Abstract
Methylation of arginine residues, catalyzed by protein arginine methyltransferases (PRMTs), is one important protein post-translational modification involved in epigenetic regulation of gene expression. A fast and effective assay for PRMT can provide valuable information for dissecting the biological functions of PRMTs, as well as for screening small-molecule inhibitors of arginine methylation. Currently, among the methods used for PRMT activity measurement, many contain laborious separation procedures, which restrict the applications of these assays for high-throughput screening (HTS) in drug discovery. The authors report here a mix-and-measure method to measure PRMT activity based on the principle of scintillation proximity assay (SPA). In this assay, 3H-AdoMet was used as methyl donor, and biotin-modified histone H4 peptide served as a methylation substrate. Following the methylation reaction catalyzed by PRMTs, streptavidin-coated SPA beads were added to the reaction solution, and SPA signals were detected by a MicroBeta scintillation counter. No separation step is needed, which simplifies the assay procedure and greatly enhances the assay speed. Particularly, the miniaturization and robustness suggest that this method is suited for HTS of PRMT inhibitors.
Keywords: protein arginine methyltransferases, PRMT, scintillation proximity assay, SPA, high-throughput screening, HTS
Introduction
Chromatin-modifying enzymes are of great significance for the epigenetic regulation of numerous DNA-based processes, such as DNA transcription, replication, and damage repair.1 Protein arginine methyltransferase (PRMT) is one type of such an important chromatin-modifying enzyme that transfers the methyl group from S-adenosyl-L-methionine (AdoMet, SAM) to their protein substrates, including nucleosomal core histones, and releases S-adenosyl-L-homocysteine (AdoHcy, SAH) as a side product. Among PRMTs, type I PRMTs (e.g., PRMT1) catalyze asymmetric dimethylation of arginine residues, and type II PRMTs (e.g., PRMT5) catalyze symmetric dimethylation. On the chromatin template, both PRMT1 and PRMT5 target arginine 3 of histone H4 (i.e., H4R3) and alter the status of transcription at target genes.2,3 Several reports show that the expression levels of PRMTs are deregulated in certain diseases. PRMT1 is upregulated in breast cancer concomitantly with a change in substrate methylation.4 PRMT1 variant 2 has been regarded as a marker of unfavorable prognosis in colon cancer patients.5 Furthermore, PRMT1 was found to be an essential element in the oncogenic mixed-lineage leukemia fusion complexes and confers an aberrant transcriptional activation property critical for the induction of leukemia.6 PRMT5, a representative type II enzyme, was also shown to play strong oncogenic roles.7 Overexpression of PRMT5 increases transformation of NIH3T3 cells, suggesting its function in promoting tumor progression.3 PRMT5 protein expression has been induced in a wide variety of human lymphoid cancer cell lines, including patient-derived chronic myelogenous leukemia (MCL) cell lines as well as MCL clinical samples.8 The multiple lines of evidence point toward that targeting PRMT1 and PRMT5 activities with small-molecule inhibitors may be an attractive therapeutic strategy in cancer chemotherapy.
Discovery of PRMT inhibitors requires efficient enzymatic assays. Currently, several methods have been reported to measure methyltransferase activities. For example, radioisotope-labeled assays have been widely used to study the enzyme kinetics of PRMTs,9 but the assay protocols have to involve postreaction separation procedures (e.g., chromatography, sodium dodecyl sulfate–polyacrylamide gel electrophoresis [SDS-PAGE], filter binding), which are labor cumbersome and restrict their application in a high-throughput format. Several groups have developed protocols to monitor the side product AdoHcy as a way of measuring the activities of PRMTs and the other types of methyltransferases.10,11 In particular, AdoHcy can be converted to homocysteine by AdoHcy hydrolases, releasing its free thiol group that can react with sulfhydryl-sensitive dyes for fluorometric or colorimetric detection.10,12,13 The methyltransferase activity of PRMTs can also be detected by the competitive fluorescence polarization immunoassay14 and enzyme-coupled luciferase assay.15 In general, these nonradioactive coupled assays overcome certain shortcomings of the radioisotope labeled methods, but the involvement of additional components could complicate the assay conditions and may produce false positives in inhibitor screening.
