all studies included in the meta-analysis (each study is represented by an open circle).
Discussion
The present study, based on the accumulated evidences from 19 cross-sectional studies, indicates that cigarette smoking is positively related to p16INK4α hypermethylation in tumor tissues from NSCLC patients. The frequency of p16INK4α methylation in smoker lung cancer patients was 2.25 times higher than that in nonsmoker patients. The association appeared to be stronger in Asian patients and in studies with a larger number of subjects, but without a histology (AC or SCC) specificity. However, this positive correlation did not exist in adjacent noncancerous tissues from NSCLC patients and in biological specimens of ‘healthy’ subjects without cancer. Given the results that cigarette smoking leading to p16INK4α hypermethylation was related to the stage progression of tumorigenesis, we speculated that p16INK4α hypermethylation might be an early marker for cancer diagnosis, particularly in cigarette smoking patients.
Recent epidemiological studies have revealed that molecular mechanisms underlying the development of lung cancers differed between nonsmokers and smokers. For instance, EGFR pathway is frequently activated by gene mutations in nonsmoker lung cancers, while mutations of KRAS often occur in smoker lung cancer. However, the mutations in either gene in lung adenocarcinomas are rarely seen although the biological consequences of KRAS and EGFR mutations share similarities in regulation of cell proliferation, survival and apoptosis [47], [48]. In the present study, we demonstrate that the frequency of p16INK4α hypermethylation is slightly but significantly higher in smoker patients than that in nonsmoker patients. It is well known that p16INK4α plays an essential role in the development of most human cancers for the reason that the p16/cyclinD1/CDK4/RB signaling pathway controls the cell cycle at the G1/S transition [49]. Hypophosphorylated RB inhibits G1/S transition by binding to E2F1 transcription factor and exerts its tumor-suppressor function. Once hyperphosphorylated by the cyclinD1/CDK4 complex, RB releases E2F1, which results in transition from G1 to S phase. p16 prevents RB from phosphorylation by inhibition of CDK4, leading to a cell cycle arrest. Suppression of p16 expression allows unregulated phosphorylation of the RB protein and leads to uncontrolled cell cycle progression and cell division [50]. p16INK4α has a low frequency of mutations in lung cancer [51]. Its inactivation is mainly through gene promoter hypermethylation [19]. For example, Nakata et al. found that in tumors with p16INK4α hypermethylation, 63.3% showed reduced expression; whereas, in tumors without p16INK4α hypermethylation, only 33.7% showed reduced expression (P = 0.0002) [29]. The positive correlation between cigarette smoking and p16INK4α hypermethylation demonstrates that cigarette smoking plays an important role in determining the molecular signatures involved in lung cancer development.
The mechanism for cigarette smoking inducing gene-specific hypermethylation, e.g. p16INK4α, remains unclear. De novo methylation uses S-adenosyl-methionine as a methyl donor and adds a methyl group to the cytosine ring to form methyl cytosine, which is catalyzed by DNA methyltransferases (DNMT) 1, 3a, or 3b [52]. It is estimated that DNMT1 is responsible for about 90% of methyltransferase activity in mammalian cells [53]. DNMT1 overexpression was found in many types of cancers including lung cancers, particularly in patients who were smokers [54]–[56]. A recent study found that DNMT1 was highly expressed in tumor tissues in a dose response manner compared with the non neoplastic stroma tissues, not only in tobacco-specific carcinogen nicotine-derived nitrosamine ketone (NNK)-induced mouse lung cancer but also in human lung cancer associated with cigarette smoking [57]. Moreover, it was demonstrated that NNK increased DNMT1 expression and activity by blocking its degradation related to ubiquitin-proteasome [57]. AKT/GSK3β/βTrCP signaling is implicated in the accumulation of nuclear DNMT1, which leads to hypermethylation of p16INK4α, fragile histidine triad gene (FHIT) and retinoic acid receptor β (RARB), and ultimately leads to tumorigenesis and poor prognosis [57]. Although the direct interaction of DNMT1 to the p16INK4α gene promoter is not yet characterized [58], these findings indicated that tobacco-induced DNMT1 overexpression might be responsible for maintaining the hypermethylation status of p16INK4α gene.
