Abstract
Muscarinic toxin 7 (MT7) is a mamba venom peptide that binds selectively to the M1 muscarinic acetylcholine receptor. We have previously shown that the second (ECL2) and third (ECL3) extracellular loops of the M1 receptor are critically involved in binding the peptide. In this study we used a mutagenesis approach on the M5 subtype of the receptor family to find out if this possesses a similar structural architecture in terms of toxin binding as the M1 receptor. An M5 receptor construct (M5-E175Y184E474), mutated at the formerly deciphered critical residues on ECL2 and 3, gained the ability to bind MT7, but with rather low affinity as determined in a functional assay (apparent Ki = 24 nM; apparent Ki for M1 = 0.5 nM). After screening for different domains and residues, we found a specific residue (P179 to L in M5) in the middle portion of ECL2 that was necessary for high affinity binding of MT7 (M5-EL179YE, apparent Ki = 0.5 nM). Mutation of P179 to A confirmed a role for the leucine side chain in the binding of MT7. Together the results reveal new binding interactions between receptors and the MT7 peptide and strengthen the hypothesis that ECL2 sequence is of utmost importance for MT binding to muscarinic receptors.
Keywords: G protein-coupled receptor, muscarinic toxin, acetylcholine receptor, ligand binding
1. Introduction
Muscarinic acetylcholine receptors (mAChRs) are G protein-coupled receptors that mediate the metabotropic effects of acetylcholine in vertebrates as well as in invertebrates. The mammalian receptor family constitutes five distinct proteins, M1-M5, with highly homologous sequences and partially overlapping tissue distribution, but with discrete physiological functions [1,2]. At the cellular level, the mAChRs exert their action by activating heterotrimeric guanine nucleotide-binding proteins (G proteins). M1, M3 and M5 receptors are coupled with the Gq protein-phospholipase C-Ca2+ mobilization pathway, whereas M2 and M4 are linked to cAMP production and ion channel regulation through Gi and Go proteins [1].
Many natural ligands have been found that target the mAChRs. Examples include the agonists muscarine from the fly agaric mushroom and pilocarpine from the Pilocarpus plants. The antagonists atropine and scopolamine can be found from the Solanaceae plant family. Most of the natural toxins found bind specifically and with relatively high affinity to muscarinic receptors, but they generally lack selectivity among subtypes. Because of the high sequence homology between receptor subtypes, especially in the orthosteric ligand-binding cavity, it has turned out very difficult to chemically produce selective compounds for each subtype [3,4,5].
An unexpected source of subtype-selective muscarinic ligands was found from mamba snakes (genus Dendroaspis) [6,7]. These snake venom components are peptide toxins that belong to a large three-finger toxin family most comprehensively characterized by the α-neurotoxins [8,9]. The muscarinic toxins (MTs) are 65-66 amino acids long, contain four disulfide bridges and, because of this structural constraint, form three characteristic loops or fingers protruding out from the disulfide core [10]. The pioneering work on MTs demonstrated that MT1 and MT2 (named after chronological order of discovery) from D. angusticeps could not displace all muscarinic receptor ligand bound to rat cortex membranes [6]. This suggested that these MTs can discriminate between subtypes of mAChRs, a fact that later became established using cloned receptor subtypes [11,12]. The MTs found in the venom of D. angusticeps seem to target M1 and M4 receptors while leaving the other subtypes untouched [9]. This might suggest that there is a close structural similarity between the M1 and M4 subtypes at the toxin-binding site and that the structure for the other subtypes thus deviates from this motif.
Although the MTs show quite high homology (65-80%) among each other, there are also toxins that target more selectively only one receptor subtype. MT3 and MT6 show low nanomolar (1-4 nM) affinities for the M4 receptor whereas M1 receptor blockade requires a hundredfold (100-300 nM) higher concentrations [9,13,14]. MT7, also called m1-toxin, is unique in the respect that it shows full selectivity for the M1 subtype, i.e., subnanomolar affinity for M1 and no detectable binding to the other subtypes at micromolar concentrations [15,16]. Since this toxin can discriminate so well among mAChR subtypes, it would be of general interest for ligand-receptor interactions to uncover the binding mechanism for MT7. It has been shown by site-directed mutagenesis of MT7 that many amino acid residues seem to make up the total binding affinity, and these residues are distributed on all three finger loops of the toxin [17]. Regarding the corresponding sites on the receptor, it can be anticipated that binding contacts with loosely conserved regions are crucially involved in the selectivity. Such nonconserved residues are frequently found in the extracellular loops (ECLs) of the receptors. In a previous study, we found that MT7 takes advantage of rather few such nonconserved residues in ECL2 and 3 for discriminating between the M1 and M3 subtypes [18]. To gain further insight into the structural requirements for MT7 binding, we report in here on the mutagenesis of the M5 subtype for gain of binding and functional inhibition effects of the toxin. The results indicate that the structural diversity of receptor ECLs could enable development of subtype-selective compounds using targeting mechanisms employed by the MTs.