We are particularly interested in developing methyltransferase assay strategies involving minimal ingredients to avoid potential complexity and ambiguity. For example, we recently designed a single-step fluorescent assay for sensitive measurement of PRMT1-mediated methylation.16 Herein, we describe a scintillation proximity assay (SPA) for the detection of PRMT activities with the aim of obtaining improved assay efficacy and throughput. In this strategy, a biotin-labeled peptide was used as the substrate of PRMT1 and PRMT5. PRMTs catalyze the transfer of the 3H-methyl group from 3H-SAM to the biotin–peptide. The methylated product is caught by streptavidin-coated SPA beads that contain scintillator molecules. The interaction between the product and the SPA beads results in the transfer of energy from tritium to scintillator, which leads to emission of luminescent signals that can be measured and quantified on a MicroBeta counter (PerkinElmer, Waltham, MA). We characterized in detail the efficacy and robustness of this method for PRMT activity measurement. The results suggest that this protocol has great application for high-throughput screening (HTS) of PRMT inhibitors.
Materials and Methods
Protein expression and purification
Recombinant His6x-tagged rat PRMT1 protein was expressed using a pET28b vector in Escherichia coli BL21(DE3) (Stratagene, La Jolla, CA) and purified on nickel–NTA affinity beads (Novagen, Madison, WI) as previously described.9 Protein concentrations were determined with a Bradford assay. Human PRMT5 protein was purchased from BPS Bioscience, Inc. (San Diego, CA).
Synthesis of the biotin-labeled H4 peptide
Solid-phase peptide synthesis (SPPS) of a biotinylated peptide containing the N-terminal 20–amino acid sequence of histone H4—that is, Ac-SGRGKGGKGLGKGGAKRHRK(Biotin)-NH2 (abbreviated as H4-BTN)—was performed on a PS3 peptide synthesizer (Protein Technologies), using an N-(9-fluorenyl) methoxycarbonyl (Fmoc) strategy. Fmoc-protected amino acids and rink amide resin were purchased from NovaBiochem (San Diego, CA). Other chemicals were purchased from Sigma-Aldrich (St. Louis, MO), unless indicated otherwise. All the synthetic reactions were performed at room temperature. For the coupling of each amino acid (AA), 4 equivalents (eq) of AA/O-benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluorophosphate (HBTU)/N-Hydroxybenzotriazole (HOBt) were used. N-methylmorpholine (NMM) was used as the catalytic base. Removal of Fmoc was performed with 20% v/v piperidine/dimethylformamide (DMF). The N-terminal amino group was acetylated with Ac2O unless indicated otherwise. The dimethyl-dioxocyclohexylidene (Dde) group on the lysine residue was removed with 2% hydrazine in DMF, and the free amino group was then reacted with biotin–NHS (Fluka, Milwaukee, WI) for biotin labeling. After the solid-phase synthesis, the resins were subsequently washed with DMF and dichloromethane and then dried in vacuum for at least 2 h before cleavage. Peptides were cleaved from the resin by treatment with 95% trifluoroacetic acid (TFA), 2.5% H2O, and 2.5% tri-isopropylsilane (TIS) (handling of TFA must be performed in a secure hood) for 3 to 4 h. Cold ether was used to precipitate the products. Crude products were collected by centrifugation and then washed with cold diethyl ether. After lyophilization, the compounds were redissolved in water and purified with reverse-phased (RP) high-performance liquid chromatography (HPLC; C18, Varian, Palo Alto, CA) on a Varian Prostar HPLC system using linear gradients of H2O/0.05% TFA (solvent A) versus acetonitrile/0.05% TFA (solvent B). Analytical HPLC and matrix-assisted laser desorption ionization–mass spectrometry (MALDI-MS) were used for characterization. The calculated MW for H4-BTN is 2260.3, and the mass detected by MALDI-MS is 2259.4.
Measurement of arginine methylation with SPA
PRMT assays were carried out in a 96-well plate (Isolate-96; PerkinElmer) at room temperature, and the reaction buffer contained 50 mM HEPES (pH 8.0), 1 mM EDTA, 50 mM NaCl, and 0.5 mM dithiothreitol (DTT). 3H-SAM (PerkinElmer; 10 or 15 Ci/mmol) was used as the methyl donor, and H4-BTN was employed as the substrate of both PRMT1 and PRMT5. Typically, 10 μL H4-BTN, 10 μL 3H-SAM, and 10 μL PRMT at different concentrations were mixed in a final volume of 30 μL in each well and then were incubated at room temperature for different periods. The reaction was quenched by dilution with 150 μL of the reaction buffer. Streptavidin-coated SPA beads (GE Healthcare, Piscataway, NJ) were weighed and suspended in the reaction buffer by vortexing. Then, 10 μL of the bead suspension was added to each quenched reaction mixture for scintillation measurement. Concentrations of each reagent were varied in different experiments. Sample-loaded plates were scanned at a speed of one well per minute on a MicroBeta2 (PerkinElmer). Each measurement was conducted in duplicates or triplicates.