Lung cancer in nonsmokers is now a prominent public health concern. However, the major causes of them have yet not been identified. Environmental tobacco smoke (ETS), for example, second-hand smoke, has been recognized as a high risk factor [59]–[62]. According to the report from International Agency for Research on Cancer (IARC), the risk for developing lung cancer from ETS exposure might reach 35% in men and 25% in women [63]. Given that cigarette smoking has a cause-effect on p16INK4α hypermethylation, ETS exposure may explain, at least partly, the variable percentage of p16INK4α hypermethylation in nonsmoker patients. Other factors such as exposed to asbestos, chromium, arsenic, cadmium, silica, or nickel, or outdoor air pollutants, previous lung disease, and dietary factors have also been implicated in non smoking-related risk [5], [6]. But so far there is still a missing link between environmental factors, p16INK4α hypermethylation, and lung cancer, which limits the use of gene-specific hypermethylation as a biomarker to detect lung cancer in early stage. Thus, the molecular mechanism underlying lung cancer, irrespective of tobacco-association, should be further elucidated.
In this study, we observed that the frequency of p16INK4α hypermethylation in NSCLC patients varied among different studies. The combined frequency in the present meta-analysis was less than 35%. The discrepancy and the relative low frequency might be due to the method used for detection of methylation, the variation in defining cigarette smokers, and insufficient information of clinical outcome. Although MSP is sufficiently sensitive, the conditions of PCR may affect the results to a large extent. The results seemed to be a little artificial particularly when PCR reaction was performed using both methylated and unmethylated primers. As for definition of cigarette smoker or nonsmoker, it lacked a consistent criterion followed by each investigation. In addition, current smokers and former smokers were not clearly distinguished, and the quantity of smoking was not calculated in the meta-analysis due to limited data. Moreover, insufficient clinical information such as the stage of NSCLC made it difficult to predict the prognosis based on the results provided.
In conclusion, cigarette smoking is suggested to be positively related to p16INK4α methylation in human NSCLC, highlighting the potential importance of p16INK4α promoter methylation in early cancer diagnosis. Furthermore, it is well known that the risk for developing lung cancer in smokers is 8 to 13 times higher than that in nonsmokers, while the risk of p16INK4α hypermethylation in lung cancer patients with smoking habits was only 2.2 times increased than that in nonsmoker patients, we speculate that many other aberrant epigenetic modifications, together with the genetic damage are involved in lung cancer development, which needs to be addressed in further investigation.
Supporting Information
Meta-analysis of cigarette smoking and p16INK4α methylation in noncancerous patients.
(TIF)
Characteristics of studies on the correlations between cigarette smoking and p16INK4α methylation in noncancerous tissue from cancer patients.
