Abstract
In the last decade hepatitis C virus (HCV) kinetics has become an important clinical tool for the optimization of therapy with (pegylated)-interferon-α (IFN) and ribavirin (RBV). Mathematical models have generated important insights into HCV pathogenesis, HCV- host dynamics, and IFN and RBV’s modes of action. Clinical trials with direct acting agents (DAAs) against various steps of the HCV life cycle have revealed new viral kinetic patterns that have not been observed with IFN±RBV. Very recently, studies have showed that single nucleotide polymorphisms (SNPs) in the IL28B gene region were associated with race/ethnicity and with response to IFN±RBV. Here we review our current knowledge of HCV kinetics and related mathematical models during IFN±RBV and/or DAA based therapies, HCV pathogenesis, and the role of IL28B polymorphism on early HCV kinetics. Better understanding of the mode of actions of drug(s) and viral kinetics may help to develop, in the near future, new individualized therapeutic regimens that include DAAs in combination with IFN+RBV.
Introduction
Hepatitis C virus (HCV) infections are a serious threat to public health with more than 180 million infected people worldwide [1]. Although HCV is the primary cause of liver cancer in the United States and the leading indication for liver transplantation, no protective vaccine exists and only a subset of patients infected with HCV genotype 1 (30% to 50%) achieve a sustained virologic response (SVR) to the standard of care (SOC), i.e., treatment with pegylated-interferon-α (PEG-IFN) plus ribavirin (RBV). Statistical models that predict the future course of HCV infection suggest that without improved anti-HCV therapeutic regimens, the total number of individuals with cirrhosis will peak in the United States at one million in 2020 [2]. As such, there is a need to develop more effective HCV therapeutic regimens.
New treatment options currently in development include direct-acting agents (DAAs) that target specific components of the HCV life-cycle (see review by TenCate et al. [3]). Although DAAs have shown promising antiviral activities in early clinical trials the selection of drug-resistant HCV variants (e.g., Figure 1, dashed line) pose a serious concern. Combination of PEG-IFN+RBV with a DAA, such as the protease inhibitors telaprevir or boceprevir, has been shown to overcome, in part, the selection of drug-resistance HCV variants and to improve the rate of SVR. Since PEG-IFN+RBV will likely be the backbone of new combination therapies with DAAs, understanding HCV kinetics during IFN+RBV treatment may help to design optimal therapeutic strategies and may reduce the risk of emergence of DAA-resistant variants. The recent identification of the role of single nucleotide polymorphisms (SNPs) upstream the interleukin 28B (IL28B) gene locus (termed here IL28B polymorphisms) on PEG-IFN±RBV treatment response may allow new personalized therapeutic approaches to treat chronic HCV-infected individuals with SOC alone and in combination with DAAs. In this review we present our current knowledge of HCV viral dynamics under SOC and/or DAA therapy, and the role of IL28B polymorphisms on early viral kinetics.
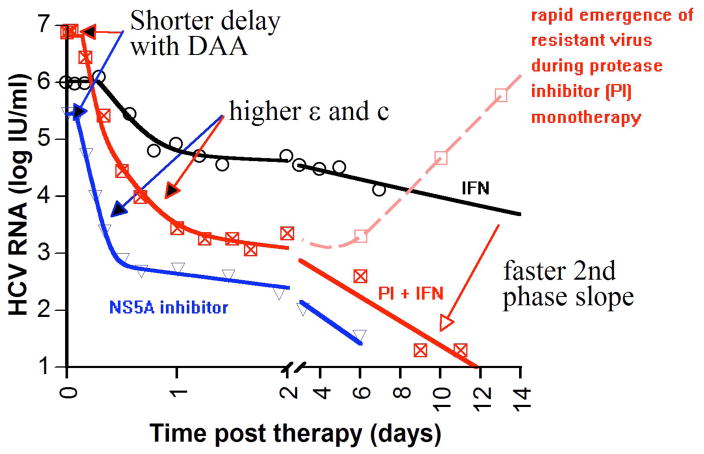
Figure 1.
Representative plasma HCV kinetics in treated individuals with daily IFN (circles), PEG-IFN+telaprevir (squares filled with x), telaprevir (empty squares) and BMS-790052 (up side down triangles). Fitting results of the biphasic decline model (Eq.1; solid lines) with these data suggest that some DAA-based treatments, in comparison to IFN-based therapies, lead to shorter delay before HCV RNA declines after initiation of treatment, t0 (e.g., Table 2), enhance viral clearance rate in serum, c [59•], and lead to higher efficacies in blocking viral production/release, ε, and faster 2nd phase slope decline or higher infected cell loss rate, δ (e.g., Table 2) [17, 22••]. The limitations of the biphasic decline model are discussed in the main text and in Table 1.