Filter binding assay of PRMT reaction
The PRMT activities were also measured using 3H and 14C radioisotope-labeled methylation assays. The assays were carried out in 0.6-mL plastic tubes at room temperature in a reaction volume of 30 μL. The reaction buffer contained 50 mM HEPES (pH 8.0), 0.5 mM DTT, 1 mM EDTA, and 50 mM NaCl. In a regular procedure, peptide substrate, 3H-AdoMet (10 Ci/mmol; PerkinElmer), 14C-AdoMet (48.8 mCi/mmol; PerkinElmer), or varied concentrations of an inhibitor were preincubated in the reaction buffer for 5 min prior to the initiation by the addition of PRMT. After it was incubated for an appropriate period of time, the reaction was quenched by spreading the reaction mixture onto anionic P81 filter paper disks (Whatman, Maidstone, Kent, UK). The paper disks were washed with 50 mM NaHCO3 (pH 9.0) and dried in air for 6 h. The amounts of methylated products were quantified by liquid scintillation on an LS 6500 multipurpose scintillation counter (Beckman Coulter, Brea, CA).
Results and Discussion
Design of SPA for arginine methylation
In arginine methylation, the co-factor SAM serves as a methyl group donor, and methylated protein or peptide and AdoHcy are products in the end. Therefore, SAM compound containing radioactive isotope labels (e.g., 3H or 14C) on the methyl group can be conveniently used for the detection of methyltransferase activity. However, in standard radioactive methylation assays, a separation step is required to separate the methylated protein or peptide product from unreacted AdoMet so that the amount of the radioactive methyl groups on the product can be quantitated by either liquid scintillation or fluorography. Typically, the separation process takes longer than the methylation reaction itself, posing a technical limitation to the throughput of the assays. We reasoned that these limitations of regular radioisotope-labeled assays could be overcome by modulating the assay protocol into the format of the SPA. 3H-labeled SAM is widely used for methylation research, and the pathway of β emission from 3H has an average value of about 1.5 μm, which is ideal for the SPA design.17 Therefore, applying SPA for PRMT activity measurement is an attractive approach. To test the suitability of this method for arginine methylation, we herein investigated the SPA strategy for the measurement of the enzymatic activities of PRMTs. A histone H4 peptide with biotin attached (i.e., H4-BTN) was synthesized as the methyl acceptor. The N-terminal H4 peptide is a common substrate of PRMT1 and PRMT5, with methylation at the arginine-3 site, and thus can be used to study the methyltransferase activities of both type I and type II PRMT enzymes. The kinetic parameters of PRMT catalysis with H4-BTN were measured for both PRMT1 (kcat = 1.1 ± 0.05 min−1 and Km = 0.31 ± 0.03 μM) and PRMT5 (kcat = 0.065 ± 0.006 min−1 and Km = 0.24 ± 0.09 μM). Clearly, the activity of PRMT5 is much lower than PRMT1. To conduct the SPA assay, the commercially available SPA beads coated with streptavidin were used for binding to methylated H4-BTN. Through the biotin–streptavidin interaction, the 3H-radiolabeled methyl group is brought into close proximity to the scintillator molecules of the SPA beads. The beta-particle emission of 3H will excite the scintillator encapsulated in SPA beads to produce luminescence that can be detected by a scintillation counter (e.g., the MicroBeta [PerkinElmer]). Any unbound 3H-labeled SAM is out of the range of scintillation proximity distance and thus will not generate SPA signals.