(DOC)
Characteristics of studies on the correlation between cigarette smoking and p16INK4α methylation in noncancerous patients.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a Distinguished Young Scholar of National Nature Science Foundation of China (NSFC) (30925029), NSFC (30771832 and 30901211), National Key Basic Research and Development Program (2010CB912803), National High Technology Research and Development Key Program of China (2008AA062504), Ministry of Health of China (200902006), the Fundamental Research Funds for the Central Universities (10ykjc05, 10lgzd10), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme GDUPS (2010). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ministry of Health of the People,s Republic of China. The cause of death in China: the third retrospective sample survey. Beijing: Peking Union Medical College Press; 2008. [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 4.Villeneuve PJ, Mao Y. Lifetime probability of developing lung cancer, by smoking status, Canada. Can J Public Health. 1994;85:385–388. [PubMed] [Google Scholar]
- 5.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers–a different disease. Nat Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 6.Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol. 2007;25:561–570. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- 7.Toh CK, Lim WT. Lung cancer in never-smokers. J Clin Pathol. 2007;60:337–340. doi: 10.1136/jcp.2006.040576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 9.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 10.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 11.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103:153–160. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- 13.Duffy MJ, Napieralski R, Martens JW, Span PN, Spyratos F, et al. Methylated genes as new cancer biomarkers. Eur J Cancer. 2009;45:335–346. doi: 10.1016/j.ejca.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 15.Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C, Soria JC, Tang X, Xu XC, Wang L, et al. Prognostic factors in resected stage I non-small-cell lung cancer: a multivariate analysis of six molecular markers. J Clin Oncol. 2004;22:4575–4583. doi: 10.1200/JCO.2004.01.091. [DOI] [PubMed] [Google Scholar]
- 17.Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 18.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 19.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez-Cespedes M, Decker PA, Doffek KM, Esteller M, Westra WH, et al. Increased loss of chromosome 9p21 but not p16 inactivation in primary non-small cell lung cancer from smokers. Cancer Res. 2001;61:2092–2096. [PubMed] [Google Scholar]
- 21.Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, et al. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]
- 22.Kim DH, Nelson HH, Wiencke JK, Zheng S, Christiani DC, et al. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001;61:3419–3424. [PubMed] [Google Scholar]
- 23.Yanagawa N, Tamura G, Oizumi H, Takahashi N, Shimazaki Y, et al. Frequent epigenetic silencing of the p16 gene in non-small cell lung cancers of tobacco smokers. Jpn J Cancer Res. 2002;93:1107–1113. doi: 10.1111/j.1349-7006.2002.tb01212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanagawa N, Tamura G, Oizumi H, Takahashi N, Shimazaki Y, et al. Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci. 2003;94:589–592. doi: 10.1111/j.1349-7006.2003.tb01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toyooka S, Suzuki M, Tsuda T, Toyooka KO, Maruyama R, et al. Dose effect of smoking on aberrant methylation in non-small cell lung cancers. Int J Cancer. 2004;110:462–464. doi: 10.1002/ijc.20125. [DOI] [PubMed] [Google Scholar]
- 26.Kim JS, Kim H, Shim YM, Han J, Park J, et al. Aberrant methylation of the FHIT gene in chronic smokers with early stage squamous cell carcinoma of the lung. Carcinogenesis. 2004;25:2165–2171. doi: 10.1093/carcin/bgh217. [DOI] [PubMed] [Google Scholar]