2. Materials and Methods
2.1. Materials
[3H]-N-methylscopolamine ([3H]NMS) (76 Ci/mmol) was purchased from Amersham Biosciences (Buckinghamshire, UK). Fura-2 acetoxymethyl ester was from Molecular Probes (Eugene, OR). Carbamoylcholine chloride (carbachol) and atropine hemisulfate were from Sigma-Aldrich (Helsinki, Finland). Recombinantly expressed MT7 was purified from insect cell medium as described previously [15] and the concentration was determined spectrophotometrically (AM = 15900 M−1cm−1). Other chemicals used were of analytical grade quality.
2.2. Receptor Constructs and Mutagenesis
The human M1 and M5 receptors have been previously described [19]. All site-directed mutagenesis constructs were generated using the PCR technique with specific oligonucleotide primers with desired mutations (TAG, Copenhagen, Denmark). Subcloning was performed using the pBluescript KS vector (Stratagene, La Jolla, CA) or the pFastBac1 vector (Invitrogen, Paisley, UK) and the correct sequences were verified by automated DNA sequencing (Turku Centre for Biotechnology, Turku, Finland). The chimeric receptor CH1 was a fusion of the M1 and M5-EYE receptor cDNAs at an ApaI site (nucleotide +474 in M1 and nucleotide +489 in M5). The chimeric receptor CH2 was a fusion of the M1 and M5-EYE receptor cDNAs between a StuI site in M1 (nucleotide +811) and a PvuII site in M5 (nucleotide +1099). All new receptor constructs were finally transferred to the pFastBac1 vector for the generation of recombinant baculoviruses with the Bac-to-Bac baculovirus expression vector system (Invitrogen).
2.3. Cell Culture and Receptor Expression
Spodoptera frugiperda Sf9 cells were grown in suspension at 27 °C in Grace’s insect medium (Invitrogen, Paisley, UK) supplemented with 8% heat-inactivated fetal bovine serum (Invitrogen), 50 µg/mL streptomycin (Invitrogen), 50 U/mL penicillin (Invitrogen) and 0.02% Pluronic F68 (Sigma-Aldrich, Helsinki, Finland). For recombinant protein expression, Sf9 cells were plated on tissue culture dishes and allowed to attach. A high-titer virus stock was then added to give a multiplicity of infection of 3-5. For the functional assay, the infections were allowed to proceed for 26 h and cultures used for radioligand binding were harvested after 48 h.
2.4. Intracellular Ca2+ Concentration Measurement
Cells were detached from plates by gentle pipetting, centrifuged 150° g for 4 min, resuspended in a smaller volume growth medium and incubated for 20 min with 4 μM fura-2 acetoxymethyl ester at room temperature. Thereafter, cells were centrifuged 150° g for 4 min, washed once in assay buffer, resuspended in assay buffer and divided into aliquotes. The aliquotes were briefly spun down in a microcentrifuge, the medium was removed and cell pellets were stored on ice until use. The assay buffer was composed of 130 mM NaCl, 5.4 mM KCl, 1 mM CaCl2, 10 mM glucose, 1.2 mM MgCl2, 4.2 mM NaHCO3, 7.3 mM NaH2PO4, 63 mM sucrose and 20 mM MES, pH adjusted to 6.3. At the beginning of the experiment, the cells were resuspended in assay buffer with or without MT7 for 5 min. Thereafter the cells were placed in a cuvette in a thermostatted (27 °C) cuvette holder with magnetic stirring in a fluorescence spectrophotometer (Photon Technology International, Ford, UK) and fluorescence recording at alternating 340 nm and 380 nm (excitation) and 510 (emission) was started. Different concentrations of carbachol were added in a cumulative fashion and calibration of each experiment was performed with 0.04% triton X-100 to measure Fmax and 11 mM EGTA to measure Fmin. Intracellular free Ca2+ concentrations ([Ca2+]i) were calculated from fluorescence obtained at 340 nm according to:
