Abstract
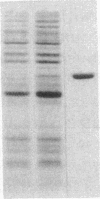
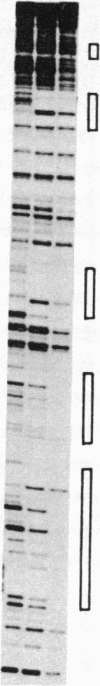
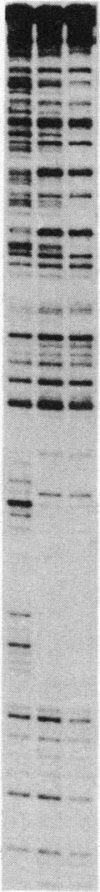
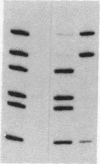
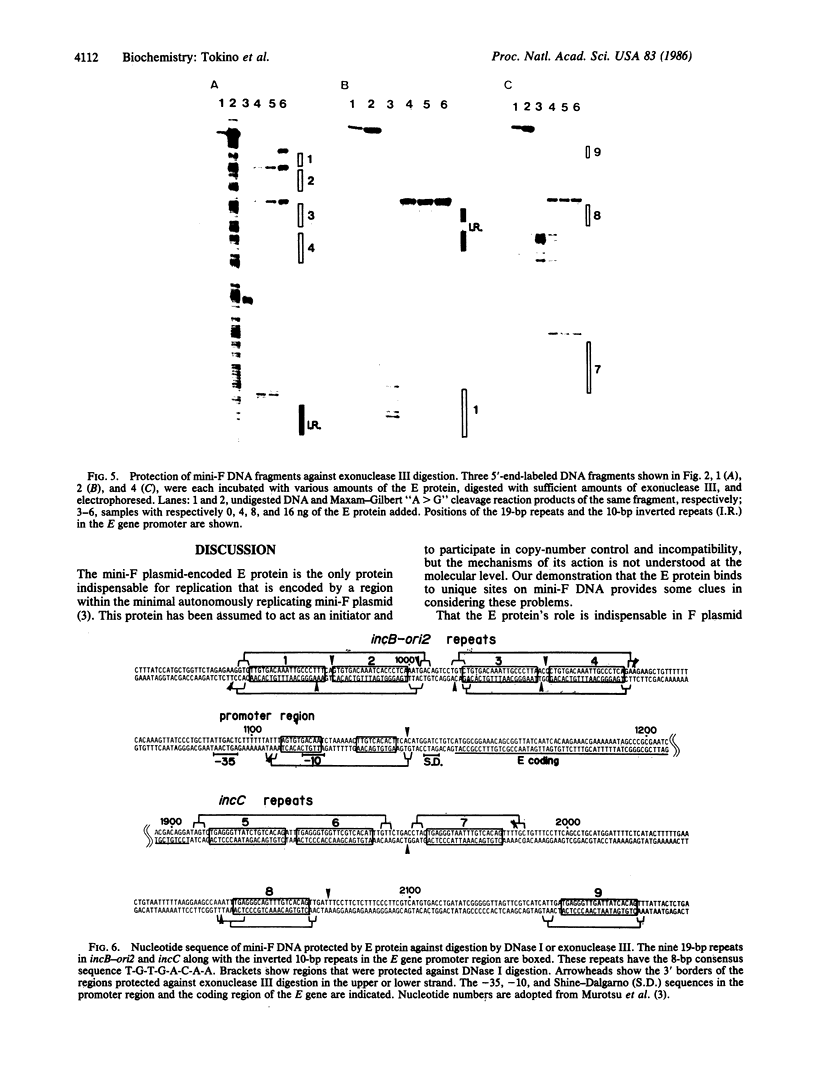
Mini-F plasmid encodes a protein, E protein, that is indispensable for its autonomous replication. We have constructed a plasmid that overproduces the E protein and have purified the protein to apparent homogeneity. Using nitrocellulose filter binding and nuclease digestion assays, we demonstrated that the E protein binds to three unique regions of the mini-F DNA sequence: the replication origin (ori2) and an incompatibility locus (incB), another incompatibility locus (incC), and the promoter for the E gene. These binding sites have a common 8-base-pair sequence. These findings suggest the direct role of the E protein in initiation of mini-F replication and copy number control. They are also in line with the in vivo evidence that the incompatibility phenotype caused by incB and incC DNA is due to titration of a factor(s) indispensable for replication and that the production of the E initiator protein of the mini-F plasmid is under autoregulatory control.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Denniston-Thompson K., Moore D. D., Kruger K. E., Furth M. E., Blattner F. R. Physical structure of the replication origin of bacteriophage lambda. Science. 1977 Dec 9;198(4321):1051–1056. doi: 10.1126/science.929187. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germino J., Bastia D. Interaction of the plasmid R6K-encoded replication initiator protein with its binding sites on DNA. Cell. 1983 Aug;34(1):125–134. doi: 10.1016/0092-8674(83)90142-3. [DOI] [PubMed] [Google Scholar]
- Gottesman M. E., Yarmolinsky M. B. Integration-negative mutants of bacteriophage lambda. J Mol Biol. 1968 Feb 14;31(3):487–505. doi: 10.1016/0022-2836(68)90423-3. [DOI] [PubMed] [Google Scholar]
- Hobom G., Grosschedl R., Lusky M., Scherer G., Schwarz E., Kössel H. Functional analysis of the replicator structure of lambdoid bacteriophage DNAs. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):165–178. doi: 10.1101/sqb.1979.043.01.023. [DOI] [PubMed] [Google Scholar]
- Kline B. C., Trawick J. Identification and characterization of a second copy number control gene in mini-F plasmids. Mol Gen Genet. 1983;192(3):408–415. doi: 10.1007/BF00392183. [DOI] [PubMed] [Google Scholar]
- Komai N., Nishizawa T., Hayakawa Y., Murotsu T., Matsubara K. Detection and mapping of six miniF-encoded proteins by cloning analysis of dissected miniF segments. Mol Gen Genet. 1982;186(2):193–203. doi: 10.1007/BF00331850. [DOI] [PubMed] [Google Scholar]
- Lovett M. A., Helinski D. R. Method for the isolation of the replication region of a bacterial replicon: construction of a mini-F'kn plasmid. J Bacteriol. 1976 Aug;127(2):982–987. doi: 10.1128/jb.127.2.982-987.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki S., Miki T., Horiuchi T. DNA sequence of an amber replication mutant indicates that a 29 kd protein is the product of the F plasmid replication gene. Mol Gen Genet. 1984;194(1-2):337–339. doi: 10.1007/BF00383537. [DOI] [PubMed] [Google Scholar]