HCV RNA kinetics before therapy
During chronic HCV infection the level of serum HCV RNA does not vary significantly (<0.5 log) on time scales of weeks to months [4,5]. The distribution of baseline HCV RNA levels in the U.S. is approximately log normal with peak values between 6.5–6.8 log10 (IU/ml) [6]. Among genotype-1 HCV treated patients, baseline HCV-RNA level is a strong independent predictor of SVR [7–10].
HCV RNA kinetics during IFN-based therapy (standard of care)
When patients chronically infected with HCV are treated with (pegylated) interferon-α (IFN) ± ribavirin, they are defined as responders or sustained virologic responders (SVR) in cases of persistent absence of serum HCV RNA for 6 months or longer after therapy. With frequent HCV RNA measurements during treatment a detailed viral kinetic picture is revealed. Neumann and colleagues [11] previously showed that after initiation of IFN therapy, HCV RNA levels generally begin declining after a 7 – 10 hour delay, which may mainly represent the time needed for IFN signal transduction and IFN stimulated gene expression [12]. The typical HCV RNA decline pattern is biphasic (Fig. 1; black solid line) with an initial rapid first phase, lasting for approximately 1 – 2 days during which HCV RNA, on average, may fall 1 to 2 logs in genotype-1 infected patients [11] and as much as 3 to 4 logs in genotype-2 infected patients [13]. Subsequently, a slower second phase of HCV RNA decline ensues. Other viral kinetic patterns such as triphasic, null response (or flat partial response) and viral rebounds have also been seen and are described in Table 1.
Table 1.
Observed plasma HCV RNA kinetics and mathematical models.
| Viral kinetic pattern | Viral kinetic definition | Model name | Comments | Model predictions | Model concerns/limitations | Ref. # |
|---|---|---|---|---|---|---|
| Biphasic | The typical viral decline seen under daily IFN or DAAs: consists of a rapid first phase (0.5 – 2 days) of viral decline followed by a slower 2nd phase decline | Biphasic (Eq.1) | Uninfected hepatocytes are at steady state during the period of analysis | High ε and δ have been shown to be important for treated patients to achieve an SVR | (i) Explain only biphasic viral decline pattern (ii) may underestimate viral clearance rate, c, and loss/death rate of infected cells, δ | [4, 11, 59•, 60, 97, 98] |
| Extended (Eq. 2) | Includes both uninfected and infected hepatocyte proliferation rates. | Low rate of SVR in subjects with high baseline viral load and/or advanced liver disease. With high ε~1, second phase viral decline is close to δ | Many unknown parameters that can only be calculated after model parameters are estimated; hepatocyte proliferation rate in HCV infected patients is not known. May underestimate viral clearance rate, c. | [19, 20, 23] | ||
| ICCI model | Explains why 2nd phase is enhanced under DAAs | Above a certain level of effectiveness, vRNA is not reduced to a new steady state but rather is declining exponentially in a treatment effectiveness dependent manner | Many unknown parameters. Lack of intracellular data | [61•] | ||
| Colombatto et al. model | Includes serum ALT levels to improve the understanding of infected cells dynamics and its relationship with treatment outcome. Allows for the simulation of the viral kinetics during the whole treatment course. | Estimate the number of infected cells at the end of therapy and accordingly the chance to achieve SVR based on ALT and HCV kinetics during the first month of SOC | Many parameters are assumed and not computed. Lack of experimental data to fully establish the relationship between the infected cells death rate and serum ALT levels | [99, 100, 106] | ||
| Triphasic | first phase (0.5 – 2 days) with rapid virus load decline followed by a shoulder phase (4 – 28 days) – in which virus load decays slowly or remains constant – and a third phase of renewed viral decay | Herrmann et al.. model | Model assumes that IFN and RBV have a delayed immunomodulatory effect | Shoulder phase is explained by very long half life of infected cells (δ~0) | Lack of frequent viral kinetic data in common clinical practice | [101] |
| Extended (Eq. 2) | Includes both uninfected and infected hepatocyte proliferation rates. | Assumes a large fraction of HCV-infected susceptible hepatocytes at baseline | Same as above | [18•, 20] | ||
| Null response or flat-partial response | Less than ~2 log decline throughout treatment | Extended (Eq. 2) or (Eq. 1) with non constant level of target cells | The definition of these viral kinetic patterns varies among studies (explained in main text) | Existence a critical drug effectiveness, εc, These viral kinetic patterns are predicted in cases that total drug effectiveness is lower than εc. | Same as above | [19, 20, 102] |