[Ca2+]i = (F − Fmin)/(Fmax − F) ° 255 nM (Kd for fura-2 − Ca2+ complex at 27 °C) (1)
2.5. Radioligand Binding
The Infected cells from monolayer culture were harvested in phosphate-buffered saline solution, centrifuged 700× g and stored as pellets at −70 °C until use. For binding experiments the cells were resuspended in binding buffer (100 mM NaCl, 20 mM HEPES, 10 mM MgCl2, 1 mM EDTA, pH adjusted to 7.4) and homogenized with an Ultra-Turrax homogenizer. The homogenates (50–100 μg of protein) were incubated with different concentrations (0.01–10 nM) of [3H]NMS in a volume of 150 μL for 45 min at room temperature to determine Kd values for the radioligand. Atropine (10 μM) was used to determine nonspecific binding. The inhibition curves for MT7 were obtained by preincubation of homogenates with different concentrations (0.001–300 nM) of MT7 diluted in binding buffer for 45 min at room temperature. Thereafter 0.5 nM [3H]NMS was added and incubation continued for another 45 min. The concentrations of MT7 reported were obtained in the final reaction volume. When measuring the radioligand dissociation rate, receptors were first preincubated with 1.5 nM [3H]NMS for 30 min. Following this, dissociation was started by addition of 10 μM atropine with or without MT7. Aliquots were removed at regular intervals from 0 to 100 min. In testing the reversibility of MT7 binding, cell homogenates were first incubated with toxin in test tubes for 45 min, thereafter centrifuged 12,000 × g for 30 min and then resuspended in fresh buffer. After 15 min the homogenates were added to the binding reactions with 0.5 nM [3H]NMS or subjected to another round of centrifugation and resuspension. All reactions were terminated by rapid filtration through prewashed GF/B filters followed by five washes with cold buffer containing 180 mM NaCl, 25 mM MgCl2, and 20 mM HEPES, pH 7.4, in a microplate harvester. Radioactivity on filters was determined in a TopCount microplate scintillation counter (PerkinElmer, Waltham, MA).
2.6. Data Analysis
Non-linear curve-fitting of the concentration-response data was performed using GraphPad Prism (GraphPad Software, La Jolla, CA). The pIC50 values were obtained through non-linear regression to fit log IC50, and the The pKx values and the cooperativity factor α were obtained through curve-fitting to the allosteric ternary complex model.
The apparent Ki values in the functional assay were obtained from data fitted to the equation:
Δ[Ca2+] = [A]Δ[Ca2+]max/([A](1 + [I]/Ki') + EC50(1 + [I]/Ki)) (2)
[A] and [I] are the concentrations of agonist and inhibitor, respectively, EC50 is the [A] producing half-maximal stimulation and Ki and Ki' are the surmountable (competitive) and the insurmountable (noncompetitive) inhibition constants, respectively [20]. The Ki value gives a measure of the right-shift of the concentration-response curve and the Ki' value gives a measure of the suppression of the maximum response. Only the apparent Ki values are given because the apparent Ki' values did not obey inhibitor concentration dependency, as previously also found when using MT7 as inhibitor [18].