- 27.Divine KK, Pulling LC, Marron-Terada PG, Liechty KC, Kang T, et al. Multiplicity of abnormal promoter methylation in lung adenocarcinomas from smokers and never smokers. Int J Cancer. 2005;114:400–405. doi: 10.1002/ijc.20761. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Lan Q, Siegfried JM, Luketich JD, Keohavong P. Aberrant promoter methylation of p16 and MGMT genes in lung tumors from smoking and never-smoking lung cancer patients. Neoplasia. 2006;8:46–51. doi: 10.1593/neo.05586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakata S, Sugio K, Uramoto H, Oyama T, Hanagiri T, et al. The methylation status and protein expression of CDH1, p16(INK4A), and fragile histidine triad in nonsmall cell lung carcinoma: epigenetic silencing, clinical features, and prognostic significance. Cancer. 2006;106:2190–2199. doi: 10.1002/cncr.21870. [DOI] [PubMed] [Google Scholar]
- 30.Toyooka S, Tokumo M, Shigematsu H, Matsuo K, Asano H, et al. Mutational and epigenetic evidence for independent pathways for lung adenocarcinomas arising in smokers and never smokers. Cancer Res. 2006;66:1371–1375. doi: 10.1158/0008-5472.CAN-05-2625. [DOI] [PubMed] [Google Scholar]
- 31.Georgiou E, Valeri R, Tzimagiorgis G, Anzel J, Krikelis D, et al. Aberrant p16 promoter methylation among Greek lung cancer patients and smokers: correlation with smoking. Eur J Cancer Prev. 2007;16:396–402. doi: 10.1097/01.cej.0000236260.26265.d6. [DOI] [PubMed] [Google Scholar]
- 32.Guzman LM, Koriyama C, Akiba S, Eizuru Y, Castillo D, et al. High frequency of p16 promoter methylation in non-small cell lung carcinomas from Chile. Biol Res. 2007;40:365–372. [PubMed] [Google Scholar]
- 33.Kim DS, Cha SI, Lee JH, Lee YM, Choi JE, et al. Aberrant DNA methylation profiles of non-small cell lung cancers in a Korean population. Lung Cancer. 2007;58:1–6. doi: 10.1016/j.lungcan.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Tessema M, Yu YY, Stidley CA, Machida EO, Schuebel KE, et al. Concomitant promoter methylation of multiple genes in lung adenocarcinomas from current, former and never smokers. Carcinogenesis. 2009;30:1132–1138. doi: 10.1093/carcin/bgp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang D, Wang J, Li Y, He Z, Zhang Y. The influence of anthracosis and p16 ink4a gene aberrant methylation on small-sized pulmonary adenocarcinoma. Exp Mol Pathol. 2010;90:131–136. doi: 10.1016/j.yexmp.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Yanagawa N, Tamura G, Oizumi H, Endoh M, Sadahiro M, et al. Inverse correlation between EGFR mutation and FHIT, RASSF1A and RUNX3 methylation in lung adenocarcinoma: relation with smoking status. Anticancer Res. 2011;31:1211–1214. [PubMed] [Google Scholar]
- 37.Zhang CY, Jin YT, Xu HY, Zhang H, Zhang WM, et al. Relationship between promoter methylation of p16, DAPK and RAR beta genes and the clinical data of non-small cell lung cancer. Chin J Med Genet. 2011;28:23–28. doi: 10.3760/cma.j.issn.1003-9406.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Konno S, Morishita Y, Fukasawa M, Shu Y, Wang D, et al. Anthracotic index and DNA methylation status of sputum contents can be used for identifying the population at risk of lung carcinoma. Cancer. 2004;102:348–354. doi: 10.1002/cncr.20643. [DOI] [PubMed] [Google Scholar]
- 39.Peng Z, Shan C, Wang H. [Value of promoter methylation of RASSF1A, p16, and DAPK genes in induced sputum in diagnosing lung cancers]. J Cent South Univ (Med Sci) 2010;35:247–253. doi: 10.3969/j.issn.1672-7347.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Zochbauer-Muller S, Lam S, Toyooka S, Virmani AK, Toyooka KO, et al. Aberrant methylation of multiple genes in the upper aerodigestive tract epithelium of heavy smokers. Int J Cancer. 2003;107:612–616. doi: 10.1002/ijc.11458. [DOI] [PubMed] [Google Scholar]
- 41.Kim H, Kwon YM, Kim JS, Lee H, Park JH, et al. Tumor-specific methylation in bronchial lavage for the early detection of non-small-cell lung cancer. J Clin Oncol. 2004;22:2363–2370. doi: 10.1200/JCO.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 42.Fujiwara K, Fujimoto N, Tabata M, Nishii K, Matsuo K, et al. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin Cancer Res. 2005;11:1219–1225. [PubMed] [Google Scholar]