| Late partial virologic response or viral breakthrough | After weeks of continuous viral decline during SOC viral load spontaneously rebounds from nadir viral load above or below assay limit, respectively. | Extended (Eq. 2) with PEG-IFN pharmacodynamic features | Hepatocytes proliferation rate constant, r, was assumed very low compared to other viral kinetic studies | The model demonstrated excellent positive (99%) and negative (97%) predictive values for SVR. | Hard to distinguish between spontaneous viral rebounds dose reductions or suboptimal drug adherence | [30•] |
| Repeated transient viral rebounds | Repeated oscillations in viral load that fall in all the above viral kinetic patterns | Eq.1 or Eq.2 with the assumption that drug effectiveness, ε, is not constant | Valid for modeling viral rebounds due to change in drug concentrations in serum | Estimate differences in pharmacodynamic parameters among individuals with different race/ethnicity and/or IL28B polymorphism | Lack of frequent drug and viral load concentration data in common clinical practice | [18•, 80•, 103, 104] |
| Rapid emergence of resistance HCV- variants | First phase (0.5 – 2 days) with rapid virus load decline followed by viral increase of resistant virus strains during monotherapy with HCV protease inhibitors | Extension of Eq.1 to two or several viral strains | Several strains of resistant virus are modeled and compete for new cell infection | The resistance emergence is supported by the rapid death of infected cells and the similarly rapid proliferation of new susceptible | Assume rapid hepatocyte turn over to support the early and rapid emergence of resistant HCV strains | [22••, 62] |
| ICCI model | Assumes that viral resistant and wild-type strains competition occurs also intracellularly | Viral mutants preexist intracellularly in most infected hepatocytes; resistance related viral breakthrough are not necessarily a marker of treatment failure; can be followed by a renewed viral decline | Same as above; the effect of deterministic and mean-field approximations is not known | [61•] |
Understanding HCV kinetics in the presence of IFN-based therapies
Biphasic Decline Model
To explain the typical biphasic HCV decline pattern observed during daily IFN treatment, Neumann and colleagues [11] developed in 1998 what we here call the biphasic decline model. This model has been very successful and extensively used and extended over the last 12 years [14–16]. The following system of differential equations describes the biphasic decline model:
| (Eq. 1) |
where I represents the density of infected cells, V represents the virus concentration in serum and To represents the target cell density at the start of therapy, which is assumed to remain constant during the first days of treatment, as previously described [11]. The model assumes that uninfected cells are infected with constant rate β, and infected cells release virions with constant rate p and are lost with a constant rate δ. Finally, virions are cleared from the serum with a constant rate c. The effectiveness of treatment in reducing the rate of new infections is described by η. In the presence of IFN, it was assumed that the average rate of viral production per cell is reduced from p to (1-ε)p, where the effectiveness of IFN in blocking virion production is described by ε, with ε=1 corresponding to a 100% effective drug. For high values of ε, as are obtained by many therapies, the effect of η on the viral load decline predicted by Eq. (1) is negligible and in many models its value is thus set to zero [11, 17•]. The value of η can also be fixed to non-zero values [18•], but in general, the available data are insufficient to estimate it. A summary of fitting results of the biphasic decline model with measured HCV RNA under high daily dose of IFN±RBV treatment is shown in Table 2.
Table 2.
Viral dynamic parameter estimates the using bi-phasic decline model (Eq.1)
| Treatment | N | Delay, t0 [hr] mean (SD) | HCV RNA clearance rate in serum, c [1/day] mean (SD) | HCV RNA half-life, t1/2 [hr] mean (SD) | Effectiveness in blocking viral production/release, ε, mean [%](SD) | Second phase slope λ2 [log10/week] mean (SD) | Loss/death rate of HCV- infected cells, δ [1/day] (SD) |
|---|---|---|---|---|---|---|---|
| 10 or 15 MIU daily IFN ± RBV [11, 105] | 31 | 8.0 (2.4) | 8.0 (4.3) | 2.7 (1.2) | 92.2 (11.2)* | 0.42 (0.36) | 0.14 (0.12) |
| telaprevir monotherapy A | 36 | 2.3 (1.6) | 13.9 (6.3) | 1.4 (0.5) | 97.7 (4.2) | 4.07 (1.97) | 1.34 (0.65) |
SD, one standard deviation.
, Model fits (unpublished data) with the same data used in [17•].