3. Results and Discussion
3.1. Mutagenesis of the M5 Receptor Subtype
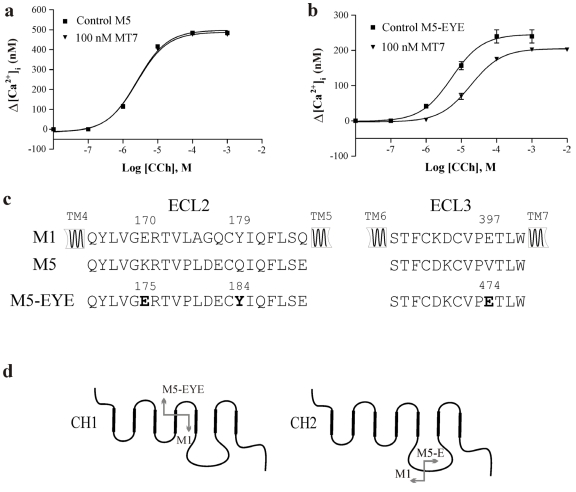
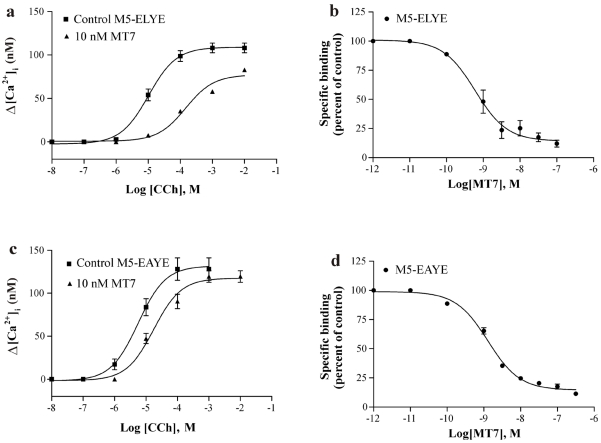
Functional inhibition of receptors by MT7 was assessed by measuring the [Ca2+]i responses in Sf9 cells expressing intact or modified receptor proteins. Activation of the M5 receptor with carbachol resulted in concentration-dependent increases in [Ca2+]i as determined from fura-2 fluorescence (Figure 1a). MT7 did not affect the activation of the intact M5 subtype. To try to gain binding of MT7, we introduced two mutations in ECL2 and one in ECL3 of M5 (M5-EYE, Figure 1c). The glutamate residue in the proximal part of ECL2 of M1 (E170 in M1, K175 in M5) and a tyrosine residue adjacent to the conserved cysteine (Y179 in M1, Q184 in M5) were both previously recognized as important for MT7 binding [18]. These residues together with the V474 to E mutation in ECL3 rendered the M5 subtype sensitive to MT7 (Figure 1b). The apparent Ki (>20 nM, Table 1) was, however, significantly higher than what we obtained with the M1 subtype (apparent Ki = 0.45 ± 0.18 nM, mean ± SD, n = 3). This prompted us to look for additional interaction sites between the toxin and the receptor. First we used chimeric M1:M5-EYE receptors to locate the region involved (Figure 1d). These chimeras (CH1 and CH2) indicated that we should refocus on the ECL2, since CH2 incorporating the ECL2 of M1 increased the affinity for MT7 about tenfold as compared to M5-EYE (Table 1). After screening for individual residues in ECL2 of M5, we found that the additional P179 to L (M5-ELYE) mutation increased significantly the affinity for MT7 (Figure 2a). This result was further confirmed in radioligand binding experiments with a calculated IC50 of 0.6 nM for MT7 (Figure 2b, Table 2). As for the M1 subtype (IC50 ≈ 0.5 nM) [18,21], MT7 does not compete directly with the orthosteric antagonist and a fraction of the radioligand binding remains even at saturating MT7 concentrations. Thus, the potencies for inhibition are presented as pIC50 values and not converted to Ki values as for competitive ligands (Table 2).
Figure 1.
The M5 receptor and the M5-EYE mutant in Sf9 cells were assayed in the absence and presence of muscarinic toxin 7 (MT7). (a and b) Carbachol (CCh) concentration-response curves for [Ca2+]i increases. Data points are means ± SD from two experiments; (c) Amino acid sequences of extracellular loops ECL2 and ECL3. TM4-7 denotes the helical transmembrane domains of the receptors. Introduced mutations in M5 are in boldface; (d) Schematic presentation of the chimeric receptors used to localize regions of toxin binding.
Table 1.
Functional characteristics of expressed receptor constructs. Values are means ± SD from three experiments.
| Construct | EC50 for CCh, μM | Max. Δ[Ca2+]i, nM | App. Ki for MT7, nM |
|---|---|---|---|
| M5 | 2.56 ± 0.34 | 498 ± 10 | − |
| M5-EYE | 5.29 ± 0.03 | 246 ± 33 | 24.06 ± 4.44 |
| CH1 | 4.60 ± 1.61 | 236 ± 10 | 11.54 ± 2.56 |
| CH2 | 6.14 ± 1.10 | 293 ± 17 | 1.85 ± 0.61 |
Figure 2.