Characterization and optimization of the assay
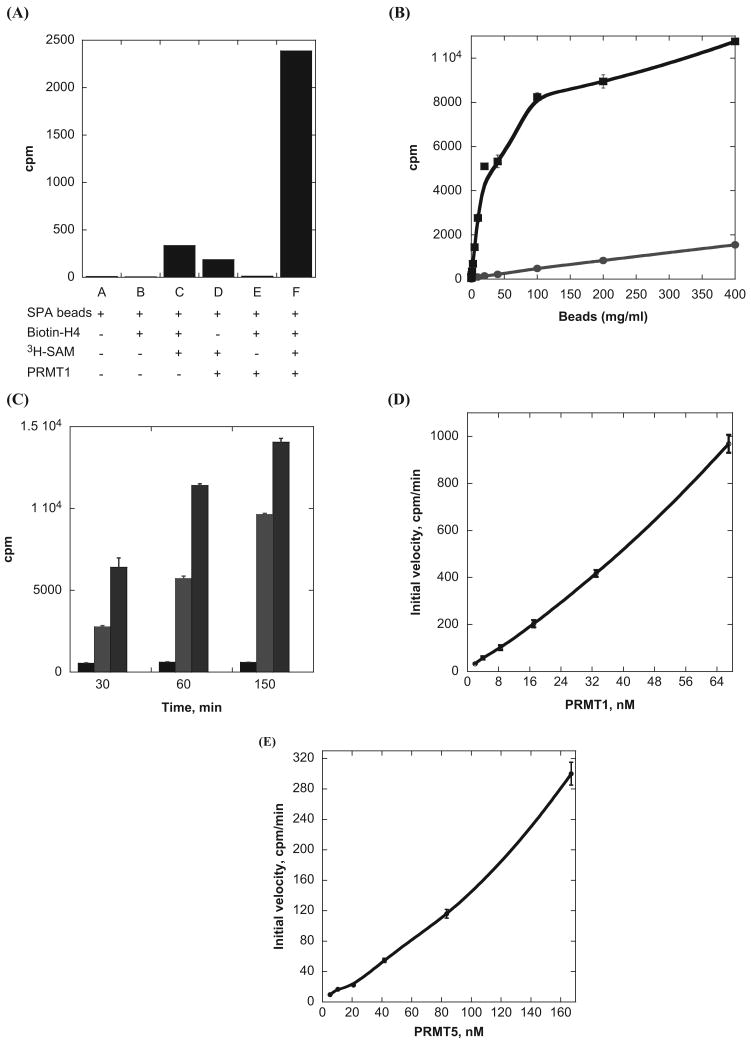
We first measured the SPA signals under different conditions to determine whether the SPA method is suited for PRMT activity detection. Assays were carried out in a 96-well plate. A reaction mixture containing 5 μM H4-BTN, 2 μM 3H-SAM, and 10 nM PRMT1 in a volume of 30 μL was made in each well and incubated at room temperature for 10 min. The reaction was quenched by dilution with 150 μL of the reaction buffer. Then 10 μL of streptavidin–SPA beads (20 mg/mL, suspended in the reaction buffer) was added to the mixture for scintillation measurement. The plate was scanned on a PerkinElmer MicroBeta2 instrument. As expected, the counts from the samples without 3H-SAM (e.g., lanes A, B, and E) were essentially nil because no radioactive materials were present (Fig. 1A). The mixtures containing 3H-SAM/H4-BTN/SPA beads or 3H-SAM/PRMT1/SPA beads produced signals at 350 and 200 cpm. These signals were likely brought about by nonspecific interactions or stochastic collisions of 3H-SAM with the SPA beads, a manifestation of the nonproximity effect (NPE).18 Signals from sample C, which lacked the PRMT1 enzyme, could be applied as the background control for the arginine methylation reaction. Strikingly, the reaction mixture containing full ingredients (PRMT1, H4-BTN, 3H-SAM, and SPA beads) produced the highest signal (lane F), and the signal ratio between samples F and C was about 10-fold. Such a large difference clearly demonstrates that the SPA signals resulted from arginine methylation of H4-BTN catalyzed by PRMT1.
Fig. 1.
Examination of the scintillation proximity assay (SPA) signals under different reaction conditions. (A) Comparison of cpm values from the samples containing different components. The reaction mixture contained 2 μM 3H-SAM, 10 nM PRMT1, and 5 μM H4-BTN. All samples were incubated 10 min, followed by dilution with the reaction buffer and addition of 10 μL of 20 mg/mL SPA beads. Lane A, only SPA beads; lane B, SPA beads and H4-BTN; lane C, beads, H4-BTN, and 3H-SAM; lane D, beads, PRMT1, and 3H-SAM; lane E, beads, PRMT1, and H4-BTN; and lane F, beads, PRMT1, and H4-BTN, and 3H-SAM. (B) Effect of the amounts of SPA beads on the SPA signals of the methylation reaction. The reaction samples contained 0.9 μM 3H-SAM, 37 nM PRMT1, and 2.5 μM H4-BTN. The blue curve is for the negative control: no PRMT1 was present. (C) The methyltransferase activity of PRMT1 and PRMT5 detected by the SPA. The reaction sample contained 1.5 μM 3H-SAM, 5 nM PRMT1 or 110 nM PRMT5, and 2 μM H4-BTN; 10 μL of 40 mg/mL SPA beads was added for scintillation counting. Black: background; blue: PRMT1 reaction; green: PRMT5 reaction. (D) Plot of initial velocity versus PRMT1 concentration. The reaction mixture contained 0.9 μM 3H-SAM and 2.5 μM H4-BTN, and 10 μL of 20 mg/mL SPA beads was added for scintillation counting. (E) Plot of initial velocity versus PRMT1 concentration. The reaction mixture contained 0.9 μM 3H-SAM and 2.5 μM H4-BTN, and 10 μL of 20 mg/mL SPA beads was added for scintillation counting.