With PEG-IFN plus ribavirin therapy, the average effectiveness ε is approximately 67%±30% [101 and even lower in HCV/HIV coinfected individuals [18•, 103].
Estimates are for subjects infected with HCV genotype 1.
A model that includes hepatocyte proliferation (Extended model)
To explain both the biphasic decline pattern and most of the aforementioned viral kinetic patterns, the biphasic model was extended to include hepatocyte proliferation [19, 20]. The following system of differential equations describes the extended model:
| (Eq. 2) |
Uninfected (T) and infected hepatocytes (I) can proliferate with maximum proliferation rates rT and rI, respectively, according to a blind homeostasis process in which there is no distinction between infected and uninfected cells in determining total liver size [21]. Due to the burdens of supporting HCV replication, Dahari and colleagues [20] assumed that infected cells may proliferate slower than uninfected cells, i.e., rI ≤ rT, a feature that is supported, in part, by in vitro data (Stanley Lemon; personal communication). Rong et al. [22••] have recently introduced the parameter N in order to describe the number of cells in the liver that are “refractory” to infection with HCV. If the total cell population, T+I+N, reaches a maximum level, Tmax, hepatocyte proliferation stops. The model assumes that target cells are produced at a constant rate s from precursors, and die at rate dT per cell. In Table 1 we summarize how the extended model (Eq. 2) and others have been used.
The notion of critical drug efficacy
A pivotal feature of models that include target cell dynamics, such as the extended model (Eq. 2), is the existence of a critical drug efficacy, εc, which determines if the virus will be eventually eradicated (as in SVR subjects) or remain detectable under treatment (such as null/flat partial responders or late viral rebounders, Table 1) [23]. If a drug blocks both infection and viral production, then the total efficacy, defined as εtot = 1 − (1 − ε)(1 − η), must be greater than εc for continuous viral decay. In cases that εtot>εc and the treatment duration is long enough then by the end of therapy the virus is predicted to be eradicated and these individuals will be SVRs. Some non-SVR individuals with continued viral decline with detectable HCV RNA and ≥2-log10 drop in viral levels at week 12 (defined as slow responders) probably would reach SVR with prolonged SOC treatment [24–28] or with more potent regimens such as SOC plus a DAA [29]. Notably, Barreiro et al. (EXTENT Team) presented in a recent meeting (HIV and Liver Disease 2010, Jackson Hole, WY, USA, September 24–26, 2010) that in HIV/HCV coinfected patients, extension of HCV therapy by 3 months improved SVR rates from 9% to 62% in subjects infected with HCV genotype 1/4 who did not achieve a rapid viral response (RVR; undetectable viral load at week 4). The same effect was reported by Barreiro et al. in subjects with genotype 2/3. Interestingly, if εtot<εc, then the theory predicts that HCV RNA levels initially decline but ultimately stabilize at a new steady state (e.g., null responders or flat-partial responders; Table 1) or rebound (e.g., viral breakthrough; Table 1) despite continued therapy and high δ. According to the model, the critical drug efficacy in each infected patient is determined by virus and host parameters such as the viral clearance rate, c, the loss/death rate of HCV-infected cells, δ, and the rates of viral production, p, and cell infection, β (see Eq. 2 in [23]). Next, we show how the notion of critical drug efficacy may explain some of the current clinical observations under SOC treatment.
Explaining why patients with high baseline viral load or advanced liver disease are difficult to treat with SOC
It is well established that patients with high baseline viral load or with advanced liver disease have poorer SVR rates under SOC compared to individuals with low viral load or without advanced liver disease. Dahari and colleagues [23] have simulated, using the extended model (Eq. 2), thousands of in silico patients using for each patient viral and host parameters picked randomly from ranges estimated in the literature. The viral load of the in silico patient population agreed with the observed distribution in US patients [6]. The model predicts that higher baseline viral load leads to a higher critical drug efficacy, εc, which reduces the chance of achieving SVR. In addition, model simulations suggest patients, such as cirrhotic patients, that have at baseline a large fraction of HCV-infected hepatocytes tend to have higher εc thus lower SVR rates. Further, Dahari and colleagues [23] showed that the predictions of the model are consistent with baseline viral loads from 245 cirrhotic and non-cirrhotic patients from the University of Illinois at Chicago.