Sf9 cells expressing mutated M5 receptor constructs were assayed for carbachol-induced [Ca2+]i increases and inhibition of [3H]NMS binding. (a and c) Functional analyses of the M5-ELYE and M5-EAYE receptor constructs in the absence and presence of MT7. Data points are means ± SD from two experiments. The apparent Ki values for MT7 were 0.52 ± 0.14 nM and 4.41 ± 1.09 nM (means ± SD, n = 4) for M5-ELYE and M5-EAYE, respectively; (b and d) Cell homogenates of each receptor construct were preincubated with different concentrations of MT7 before addition of 0.5 nM [3H]NMS. Bound radioactivity was converted to receptor concentration and presented as percent of control values in the absence of toxin. Data points are means ± SEM from three experiments.
Table 2.
Binding characteristics of expressed receptor constructs. Values are means ± SEM from three experiments.
| Construct | Kd for [3H]NMS, nM | pIC50 for MT7 | p Kx for MT7 | Cooperativity factor α |
|---|---|---|---|---|
| M5 | 0.79 ± 0.16 | − | − | − |
| M5-ELYE | 0.54 ± 0.09 | 9.22 ± 0.11 | 9.48 ± 0.10 | 0.08 ± 0.02 |
| M5-EAYE | 0.49 ± 0.08 | 8.89 ± 0.04 | 9.16 ± 0.04 | 0.08 ± 0.01 |
| M5-ELY | 0.73 ± 0.09 | 8.21 ± 0.05 | 8.38 ± 0.04 | 0.21 ± 0.01 |
The P179t to L mutation in the middle of ECL2 of the M5 receptor appeared to have an important role in the binding of MT7. The corresponding residue of the M1 receptor we did not specifically investigate in our previous study, and the M3 double mutant with the corresponding P217 to L and the additional P218 to A mutations did not show an inhibitory effect of MT7 [18]. To further investigate whether the aliphatic side chain of the leucine is important for the binding or if the proline only postures a steric hindrance for other interactions to take place, we mutated the P179 to A and analyzed the receptor construct in the functional assay (Figure 2c) and with radioligand binding (Figure 2d, Table 2). MT7 was found to possess lower potency for M5-EAYE in both assays when compared to M5-ELYE, indicating a contribution to the overall binding for the leucine side chain.
3.2. The Role of the ECL3 Glutamate
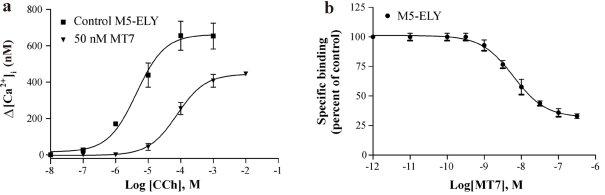
In our previous study using chimeric M1:M3 receptors, we found that a lysine residue (K523) in M3 ECL3 sequence resulted in a tenfold loss in the apparent Ki value for MT7 [18]. It was not clear, however, whether this was due to a loss of binding contact with the corresponding negatively charged glutamate (E397) of M1 or if the long acyl chain and positively charged head group of the M3 lysine somehow precluded other types of interactions between the receptor and the toxin. In the M5 receptor, the corresponding residue is a valine (V474), which should at least not exhibit any charge repulsion. We thus exchanged the mutated ECL3 of M5-ELYE for the intact M5 sequence and measured the binding characteristics of MT7 (Figure 3). In the functional assay, the M5-ELY construct lost an approximately fourfold blocking potency (apparent Ki = 2.06 ± 0.86 nM, mean ± SD, n = 3) as compared to M5-ELYE, and about tenfold loss in binding affinity (Table 2). In addition to the loss in binding affinity, we noticed that the inhibitory capacity of MT7 in terms of displacement of radioligand was significantly lower than for the other receptor constructs, i.e., the remaining bound radioligand in the presence of saturating concentrations of MT7 mounted to about 30% for M5-ELY (Figure 3b), whereas for M5-ELYE (Figure 2b) and for M1 the corresponding level was about 15%. We do not currently have an explanation for these different behaviors.
Figure 3.
Sf9 cells expressing the M5-ELY construct were assayed for carbachol-induced [Ca2+]i increases and inhibition of [3H]NMS binding. (a) Representative concentration-dependent increases in [Ca2+]i for control and in the presence of 50 nM MT7. The apparent Ki was determined to 2.06 ± 0.86 nM (mean ± SD, n = 3); (b) Titration of MT7 against [3H]NMS with M5-ELY. Data points are means ± SEM from three experiments.