Next, we examined how the amounts of SPA beads affect the scintillation output of the SPA measurement. A methylation reaction sample was prepared containing 0.9 μM 3H-SAM, 37 nM PRMT1, and 2.5 μM H4-BTN and was incubated for 20 min at room temperature. Following a dilution with the reaction buffer, 10 μL of SPA beads at different concentrations, ranging from 0 to 400 mg/mL, was added to the reaction mixture and scintillation counts were measured. As shown in Figure 1B, the cpm values quickly increased as more SPA beads were added. This reflected that increasing numbers of methylated H4-BTN molecules bound to streptavidin-coated SPA beads, proportional to the level of SPA readout. At 100 mg/mL or higher concentrations of SPA beads, the rate of cpm growth slowed, and a plateau was reached. Likely, at this stage, all H4-BTN molecules were bound by SPA beads, and thus the addition of more SPA beads had minor or no effect on signal readouts. A control experiment was also performed, which contained 3H-SAM, H4-BTN, and SPA beads, with the purpose of testing how the background counts changed as the amounts of SPA beads increased. The data showed a gradual increase of the background signal with growing amounts of SPA beads. As mentioned above, this is likely caused by NPE18,19—namely, excitation of the SPA beads by unbound 3H-SAM in the reaction mixture contributed to the observed background signals. To minimize the NPE effect, we applied a dilution step after each methylation reaction. The sixfold dilution with the reaction buffer not only reduced the background but also sharply decreased the rate of arginine methylation. Overall, the signal to background (S/B) performance was excellent in a wide range of concentrations of SPA beads. In practical assays, it is not obligatory to select nonlimiting amounts of SPA beads for maximum signal readout. The high cost of SPA beads is a limiting factor for consideration. In addition, it is not always beneficial to choose high bead amounts. As the concentration of the beads increased, the SPA signal went higher, but the background counts also went higher because of NPE. We found that an optimal S/B ratio of 35 was reached at ∼20 mg/mL of bead concentration.
We further tested whether the SPA can be used for detecting the activity of type II PRMTs (e.g., PRMT5). Because H4 is a common substrate of PRMT1 and PRMT5, we directly employed H4-BTN to analyze PRMT5 activity with the SPA method. Methyltransferase activities at three selected time points, 30, 60, and 150 min, were measured for both PRMT1 and PRMT5. Concentration of PRMT5 (110 nM) used was higher than that of PRMT1 (5 nM) because the former one has weaker specific activity. As seen in Figure 1C, the SPA signals generated from both the PRMT1 and PRMT5 reaction mixtures increased significantly as a function of time, supporting that SPA signals resulted from the enzymatic activities of the two proteins. In comparison with the reaction samples, the background counts from the control sample that lacked enzymes were significantly lower and did not increase over incubation time. These data demonstrate that the SPA is an efficacious method for measuring activities of both type I and type II PRMTs. We also measured the relationship between the rates of H4-BTN methylation and the concentration of PRMT (Fig. 1D,E). The activity of PRMT1 and PRMT5 was not exactly linear with respect to their concentration. This biochemical feature is unique for PRMT catalysis, likely reflective of the effect of protein oligomerization.
Correlation between the SPA and the filter binding assay
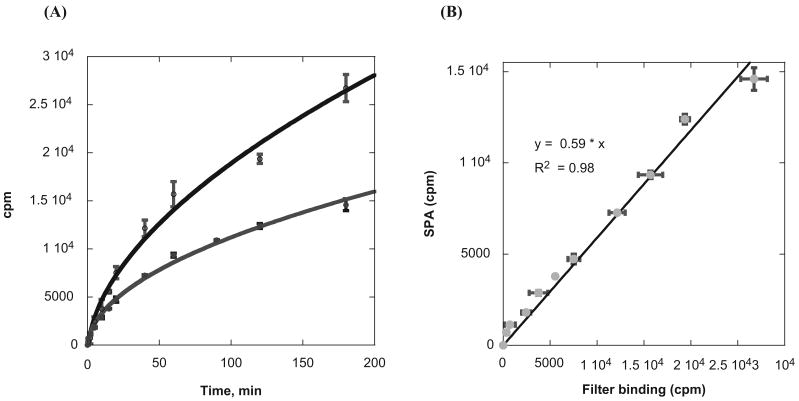
It is of interest to investigate whether the activity of PRMTs measured with the SPA method correlates well with that of the standard radioisotope-labeled assays. Because the scintillation counts measured in the SPA depend on the energy transfer efficiency between the 3H isotope and the scintillator molecules encapsulated in the SPA beads, technically the cpm data from the SPA will be smaller than that of a regular liquid scintillation method. In general, streptavidin-coated SPA beads will give approximately 30% to 45% of the cpm reading compared with the conventional liquid scintillation counting (SPA bead manual; GE Healthcare). To assess the quantitative relationship between the SPA and the regular liquid scintillation method for PRMT activity measurement, we compared the cpm counts obtained with the two methods under the same experimental condition: 0.8 μM 3H-SAM, 10 nM PRMT1, 1 μM H4-BTN, and 40 mg/mL SPA beads (for the SPA). From the reaction course curve, the scintillation counts of both the filter binding assay and the SPA increased with the reaction time (Fig. 2A). The shapes of both curves were similar, and the cpm signals of the filter binding assay were higher than the SPA. The curve displayed an initial linear phase in the first 10 to 15 min and then extended to an asymptotic plateau phase. The cpm value of the SPA was about half of that of the filter binding assay at the reaction time of 180 min. We plotted the SPA and the filter binding assay data in one chart with the individual counts from the filter binding assay along the x-axis and the SPA counts on the y-axis (Fig. 2B). The two sets of data show a linear relationship (the correlation coefficient is 0.99), demonstrating that the SPA data and the filter binding assay data are well correlated. The obtained correlation equation (Y = 0.59 × X) can be used as the conversion factor to transform the data from the SPA measurement to equivalent values of the filter binding assay and vice versa.