- 43.de Fraipont F, Moro-Sibilot D, Michelland S, Brambilla E, Brambilla C, et al. Promoter methylation of genes in bronchial lavages: a marker for early diagnosis of primary and relapsing non-small cell lung cancer? Lung Cancer. 2005;50:199–209. doi: 10.1016/j.lungcan.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 44.Belinsky SA, Klinge DM, Dekker JD, Smith MW, Bocklage TJ, et al. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res. 2005;11:6505–6511. doi: 10.1158/1078-0432.CCR-05-0625. [DOI] [PubMed] [Google Scholar]
- 45.Saatci C, Caglayan AO, Ozkul Y, Tahiri S, Turhan AB, et al. Detection of p16 promotor hypermethylation in “Maras powder” and tobacco users. Cancer Epidemiol. 2009;33:47–50. doi: 10.1016/j.canep.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 46.Brait M, Ford JG, Papaiahgari S, Garza MA, Lee JI, et al. Association between lifestyle factors and CpG island methylation in a cancer-free population. Cancer Epidemiol Biomarkers Prev. 2009;18:2984–2991. doi: 10.1158/1055-9965.EPI-08-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 48.Liu G, Zhou W, Christiani DC. Molecular epidemiology of non-small cell lung cancer. Semin Respir Crit Care Med. 2005;26:265–272. doi: 10.1055/s-2005-871983. [DOI] [PubMed] [Google Scholar]
- 49.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 50.Liggett WH, Jr, Sidransky D. Role of the p16 tumor suppressor gene in cancer. J Clin Oncol. 1998;16:1197–1206. doi: 10.1200/JCO.1998.16.3.1197. [DOI] [PubMed] [Google Scholar]
- 51.Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 52.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 53.Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, et al. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 54.Belinsky SA, Nikula KJ, Baylin SB, Issa JP. Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc Natl Acad Sci USA. 1996;93:4045–4050. doi: 10.1073/pnas.93.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT, et al. Alteration of DNA methyltransferases contributes to 5′CpG methylation and poor prognosis in lung cancer. Lung Cancer. 2007;55:205–213. doi: 10.1016/j.lungcan.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 57.Lin RK, Hsieh YS, Lin P, Hsu HS, Chen CY, et al. The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation and tumor suppressor gene hypermethylation in mice and lung cancer patients. J Clin Invest. 2010;120:521–532. doi: 10.1172/JCI40706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato M, Horio Y, Sekido Y, Minna JD, Shimokata K, et al. The expression of DNA methyltransferases and methyl-CpG-binding proteins is not associated with the methylation status of p14(ARF), p16(INK4a) and RASSF1A in human lung cancer cell lines. Oncogene. 2002;21:4822–4829. doi: 10.1038/sj.onc.1205581. [DOI] [PubMed] [Google Scholar]
- 59.Zhong L, Goldberg MS, Parent ME, Hanley JA. Exposure to environmental tobacco smoke and the risk of lung cancer: a meta-analysis. Lung Cancer. 2000;27:3–18. doi: 10.1016/s0169-5002(99)00093-8. [DOI] [PubMed] [Google Scholar]
- 60.Brennan P, Buffler PA, Reynolds P, Wu AH, Wichmann HE, et al. Secondhand smoke exposure in adulthood and risk of lung cancer among never smokers: a pooled analysis of two large studies. Int J Cancer. 2004;109:125–131. doi: 10.1002/ijc.11682. [DOI] [PubMed] [Google Scholar]
- 61.Vineis P, Alavanja M, Buffler P, Fontham E, Franceschi S, et al. Tobacco and cancer: recent epidemiological evidence. J Natl Cancer Inst. 2004;96:99–106. doi: 10.1093/jnci/djh014. [DOI] [PubMed] [Google Scholar]
- 62.Kurahashi N, Inoue M, Liu Y, Iwasaki M, Sasazuki S, et al. Passive smoking and lung cancer in Japanese non-smoking women: a prospective study. Int J Cancer. 2008;122:653–657. doi: 10.1002/ijc.23116. [DOI] [PubMed] [Google Scholar]
- 63.IARC. International Agency for Research on Cancer: Tobacco smoke and involuntary smoking. IARC monographs on the evaluation of the carcinogenic risk of chemicals to humans, vol 83. Lyon: IARC; 2004. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Meta-analysis of cigarette smoking and p16INK4α methylation in noncancerous patients.
(TIF)
Characteristics of studies on the correlations between cigarette smoking and p16INK4α methylation in noncancerous tissue from cancer patients.
(DOC)
Characteristics of studies on the correlation between cigarette smoking and p16INK4α methylation in noncancerous patients.
(DOC)