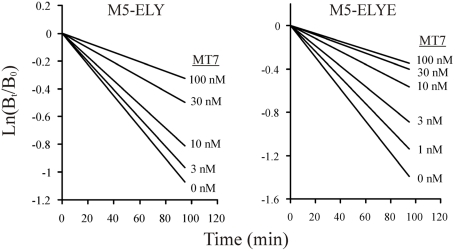
3.3. Allosteric Interactions and Irreversibility of Binding
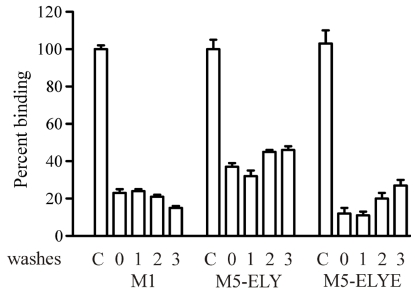
In addition to the high affinity binding of MT7 to the M1 receptor, the characteristics of this toxin−receptor complex also include an allosteric slowing of antagonist dissociation [22,23], and a pseudo-irreversibility of binding, i.e., the toxin does not dissociate from the receptor in spite of extensive washing [7,24]. To find out if our M5 constructs exhibited similar properties, we first tested the ability to slow down the dissociation of prebound [3H]NMS. With both M5-ELY and M5-ELYE there was a concentration-dependent deceleration of antagonist dissociation (Figure 4). The pIC50,diss values (affinity for [3H]NMS-occupied receptor) were 7.73 ± 0.06 and 8.47 ± 0.06 for M5-ELY and M5-ELYE, respectively. We also tested the irreversibility of MT7 binding at M1 and the mutated M5 constructs. We have previously used a protocol with repeated centrifugations to demonstrate the reversibility of MTα binding to the α2B adrenergic receptor [25]. Using the same protocol for MT7, we could not recover control binding sites with either M1 or the mutated M5 constructs (Figure 5). These data indicate that the M5 constructs have been converted to M1 phenotypes in the known pharmacological aspects of MT7 binding.
Figure 4.
Deceleration of [3H]NMS dissociation by MT7. The M5-ELY and M5-ELYE constructs were preincubated with [3H]NMS and dissociation initiated by addition of atropine with or without different concentrations of MT7. Samples were collected at different time points and [3H]NMS binding (Bt) determined. Data presented are from one experiment performed with triplicate samples. Two additional experiments with 10 and 100 nM MT7 gave similar results. Only the trendlines are shown for clarity. The control rate for M5-ELY was 0.113 min−1 and 0.003 min−1 with 100 nM MT7. The control rate for M5-ELYE was 0.147 min−1 and 0.004 min−1 with 100 nM MT7.
Figure 5.
Reversibility test for MT7. Receptor constructs were preincubated with MT7 (30 nM with M1 and M5-ELYE, 100 nM with M5-ELY) for 45 min and then subjected to [3H]NMS binding (control (C) and zero washes) or centrifuged and washed one to three times before [3H]NMS binding. Data are means ± SEM from three experiments.
4. Conclusions
The MT7 toxin shows an astonishing selectivity for the M1 receptor, making it an attractive template for how to develop receptor subtype-selective compounds. To accomplish such a task, we need to understand how MT7 binds to the receptor and which receptor epitopes are chosen by the toxin. We have in this study converted by mutagenesis the M5 receptor to bind MT7 with high affinity, without interfering to any larger extent with orthosteric antagonist binding (as determined from measured affinities of [3H]NMS) or agonist activation of the receptors (as determined from the functional assays). The importance of the leucine residue found in the middle of ECL2 of the M1 receptor expands the list of critical residues involved in the binding of MT7.
During the preparation of this manuscript, Marquer et al. reported on the mutation of the M1 receptor and MT7. They also identified the leucine in the middle of ECL2 as a player in the toxin interactions [26]. Mutation of the leucine to proline, as in M3, decreased the binding affinity for MT7. On the other hand, the leucine to alanine mutation did not appear to affect toxin binding. This is in contrast to what we found with the M5 receptor. It is possible that MT7 binds with slightly altered conformation to the mutated M5 receptor than to the M1 receptor and thus utilizes receptor residues that have a less important role in M1. It may also be that our approach to mutate other receptor subtypes for gain of binding reveal more subtle interactions if there are less overall interaction sites as compared to the MT7−M1 receptor complex.