Fig. 2.
Correlation of the scintillation proximity assay (SPA) with the standard filter binding assay. (A) Time course of H4-BTN methylation measured by the SPA (blue) and the filter binding assay (black). The reaction mixture contained 0.8 μM 3H-SAM, 10 nM PRMT1, and 1 μM H4-BTN. In the SPA measurement, 10 μL of 40 mg/mL SPA beads was used. (B) Correlation of the data of the SPA with that of the filter binding assay. The cpm of the filter binding assay is on the x-axis and the cpm of SPA is on the y-axis.
Evaluation of PRMT inhibitors with the SPA
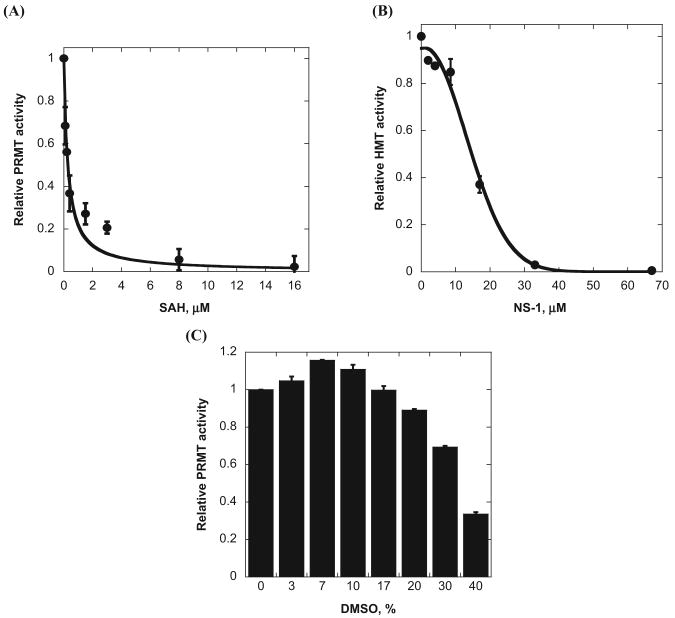
We next tested the efficacy of the SPA for studying PRMT inhibitors. The activity of PRMT1 was measured in a range of concentrations of SAH, a feedback inhibitor of PRMTs, and NS-1, which is a previously reported arginine methylation inhibitor.9 The reaction was initiated with the addition of 37 nM PRMT1 after the other components (0.9 μM 3H-SAM, 2.5 μM H4-BTN, and the inhibitor) were premixed and equilibrated for 5 min. At the end of the methylation reaction, 20 mg/mL of beads was added for the SPA measurement. The SPA signals of the reaction mixture decreased as the concentrations of SAH and NS-1 increased, consistent with enhanced inhibition by the compounds (Fig. 3A,B). The concentration-dependent inhibition data yielded an IC50 of 0.3 μM for SAH and 14.7 μM for NS-1, which matched the values tested by the filter binding assay (i.e., 0.4 μM for sah and 12.7 μM for NS-1).9 Together, these data support that the SPA method is well suited for the study of arginine methylation inhibitors.
Fig. 3.
Use of the scintillation proximity assay (SPA) for type I protein arginine methyltransferase (PRMT1) inhibitor characterization. (A) Concentration-dependent inhibition of PRMT1 activity by S-adenosyl-L-homocysteine (SAH). (B) Concentration-dependent inhibition of PRMT1 activity by NS-1. (C) Effect of DMSO on PRMT1 activity. Each reaction mixture contained 0.9 μM 3H-SAM, 37 nM PRMT1, and 2.5 μM H4-BTN. Then, 20 mg/mL of beads was added after the reaction.