Predicting late viral rebounds during therapy and SVR
In a recent modeling paper Snoeck et al. [30•], analyzed HCV kinetics in 2000 subjects that were treated with SOC and showed that the extended model predicts spontaneous (i.e., without suboptimal compliance to therapy or dose reductions) late rebounds (after 8 to 40 weeks from initiation of therapy). Such late rebounds occurred in subjects from this study who had undetectable HCV concentrations and were termed viral breakthrough. These observations are interesting as they suggest that in these patients the total drug efficacy is lower than the critical drug efficacy and eventually the viral load will increase spontaneously. Snoeck et al. assumed a very small maximum hepatocyte proliferation rate (doubling time=123 days) in all patients, which in part, allowed the prediction of late rebounds [31]. Other important aspects of the Snoeck et al. approach in using Eq. 2 were the implementation of a PEG-IFN pharmacodynamic model, a cure/viral eradication boundary, the employment of a lower limit of quantification for viral load and implementation of a population modeling approach (see more on population modeling in [14]. Lastly, the authors showed that the model demonstrated excellent predictive values for SVR as well as high sensitivity and specificity.
HCV RNA kinetics under DAA-based treatment
The development of in vitro systems such as HCV replicons [32], and the infectious HCV cell culture system [33–35] have advanced our understanding of the viral lifecycle leading to the identification of a number of putative direct acting agents against both virus and host targets (see review by TenCate et al. [3]). They have also allowed the development of screening assays for chemical libraries resulting in the identification of new targets for antiviral drug discovery (e.g., [36, 37]). Examples of this approach are the discovery of HCV NS4B inhibitor, clemizole [38], and the HCV NS5A inhibitor BMS-790052 [39•].
Many compounds are at the preclinical developmental stage, and some have entered clinical trials. However, concerns have been raised about tolerance and the emergence of drug resistant HCV variants during treatment with DAAs [40, 41]. Based on the lessons learned from HIV therapy, the near future of anti-HCV therapy will likely be a combination approach using a DAA with PEG-IFN/RBV, which will allow for reduced DAA dosing and treatment duration while reducing the chance of drug resistance emergence observed during monotherapy with single DAAs (e.g., [42, 43]; Figure 1, dashed line).
Strategies with DAA+SOC
Final results of the phase III trials (ADVANCE [44] and ILLUMINATE [45]) with the HCV NS3/4A protease inhibitor telaprevir (T) and PEG-IFN-α-2a+RBV (PR) were recently published.. Two regimens using telaprevir in combination with PEG-IFN plus RBV (TPR) were tested (i.e., 8 weeks vs. 12 weeks) in the ADVANCE study, followed by 24 or 48 weeks with PR in eRVR (defined as undetectable HCV RNA at weeks 4 and 12) patients and non-eRVR patients, respectively. In the ILLUMINATE study TPR treatment of 12 weeks was followed by 24 or 48 weeks of PR. Patients who achieved eRVR were randomized at week 20 to continue receiving PR for 24 or 48 weeks of total treatment; non-eRVR patients were assigned for 48 weeks of treatment. Both phase III studies had a control arm, i.e., PR for 48 weeks. Among patients who achieved eRVR (ILLUMINATE study), a 24-week telaprevir-based regimen was non-inferior to 48-week telaprevir-based regimen (92% SVR compared to 88%) and overall 72% of patients achieved SVR. In the ADVANCE study a significantly (P<0.0001) greater proportion of patients achieved SVR with 12-week and 8-week TPR regimens (75% and 69%, respectively) compared with PR48 control arm (44%). Kieffer et al. [46] performed viral dynamic analyses in a subset of patients in whom viral genome sequencing data was available (N=91). They suggest that week-12 regimen of TPR is necessary to exert an optimal antiviral pressure on wild-type virus and lower-level telaprevir resistant variants, thus enhancing response rates.
Lead-in strategy with SOC
While with telaprevir-based regimens (i.e., SOC+telaprevir) were followed with SOC alone others have tested a lead-in strategy in which SOC is given for 3 days to 4 weeks after which a DAA is added to the drug cocktail [47–50]. This approach has three theoretical advantages. First, it allows one to identify patients with null or a very limited response to SOC; for these patients, addition of a single DAA may increase the risk of drug resistance and viral breakthrough because the triple therapy is like a form of DAA monotherapy [51]. Second, the lead-in allows PEG-IFN and RBV to reach steady state levels so that when a DAA is added to the SOC it is already highly active against resistant virus. Lastly, SOC will lead to a lower baseline viral load at DAA initiation, and thus will reduce the chance of resistance emergence [22••].
