Abstract
Photolysis of rhodopsin leads to the formation of an activated intermediate that activates a G protein, thus beginning the visual cascade. This activated form of rhodopsin appears coincident in time with the spectroscopically defined intermediate, metarhodopsin II. Metarhodopsin I, the precursor of metarhodopsin II, contains a protonated Schiff base, whereas metarhodopsin II does not. The question of whether the deprotonation of the protonated Schiff base is obligate in the formation of activated rhodopsin was addressed by monomethylating the active-site lysine of permethylated rhodopsin and determining whether this pigment can activate the G protein upon photolysis. The photolysis of the new pigment, which absorbs at 520 nm, led to the formation of a relatively stable metarhodopsin I-like intermediate with a lambda max of approximately equal to 485 nm, with no apparent formation of either metarhodopsin II- or metarhodopsin III-like intermediates. The only probe available to detect formation of the active form of rhodopsin is G protein activation. Photolysis of the pigment in the presence of the G protein did not lead to measurable activation of the GTPase activity of the latter. These studies establish a functional link between Schiff base deprotonation and activation of the G protein. It is concluded that proton transfer from the protonated Schiff base of rhodopsin is obligate for the initiation of visual transduction.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahamson E. W., Ostroy S. E. The photochemical and macromolecular aspects of vision. Prog Biophys Mol Biol. 1967;17:179–215. doi: 10.1016/0079-6107(67)90007-7. [DOI] [PubMed] [Google Scholar]
- Applebury M. L., Zuckerman D. M., Lamola A. A., Jovin T. M. Rhodopsin. Purification and recombination with phospholipids assayed by the metarhodopsin I leads to metarhodopsin II transition. Biochemistry. 1974 Aug 13;13(17):3448–3458. doi: 10.1021/bi00714a005. [DOI] [PubMed] [Google Scholar]
- Bennett N., Michel-Villaz M., Kühn H. Light-induced interaction between rhodopsin and the GTP-binding protein. Metarhodopsin II is the major photoproduct involved. Eur J Biochem. 1982 Sep;127(1):97–103. doi: 10.1111/j.1432-1033.1982.tb06842.x. [DOI] [PubMed] [Google Scholar]
- Busch G. E., Applebury M. L., Lamola A. A., Rentzepis P. M. Formation and decay of prelumirhodopsin at room temperatures. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2802–2806. doi: 10.1073/pnas.69.10.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoon R. D., Rando R. R. all-trans-retinoids and dihydroretinoids as probes of the role of chromophore structure in rhodopsin activation. Biochemistry. 1985 Nov 5;24(23):6446–6452. doi: 10.1021/bi00344a021. [DOI] [PubMed] [Google Scholar]