Organic solvents such as DMSO, acetonitrile, and 1-methyl-2-pyrrolidinone (NMP; e.g., 1%–5% v/v) are commonly used to dissolve drug candidates prior to HTS. We tested the tolerance of the SPA to different concentrations of DMSO. As seen in Figure 3C, the response from the SPA beads remained at high levels in the range of 0% to 17% (v/v) and only began to decrease when DMSO concentration reached >10%. The decrease in the SPA counts at high DMSO concentrations could be caused by the toxicity of DMSO to either the intrinsic activity of PRMT1 or the scintillation proximity detection. In a parallel study, we determined the DMSO effect with the filter binding method, and very similar results were obtained (data not shown). Thus, the gradual diminishing of the SPA readouts at higher concentrations of DMSO was due to its inhibitory effect on PRMT1 activity, likely by altering the structure of the enzyme active site. Overall, the SPA method tolerates well the presence of DMSO, and PRMT1 retains its high activity up to 17% DMSO, providing a wide range of choice for using this solvent in inhibitor discovery.
Evaluation of the SPA for screening PRMT inhibitors
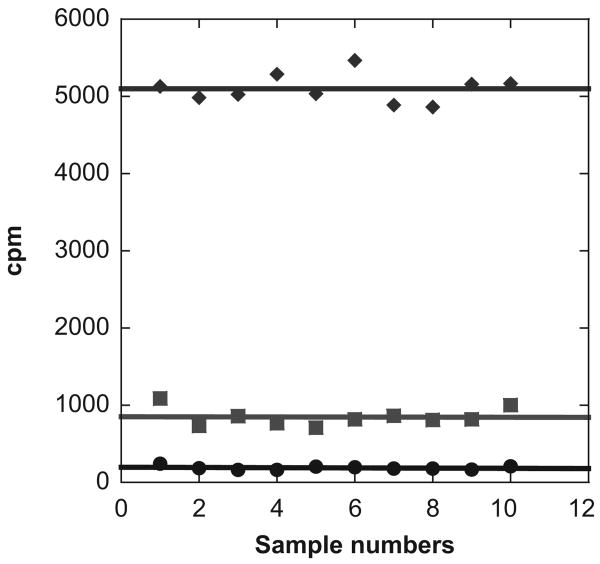
A particular advantage of the SPA for PRMT activity measurement is no need for postreaction separation procedures, which makes it an attractive approach for screening arginine methylation inhibitors in the HTS format. We tested the suitability of the SPA assay for HTS by measuring the Z and Z′ factors, which are statistical parameters for the evaluation of the qualification and robustness of HTS assays.20 The experiments were carried out in a 96-well plate format. Three sets of samples were prepared: The first one was for the positive control (containing PRMT1, 3H-SAM, and H4-BTN), the second one was for the negative control (containing 3H-methyl-SAM and H4-BTN), and the last set included the inhibition mixtures (containing PRMT1, 3H-SAM, H4-BTN, and SAH). Twelve data points were collected for each set of samples (Fig. 4) and used for calculation of statistic parameters Z and Z′. In practice, the values of Z or Z′ factor range from −∞ to 1, and a qualified and robust assay typically has Z or Z′ values higher than 0.5.20 Calculation from the data of the sampling measurements offers a Z value of 0.65 and a Z′ value of 0.80, which validates the robustness of the SPA for arginine methylation study. Together, our data support that the SPA method is of great suitability for screening small-molecule inhibitors of PRMTs in the HTS format.
Fig. 4.
A scatter plot of the scintillation proximity assay (SPA) signals displaying the levels of methyltransferase activities of RPMT1 compared with the background signal level. Green line: the positive controls containing 0.9 μM 3H-SAM, 37 nM PRMT1, and 2.5 μM H4-BTN. Black line: the negative controls containing 0.9 μM 3H-SAM and 2.5 μM H4-BTN. Blue line: the inhibitor samples containing 0.9 μM 3H-SAM, 37 nM PRMT1, 2.5 μM H4-BTN, and 3 μM SAH. All reaction samples were incubated for 20 min, and following the reaction, 10 μL of 20 mg/mL SPA beads was added to each sample for scintillation counting.
In conclusion, a sensitive SPA method has been developed and evaluated for the measurement of PRMT-mediated arginine methylation. High S/B ratios can be reached to 35 under the optimized reaction conditions. In comparison with the classic radioactive assay, the mix-and-measure SPA approach does not require an extra separation procedure for product isolation and has the advantages of producing low volumes of radioactive wastes, generating results at a faster assay speed, and being less labor intensive. The robustness of the assay suggests that the SPA approach is readily adaptable to the HTS format for the discovery of PRMT modulators.