Acknowledgments
This study was funded by The Magnus Ehrnrooth Foundation and The Academy of Finland.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Caulfield M.P., Birdsall N.J. International Union of Pharmacology. XVII. Classification of muscarinic acetylcholine receptors. Pharmacol. Rev. 1998;50:279–290. [PubMed] [Google Scholar]
- 2.Wess J. Muscarinic acetylcholine receptor knockout mice: Novel phenotypes and clinical implications. Annu. Rev. Pharmacol. Toxicol. 2004;44:423–450. doi: 10.1146/annurev.pharmtox.44.101802.121622. [DOI] [PubMed] [Google Scholar]
- 3.Eglen R.M., Choppin A., Dillon M.P., Hegde S. Muscarinic receptor ligands and their therapeutic potential. Curr. Opin. Chem. Biol. 1999;3:426–432. doi: 10.1016/S1367-5931(99)80063-5. [DOI] [PubMed] [Google Scholar]
- 4.Eglen R.M., Choppin A., Watson N. Therapeutic opportunities from muscarinic receptor research. Trends Pharmacol. Sci. 2001;22:409–414. doi: 10.1016/s0165-6147(00)01737-5. [DOI] [PubMed] [Google Scholar]
- 5.Heinrich J.N., Butera J.A., Carrick T., Kramer A., Kowal D., Lock T., Marquis K.L., Pausch M.H., Popiolek M., Sun S.C., Tseng E., Uveges A.J., Mayer S.C. Pharmacological comparison of muscarinic ligands: Historical versus more recent muscarinic M1-preferring receptor agonists. Eur. J. Pharmacol. 2009;605:53–56. doi: 10.1016/j.ejphar.2008.12.044. [DOI] [PubMed] [Google Scholar]
- 6.Adem A., Åsblom A., Johansson G., Mbugua P.M., Karlsson E. Toxins from the venom of the green mamba Dendroaspis angusticeps that inhibit the binding of quinuclidinyl benzilate to muscarinic acetylcholine receptors. Biochim. Biophys. Acta. 1988;968:340–345. doi: 10.1016/0167-4889(88)90025-0. [DOI] [PubMed] [Google Scholar]
- 7.Max S.I., Liang J.S., Potter L.T. Purification and properties of m1-toxin, a specific antagonist of m1 muscarinic receptors. J. Neurosci. 1993;13:4293–4300. doi: 10.1523/JNEUROSCI.13-10-04293.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsetlin V. Snake venom alpha-neurotoxins and other “three-finger” proteins. Eur. J. Biochem. 1999;264:281–286. doi: 10.1046/j.1432-1327.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- 9.Karlsson E., Jolkkonen M., Mulugeta E., Onali P., Adem A. Snake toxins with high selectivity for subtypes of muscarinic acetylcholine receptors. Biochimie. 2000;82:793–806. doi: 10.1016/s0300-9084(00)01176-7. [DOI] [PubMed] [Google Scholar]
- 10.Ségalas I., Roumestand C., Zinn-Justin S., Gilquin B., Ménez R., Ménez A., Toma F. Solution structure of a green mamba toxin that activates muscarinic acetylcholine receptors, as studied by nuclear magnetic resonance and molecular modeling. Biochemistry. 1995;34:1248–1260. doi: 10.1021/bi00004a019. [DOI] [PubMed] [Google Scholar]
- 11.Kornisiuk E., Jerusalinsky D., Cervenansky C., Harvey A.L. Binding of muscarinic toxins MTx1 and MTx2 from the venom of the green mamba Dendroaspis angusticeps to cloned human muscarinic cholinoceptors. Toxicon. 1995;33:11–18. doi: 10.1016/0041-0101(94)00161-Z. [DOI] [PubMed] [Google Scholar]
- 12.Kornisiuk E., Jerusalinsky D., Cervenansky C., Harvey A.L. Corrigendum. Toxicon. 1995;33:1111. doi: 10.1016/0041-0101(94)00161-z. [DOI] [PubMed] [Google Scholar]
- 13.Jolkkonen M., van Giersbergen P.L., Hellman U., Wernstedt C., Karlsson E. A toxin from the green mamba Dendroaspis angusticeps: Amino acid sequence and selectivity for muscarinic m4 receptors. FEBS Lett. 1994;352:91–94. doi: 10.1016/0014-5793(94)00933-3. [DOI] [PubMed] [Google Scholar]