- Cooper A. Energy uptake in the first step of visual excitation. Nature. 1979 Nov 29;282(5738):531–533. doi: 10.1038/282531a0. [DOI] [PubMed] [Google Scholar]
- Doukas A. G., Aton B., Callender R. H., Ebrey T. G. Resonance Raman studies of bovine metarhodopsin I and metarhodopsin II. Biochemistry. 1978 Jun 13;17(12):2430–2435. doi: 10.1021/bi00605a028. [DOI] [PubMed] [Google Scholar]
- Emeis D., Kühn H., Reichert J., Hofmann K. P. Complex formation between metarhodopsin II and GTP-binding protein in bovine photoreceptor membranes leads to a shift of the photoproduct equilibrium. FEBS Lett. 1982 Jun 21;143(1):29–34. doi: 10.1016/0014-5793(82)80266-4. [DOI] [PubMed] [Google Scholar]
- Fesenko E. E., Kolesnikov S. S., Lyubarsky A. L. Induction by cyclic GMP of cationic conductance in plasma membrane of retinal rod outer segment. Nature. 1985 Jan 24;313(6000):310–313. doi: 10.1038/313310a0. [DOI] [PubMed] [Google Scholar]
- Fung B. K., Hurley J. B., Stryer L. Flow of information in the light-triggered cyclic nucleotide cascade of vision. Proc Natl Acad Sci U S A. 1981 Jan;78(1):152–156. doi: 10.1073/pnas.78.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrave P. A., McDowell J. H., Curtis D. R., Wang J. K., Juszczak E., Fong S. L., Rao J. K., Argos P. The structure of bovine rhodopsin. Biophys Struct Mech. 1983;9(4):235–244. doi: 10.1007/BF00535659. [DOI] [PubMed] [Google Scholar]