- Matsubara K. Preparation of plasmid lambda dv from bacteriophage lambda: role of promoter-operator in the plasmid replicon. J Virol. 1974 Mar;13(3):596–602. doi: 10.1128/jvi.13.3.596-602.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K. Replication control system in lambda dv. Plasmid. 1981 Jan;5(1):32–52. doi: 10.1016/0147-619x(81)90076-7. [DOI] [PubMed] [Google Scholar]
- Murotsu T., Tsutsui H., Matsubara K. Identification of the minimal essential region for the replication origin of miniF plasmid. Mol Gen Genet. 1984;196(2):373–378. doi: 10.1007/BF00328075. [DOI] [PubMed] [Google Scholar]
- Shalloway D., Kleinberger T., Livingston D. M. Mapping of SV40 DNA replication origin region binding sites for the SV40 T antigen by protection against exonuclease III digestion. Cell. 1980 Jun;20(2):411–422. doi: 10.1016/0092-8674(80)90627-3. [DOI] [PubMed] [Google Scholar]
- Shirakawa M., Tsurimoto T., Matsubara K. Plasmid vectors designed for high-efficiency expression controlled by the portable recA promoter-operator of Escherichia coli. Gene. 1984 Apr;28(1):127–132. doi: 10.1016/0378-1119(84)90096-9. [DOI] [PubMed] [Google Scholar]
- Søgaard-Andersen L., Rokeach L. A., Molin S. Regulated expression of a gene important for replication of plasmid F in E. coli. EMBO J. 1984 Feb;3(2):257–262. doi: 10.1002/j.1460-2075.1984.tb01794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis K., Cabello F., Cohen S. N. Cloning, isolation, and characterization of replication regions of complex plasmid genomes. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2242–2246. doi: 10.1073/pnas.72.6.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolun A., Helinski D. R. Direct repeats of the F plasmid incC region express F incompatibility. Cell. 1981 Jun;24(3):687–694. doi: 10.1016/0092-8674(81)90095-7. [DOI] [PubMed] [Google Scholar]
- Tolun A., Helinski D. R. Separation of the minimal replication region of the F plasmid into a replication origin segment and a trans-acting segment. Mol Gen Genet. 1982;186(3):372–377. doi: 10.1007/BF00729456. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Hase T., Matsubara H., Matsubara K. Bacteriophage lambda initiators: preparation from a strain that overproduces the O and P proteins. Mol Gen Genet. 1982;187(1):79–86. doi: 10.1007/BF00384387. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Matsubara K. Purification of bacteriophage lambda O protein that specifically binds to the origin of replication. Mol Gen Genet. 1981;181(3):325–331. doi: 10.1007/BF00425606. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Matsubara K. Purified bacteriophage lambda O protein binds to four repeating sequences at the lambda replication origin. Nucleic Acids Res. 1981 Apr 24;9(8):1789–1799. doi: 10.1093/nar/9.8.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H., Fujiyama A., Murotsu T., Matsubara K. Role of nine repeating sequences of the mini-F genome for expression of F-specific incompatibility phenotype and copy number control. J Bacteriol. 1983 Jul;155(1):337–344. doi: 10.1128/jb.155.1.337-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocke C., Bastia D. DNA-protein interaction at the origin of DNA replication of the plasmid pSC101. Cell. 1983 Dec;35(2 Pt 1):495–502. doi: 10.1016/0092-8674(83)90183-6. [DOI] [PubMed] [Google Scholar]
- Watson L. A., Phua S. H., Bergquist P. L., Lane H. E. An Mr 29000 protein is essential for mini-F maintenance in E. coli. Gene. 1982 Sep;19(2):173–178. doi: 10.1016/0378-1119(82)90003-8. [DOI] [PubMed] [Google Scholar]