Acknowledgments
This work is supported in part by AHA grant 09BGIA2220207 and NIH grant R01GM086717. JW is supported by a Molecular Basis of Disease Fellowship at Georgia State University.
References
- 1.Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al. Methylation of Histone H4 at Arginine 3 Facilitating Transcriptional Activation by Nuclear Hormone Receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 3.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-Associated PRMT5 Methylates Histone H3 Arginine 8 and Negatively Regulates Expression of ST7 and NM23 Tumor Suppressor Genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goulet I, Gauvin G, Boisvenue S, Cote J. Alternative Splicing Yields Protein Arginine Methyltransferase 1 Isoforms with Distinct Activity, Substrate Specificity, and Subcellular Localization. J Biol Chem. 2007;282:33009–33021. doi: 10.1074/jbc.M704349200. [DOI] [PubMed] [Google Scholar]
- 5.Papadokostopoulou A, Mathioudaki K, Scorilas A, Xynopoulos D, Ardavanis A, Kouroumalis E, Talieri M. Colon Cancer and Protein Arginine Methyltransferase 1 Gene Expression. Anticancer Res. 2009;29:1361–1366. [PubMed] [Google Scholar]
- 6.Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein Arginine-Methyltransferase-Dependent Oncogenesis. Nat Cell Biol. 2007;9:1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Pal S, Sif S. Protein Arginine Methyltransferase 5 Suppresses the Transcription of the RB Family of Tumor Suppressors in Leukemia and Lymphoma Cells. Mol Cell Biol. 2008;28:6262–6277. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low Levels of miR-92b/96 Induce PRMT5 Translation and H3R8/H4R3 Methylation in Mantle Cell Lymphoma. EMBO J. 2007;26:3558–3569. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Y, Li M, Wang B, Zheng YG. Discovery and Mechanistic Study of a Class of Protein Arginine Methylation Inhibitors. J Med Chem. 2010;53:6028–6039. doi: 10.1021/jm100416n. [DOI] [PubMed] [Google Scholar]
- 10.Dorgan KM, Wooderchak WL, Wynn DP, Karschner EL, Alfaro JF, Cui Y, Zhou ZS, Hevel JM. An Enzyme-Coupled Continuous Spectrophotometric Assay for S-Adenosylmethionine-Dependent Methyltransferases. Anal Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Collazo E, Couture JF, Bulfer S, Trievel RC. A Coupled Fluorescent Assay for Histone Methyltransferases. Anal Biochem. 2005;342:86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS. An Enzyme-Coupled Colorimetric Assay for S-Adenosylmethionine-Dependent Methyltransferases. Anal Biochem. 2004;326:100–105. doi: 10.1016/j.ab.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 13.Wang C, Leffler S, Thompson DH, Hrycyna CA. A General Fluorescence-Based Coupled Assay for S-Adenosylmethionine-Dependent Methyltransferases. Biochem Biophys Res Commun. 2005;331:351–356. doi: 10.1016/j.bbrc.2005.03.170. [DOI] [PubMed] [Google Scholar]
- 14.Graves TL, Zhang Y, Scott JE. A Universal Competitive Fluorescence Polarization Activity Assay for S-Adenosylmethionine Utilizing Methyltransferases. Anal Biochem. 2008;373:296–306. doi: 10.1016/j.ab.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibanez G, McBean JL, Astudillo YM, Luo M. An Enzyme-Coupled Ultrasensitive Luminescence Assay for Protein Methyltransferases. Anal Biochem. 2010;401:203–210. doi: 10.1016/j.ab.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y, Xie N, Wu J, Yang C, Zheng YG. Inhibitory Study of Protein Arginine Methyltransferase 1 Using a Fluorescent Approach. Biochem Biophys Res Commun. 2009;379:567–572. doi: 10.1016/j.bbrc.2008.12.119. [DOI] [PubMed] [Google Scholar]
- 17.Cook ND. Scintillation Proximity Assay: A Versatile High-Throughput Screening Technology. Drug Discov Today. 1996;1:287–294. [Google Scholar]
- 18.Glickman JF, Schmid A, Ferrand S. Scintillation Proximity Assays in High-Throughput Screening. Assay Drug Dev Technol. 2008;6:433–455. doi: 10.1089/adt.2008.135. [DOI] [PubMed] [Google Scholar]
- 19.Baker MR, Zarubica T, Wright HT, Rife JP. Scintillation Proximity Assay for Measurement of RNA Methylation. Nucleic Acids Res. 2009;37:e32. doi: 10.1093/nar/gkn1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]