In a recent publication, Kwo et al. [47] described the final results of the open-label phase II trial (SPRINT-1) with boceprevir, an HCV NS3/4A protease inhibitor. Five hundred and twenty treatment-naïve patients with HCV genotype-1 were randomly assigned to PEG-IFN-α-2b+ribavirin treatment (PR) for 48 weeks (PR48 or SOC); PR for 4 weeks, followed by PR+boceprevir for 24 weeks (PR4/PRB24) or 44 weeks (PR4/PRB44); or PRB for 28 weeks (PRB28) or 48 weeks (PRB48). All four boceprevir groups (PRB28, PR4/PRB24, PRB48, and PR4/PRB44) had significantly higher SVR rates (54%, 56%, 67% and 75%, respectively) than under SOC (38%), with the highest SVR rate with PR4/PR44. The investigators of SPRINT-1 advocate for the use of a lead-in approach before the addition of boceprevir over a 48-week duration of treatment as recently tested in the phase-III (SPRINT-2) trial [109]. In another part of the SPRINT-1 trial 75 patients were randomly assigned to receive either PRB48 (N=16) or low-dose ribavirin plus PB for 48 weeks. The low-dose ribavirin arm was associated with higher rates of viral breakthrough and relapse compared to PRB48 and similar to SOC alone.
Understanding HCV RNA kinetics under DAA-based treatment
In most patients, viral kinetics after initiation of DAA treatment is characterized by a short delay, followed by a biphasic decline that consists of first rapid viral decline phase followed by a slower phase slope (Figure 1, red or blue lines) [17•, 39•, 52, 53]. Interestingly, both the duration and the kinetics of each phase differ significantly from what is observed during IFN-based therapy (Figure 1 and Table 2). Moreover, some patients treated with protease inhibitor monotherapy have experienced a resistance related viral breakthrough [43, 54] as illustrated in Figure 1 (dashed line).
The delay before HCV RNA declines after initiation of treatment (t0)
After initiation of IFN-based therapy a delay, t0, is observed before viral RNA begins declining. This delay was estimated as ~8 hr using the biphasic decline model (Table 2). Recently, modeling IFN inhibition kinetics in replicons showed that the intracellular subgenomic HCV RNA concentration drops from baseline levels about 10 hr after administration of IFN [12]. The in vitro data suggest that IFN signaling and subsequent interferon stimulated gene induction are the main causes of the observed delay in vivo rather than (PEG)-IFN pharmacokinetics. It is expected that DAAs will lead to dramatically shorter lags, due to their direct affect on the HCV life cycle. In fact, drops in viremia have been observed as early as two hours after telaprevir (Table 2) BILN-2061 [55] and BMS-790052 [39•] initiation.
Magnitude of the first phase of viral decline
After this initial delay, the profound antiviral effect of DAAs gives rise in most patients to a first phase viral decline of 2–4 log IU ml−1 from baseline (equivalent to 99%<ε<99.99%) with HCV NS3 protease inhibitors [17•, 52, 56], or BMS-790052, an HCV NS5A inhibitor [39•], and somewhat lower declines with other DAAs such as the nucleoside and non-nucleoside HCV polymerase inhibitors among others (see Table 1 in [51]).
Plasma HCV clearance rate, c, estimates
By fitting the biphasic decline model (Eq.1) to HCV RNA measurements obtained from patients treated with IFN, c was estimated for genotype 1 virus as 8.0 d−1 on average, i.e., t1/2=2.7 hours (Table 2). Interestingly, this estimated t1/2 is significantly larger than what has been estimated for HIV [57], a similar sized virus. Since c represents a physiological quantity, it is expected that the value of c estimated from data obtained during DAA therapy would be remain unchanged, as has been reported with the protease inhibitors BILN-2061 [55], TMC-435 [58], MK-7009 [56] and R7227 in combination with PEG-IFN/RBV or with R7128, an HCV nucleoside polymerase inhibitor [53]. Interestingly, the value of c estimated from genotype 1 patients treated with telaprevir has been reported as c = 13.9 d−1 [17•], which corresponds to a t1/2 = 1.4 hours (Table 2) and the NS5A inhibitor BMS-790052 [39•] appears to generate an even more rapid HCV RNA decline, i.e., c=22.9 d−1 which corresponds to a t1/2 = 0.74 hours [59•]. The reasons underlying these discrepancies in the estimated values of c remain to be identified.