- 14.Liang J.S., Carsi-Gabrenas J., Krajewski J.L., McCafferty J.M., Purkerson S.L., Santiago M.P., Strauss W.L., Valentine H.H., Potter L.T. Anti-muscarinic toxins from Dendroaspis angusticeps. Toxicon. 1996;34:1257–1267. doi: 10.1016/s0041-0101(96)00109-2. [DOI] [PubMed] [Google Scholar]
- 15.Näsman J., Jolkkonen M., Ammoun S., Karlsson E., Åkerman K.E. Recombinant expression of a selective blocker of M(1) muscarinic receptors. Biochem. Biophys. Res. Commun. 2000;271:435–439. doi: 10.1006/bbrc.2000.2657. [DOI] [PubMed] [Google Scholar]
- 16.Mourier G., Dutertre S., Fruchart-Gaillard C., Menez A., Servent D. Chemical synthesis of MT1 and MT7 muscarinic toxins: Critical role of Arg-34 in their interaction with M1 muscarinic receptor. Mol. Pharmacol. 2003;63:26–35. doi: 10.1124/mol.63.1.26. [DOI] [PubMed] [Google Scholar]
- 17.Fruchart-Gaillard C., Mourier G., Marquer C., Stura E., Birdsall N.J., Servent D. Different interactions between MT7 toxin and the human muscarinic M1 receptor in its free and N-methylscopolamine-occupied states. Mol. Pharmacol. 2008;74:1554–1563. doi: 10.1124/mol.108.050773. [DOI] [PubMed] [Google Scholar]
- 18.Kukkonen A., Peräkylä M., Åkerman K.E., Näsman J. Muscarinic toxin 7 selectivity is dictated by extracellular receptor loops. J. Biol. Chem. 2004;279:50923–50929. doi: 10.1074/jbc.M406424200. [DOI] [PubMed] [Google Scholar]
- 19.Kukkonen J.P., Näsman J., Ojala P., Oker-Blom C., Åkerman K.E.O. Functional properties of muscarinic receptor subtypes Hm1, Hm3 and Hm5 expressed in Sf9 cells using the baculovirus expression system. J. Pharmacol. Exp. Ther. 1996;279:593–60. [PubMed] [Google Scholar]
- 20.Kukkonen J., Åkerman K.E. Apparent noncompetitive antagonism of muscarinic receptor mediated Ca2+ mobilization by some muscarinic antagonists. Biochem. Biophys. Res. Commun. 1992;189:919–924. doi: 10.1016/0006-291X(92)92291-5. [DOI] [PubMed] [Google Scholar]
- 21.Näreoja K., Kukkonen J., Rondinelli S., Toivola D., Meriluoto J., Näsman J. Adrenoceptor activity of muscarinic toxins identified from mamba venoms. Br. J. Pharmacol. 2011;164:538–550. doi: 10.1111/j.1476-5381.2011.01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Max S.I., Liang J.S., Potter L.T. Stable allosteric binding of m1-toxin to m1 muscarinic receptors. Mol. Pharmacol. 1993;44:1171–1175. [PubMed] [Google Scholar]
- 23.Fruchart-Gaillard C., Mourier G., Marquer C., Menez A., Servent D. Identification of various allosteric interaction sites on M1 muscarinic receptor using 125I-Met35-oxidized muscarinic toxin 7. Mol. Pharmacol. 2006;69:1641–1651. doi: 10.1124/mol.105.020883. [DOI] [PubMed] [Google Scholar]
- 24.Olianas M.C., Maullu C., Adem A., Mulugeta E., Karlsson E., Onali P. Inhibition of acetylcholine muscarinic M(1) receptor function by the M(1)-selective ligand muscarinic toxin 7 (MT-7) Br. J. Pharmacol. 2000;131:447–452. doi: 10.1038/sj.bjp.0703606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koivula K., Rondinelli S., Näsman J. The three-finger toxin MTalpha is a selective alpha(2B)-adrenoceptor antagonist. Toxicon. 2010;56:440–447. doi: 10.1016/j.toxicon.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Marquer C., Fruchart-Gaillard C., Letellier G., Marcon E., Mourier G., Zinn-Justin S., Menez A., Servent D., Gilquin B. Structural model of ligand-G protein-coupled receptor (GPCR) complex based on experimental double mutant cycle data: MT7 snake toxin bound to dimeric hM1 muscarinic receptor. J. Biol. Chem. 2011;286:31661–31675. doi: 10.1074/jbc.M111.261404. [DOI] [PMC free article] [PubMed] [Google Scholar]