- Hayward G., Carlsen W., Siegman A., Stryer L. Retinal chromophore of rhodopsin photoisomerizes within picoseconds. Science. 1981 Feb 27;211(4485):942–944. doi: 10.1126/science.7466366. [DOI] [PubMed] [Google Scholar]
- Kwok-Keung Fung B., Stryer L. Photolyzed rhodopsin catalyzes the exchange of GTP for bound GDP in retinal rod outer segments. Proc Natl Acad Sci U S A. 1980 May;77(5):2500–2504. doi: 10.1073/pnas.77.5.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
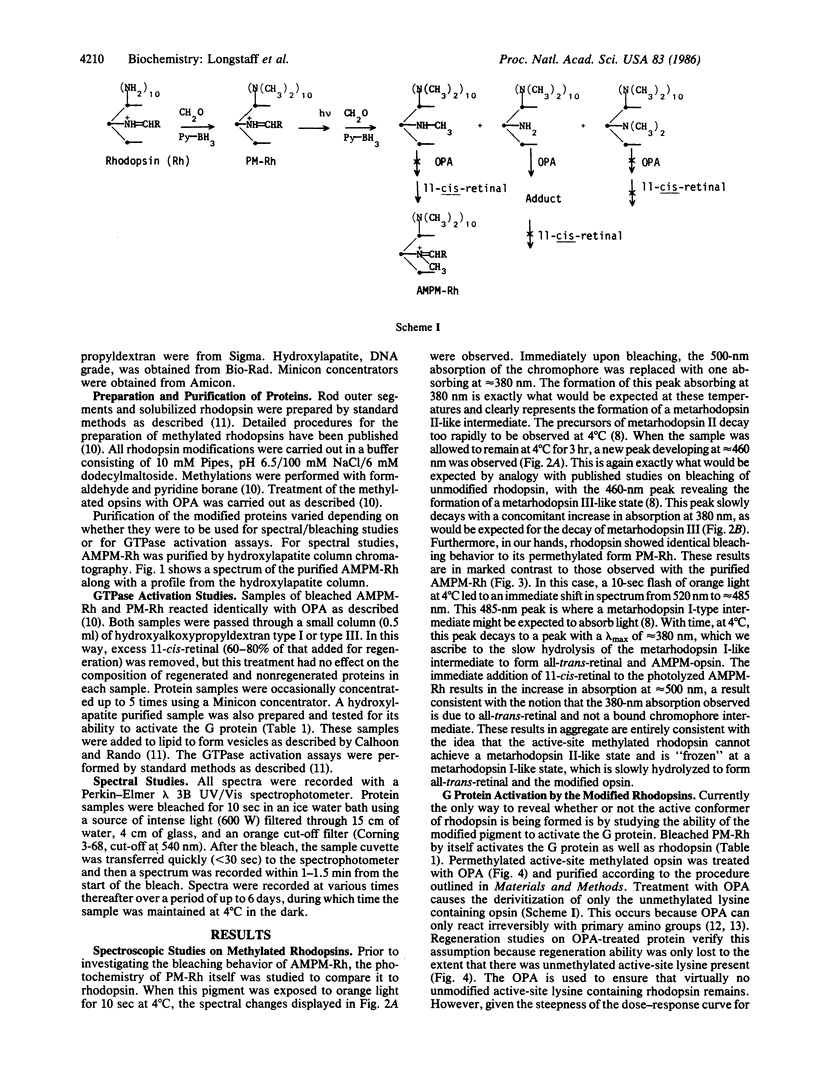
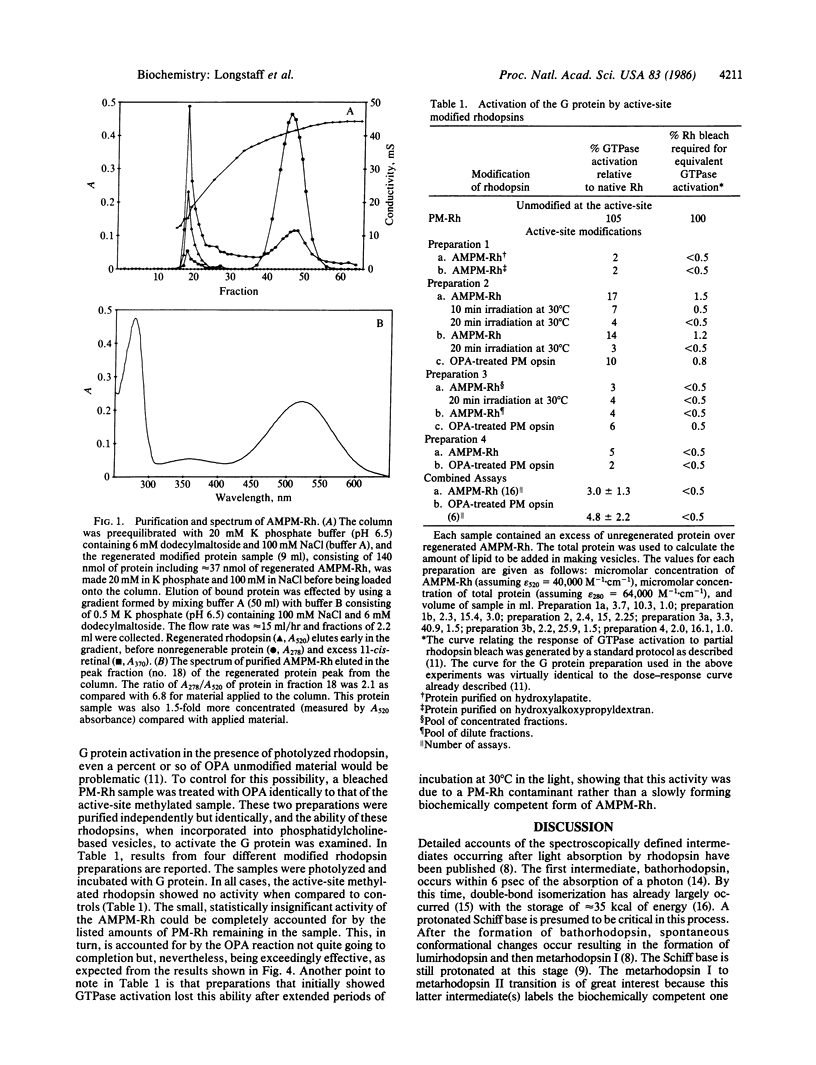
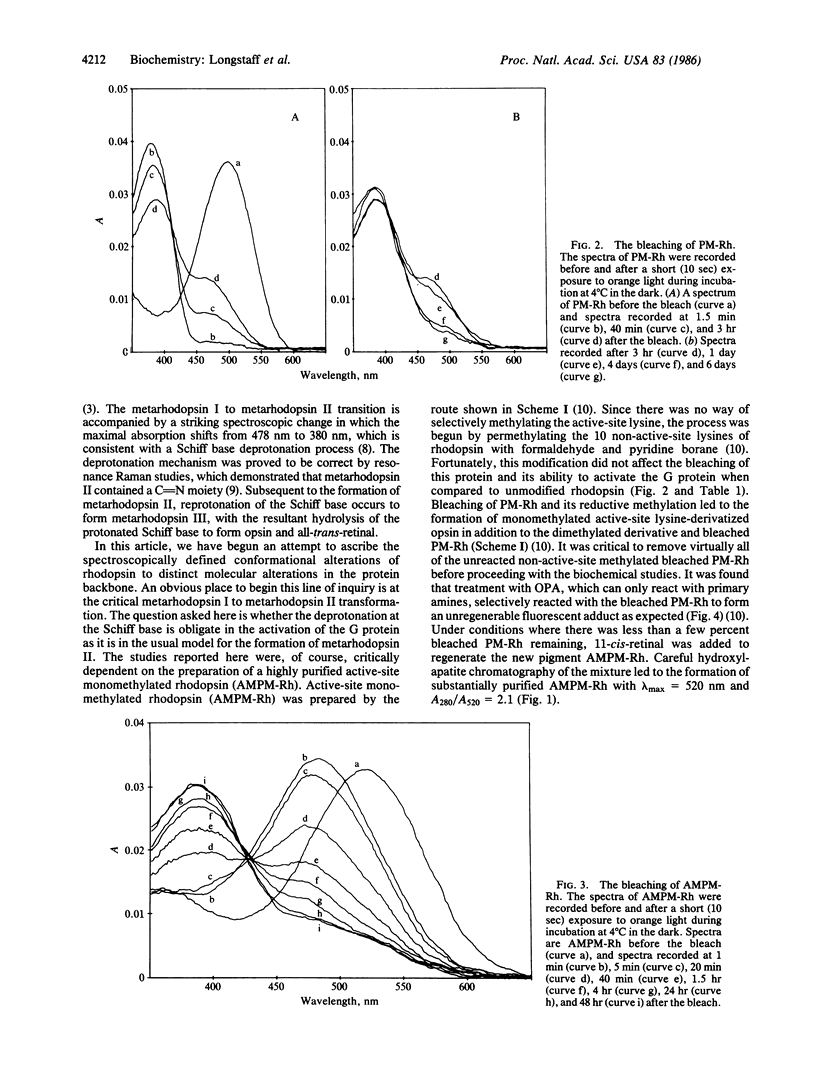
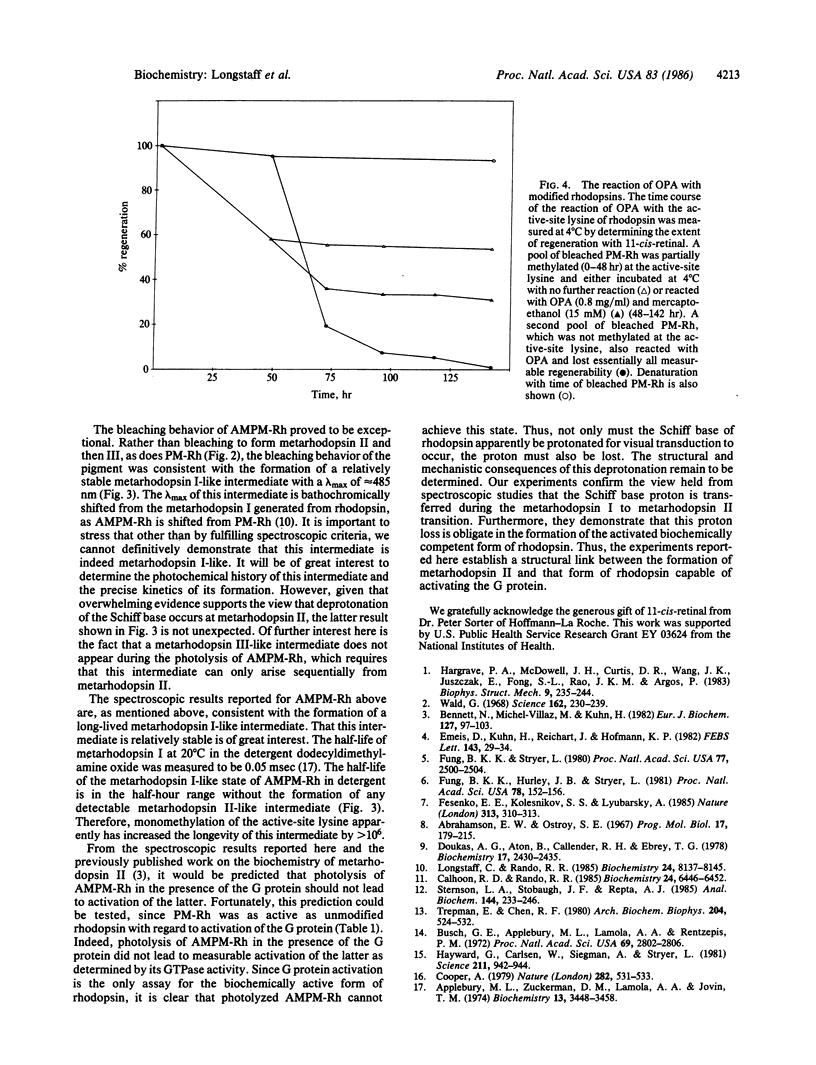
- Longstaff C., Rando R. R. Methylation of the active-site lysine of rhodopsin. Biochemistry. 1985 Dec 31;24(27):8137–8145. doi: 10.1021/bi00348a045. [DOI] [PubMed] [Google Scholar]
- Sternson L. A., Stobaugh J. F., Repta A. J. Rational design and evaluation of improved o-phthalaldehyde-like fluorogenic reagents. Anal Biochem. 1985 Jan;144(1):233–246. doi: 10.1016/0003-2697(85)90111-3. [DOI] [PubMed] [Google Scholar]
- Trepman E., Chen R. F. Fluorescence stopped-flow study of the o-phthaldialdehyde reaction. Arch Biochem Biophys. 1980 Oct 15;204(2):524–532. doi: 10.1016/0003-9861(80)90064-8. [DOI] [PubMed] [Google Scholar]
- Wald G. Molecular basis of visual excitation. Science. 1968 Oct 11;162(3850):230–239. doi: 10.1126/science.162.3850.230. [DOI] [PubMed] [Google Scholar]