The second phase of viral decline and the net loss of infected cells, δ
Under high treatment effectiveness (ε~1), both the standard and extended models attribute the rate of second phase viral decline, λ2, to the net loss of infected cells, δ [60]. Yet, protease inhibitors result in most patients in a second phase viral decline λ2> 1 log10/week [17•, 39•, 52, 55] as compared to λ2<1 log10/week in most patients treated with IFN (Figure 1 and Table 2). To understand these differences, models by Dahari et al. [12] and Guedj and Neumann [61•] have suggested that intracellular HCV RNA may undergo a biphasic decline during potent therapy. Under such circumstances, the rate of the in vivo second phase could be the combined result of the rate of death of infected and the rate of loss of the ability of the remaining infected cells to produce virus as their intracellular RNA degrades [61•]. Guedj and Neumann further assumed [61•] that the rate of loss of the ability to produce virus depends on the treatment effectiveness, ε, thus providing an explanation for the viral kinetics observed in patients who were treated with telaprevir [62]. More details on this issue can be found in a recent modeling review [14].
Models predict the existence of HCV drug resistance variants before therapy
Whereas the complex intracellular signaling pathways involved with the response to IFN appears to limit the emergence of resistant virus, only a single nucleotide change may be associated with resistance to DAAs. Given the large number of virions produced every day and the error prone nature of the HCV RNA-dependent RNA polymerase, it has been predicted that all possible single and double mutants are generated multiple times each day [22••]. Hence all viable single and double mutants that confer drug resistance should preexist and may compete with the wild-type virus during therapy [62]. This has been discussed extensively in a recent review [51].
Modeling the emergence of drug-resistant variants during therapy
In order to model the emergence of drug resistant virus, the model given by Eq. (1) or Eq. (2) has been extended to include both drug sensitive or wild-type (WT) virus and other drug resistant strains of virus. Rong et al. [22••] considered only a single strain of resistant virus, where Bambang et al. [62] considered multiple strains. Both models fit data, but found that in order to generate high levels of resistant virus they had to be a high rate of hepatocyte turnover. Guedj and Neumann [61•] in a theoretical model explored the possibility that intracellular competition between WT and drug resistant HCV RNA could occur within infected cells and lead to viral breakthrough without resistant virus infecting new cells. Theoretically, if the latter hypothesis is true, under treatment with an HCV NS3 inhibitor (such as telaprevir) in combination with a potent HCV entry inhibitor (that are under development [3]), the rapid emergence of resistant virus observed during protease inhibitor monotherapy would still occur.
Also, models predict that resistant virus may take over the viral population without giving rise to a viral rebound. In this case the rapid decline of WT virus may be smoothly replaced by a slower decline of resistant virus due to its lower sensitivity to treatment. This switch may be unnoticeable if the viral load is already under the level of detection. Hence, a shortening of the treatment duration based on a rapid virological response could be misleading and give rise to a post-treatment relapse with resistant virus, as observed in some patients receiving one DAA and the standard of care [63, 64].
IL28B polymorphisms, viral kinetics/dynamics and outcome of therapy
From September 2009 to January 2010 four large independent genome-wide association studies (GWAS) described a novel association between single nucleotide polymorphisms (SNPs) near the interleukin 28B (IL28B) gene locus and the response to HCV treatment with the standard of care [65–68]. Suppiah et al. [67] and Tanaka et al. [66] described rs8099917 (8 kb upstream of IL28B) as the SNP with the strongest association with SVR in subjects of European and Japanese ancestry, respectively. Ge et al. [65] and Rauch et al. [68] identified rs12979860 (3 kb upstream of IL28B) as the SNP with the strongest association with treatment response in individuals from North America [69–71]. Recently, a high-sensitivity technique based on DNA genotyping in serum has been developed [72].
Favorable alleles and their association with outcome of therapy
Ge et al. [65] found that individuals with the CC allele (SNP rs12979860) had a 2-fold greater rate of SVR than those with the TT allele. Rauch et al. [68] found a 2-fold greater rate of treatment failure in individuals carrying one or two copies of the rs8099917 risk G-allele (the favorable allele was termed TT allele). Similar trends were found by the other two GWAS. McCarthy et al. [73] performed a multivariate analysis that included age, gender, HCV genotype, treatment history, and fibrosis and demonstrated that the CC allele (rs12979860) conferred almost a 6-fold increased odds ratio for SVR. In addition, the CC allele predicted SVR with 78% specificity and 65% sensitivity in patients infected with HCV genotype 1. Recently, Stattermayer et al. [74] showed that the favorable alleles (TT (rs8099917) and CC (rs12979860)), in Austrian subjects infected with HCV genotype 1, had positive predictive values (PPV) of 81% and 72%, respectively and negative predictive values (NPV) of 59% and 58%, respectively, for SVR. It should be noted that none of the studies published to date have found high NPV for non-SVR. Therefore, to increase the prediction rates of nonresponse, additional markers need to be found and combined with IL28B polymorphisms. Interestingly, Lagging et al. (Oral presentation (O-35) at the 17th International Meeting on HCV and Related Viruses, September 10–14, 2010, Yokohama, Japan) reported an association between baseline plasma levels of interferon gamma-induced protein 10 kDa, IP-10 [75] and IL28B polymorphisms in 253 Caucasian patients infected with HCV genotype 1 or 2; and the favorable alleles were significantly correlated with lower baseline IP-10. They also reported that the combination of baseline IP-10<150 pg/ml, the decline in HCV RNA at day 4 and the favorable allele achieved increased rates of SVR rates (75% to 85%) among subjects infected with HCV genotype-1. Another recent report supports this approach [76]. Interestingly, Sarrazin et al. [107] have recently reported that IL28B genes were not associated with SVR in patients who achieved an RVR, i.e., had undetectable viral load at week 4.
Association between viral kinetics, pharmacodynamic parameters and IL28B polymorphisms
Detailed HCV kinetic analyses provided new and important information regarding the impact of IL28B genotype on early viral response to PEG-IFN plus RBV in treated patients. The favorable alleles (TT (rs8099917) or CC (rs12979860)) were most strongly associated with a higher first phase (during the first days from initiation of treatment) viral decline from baseline than in subjects with risk alleles (mean difference of ~0.5 log) [74, 77–79]. The same pattern was found [80•] in a small cohort of HIV/HCV(genotype 1/3) coinfected patients from Brazil [18•, 81] treated with SOC for 48 weeks. In the latter study, detailed viral kinetic and pharmacodynamic parameters, estimated via mathematical modeling (Eqs. 1 and 2), suggest that the average PEG-IFN- α-2a effectiveness in blocking production/release of virions from infected cells, ε, during the first week of therapy was significantly higher in patients with CC allele compared with patients with T alleles (92% vs. 77%, respectively). In addition, the PEG-IFN-α-2a concentration at which the drug’s effectiveness in blocking viral production is half its maximum, EC50, was lower in patients with the CC allele compared in patients with T alleles, indicating that the CC allele confers a higher sensitivity to IFN treatment. The mechanism of action of the IL28B polymorphism is not known, however, since these SNPs are located near the IL28B gene they might mediate endogenous production of IFN-λ, which in turn could contribute to the first phase response by stimulating IFN signaling and reducing viral production [82]. The interesting association among baseline interferon stimulated genes levels in the liver, IL28B polymorphisms and treatment response will require further study [83].
IL28B polymorphism and the rate of second phase viral decline
Differences in the viral decline slope (beyond the first phase) between patients carrying the favorable allele and risk alleles were found by Thompson et al. [84]. Having very frequent viral load sampling during 4 weeks of SOC, Araujo et al. [80•] found that the second phase slope of viral decline (λ2, measured between days 2–29) and the infected cell loss rate, δ, were larger in HIV/HCV(genotype-1) patients with the CC (rs12979860) allele than in patients with T alleles (λ2= 0.54 and 0.22 log/wk, respectively; δ=0.20 and 0.11 d−1, respectively). Mangia et al. [108] reported that the CC (rs12979860) allele was highly predictive of RVR which reflects, in part, high λ2. Although there is no direct evidence that δ estimated from the second phase of HCV RNA decline is the loss rate of infected cells, Neumann et al. [11] and Zeuzem et al. [85] noted that δ was correlated with baseline alanine aminotransferase (ALT), a surrogate marker of liver cell necrosis. In addition, Pilli et al. [86] found that patients with rapid viral declines were associated with high HCV-specific CD8+ T cell proliferative responses at baseline. Interestingly, in a large cohort study (N=1364) with subjects infected with HCV genotype-1, higher baseline ALT levels and more common necro-inflammatory activity (METAVIR; A2-3) [87], but not advanced hepatic fibrosis [88], were associated with patients with the CC (rs12979860) allele. However, in 304 HIV/HCV-coinfected individuals the IL28B rs12979860 SNP was associated with a higher prevalence of cirrhosis, suggesting that IL28B CC genotype is associated with more rapid progression of fibrosis in patients with chronic HCV infection, perhaps by increasing liver inflammation [89]. These observations might support the concept that subjects with the CC allele have a higher second phase slope of viral decline (or δ), predominately due to increased cellular immune responses, which would explain the observed higher ALT levels, higher rates of SVR and increased necro-inflammatory activity.
Lastly, Chevaliez et al. (poster presentation (P-280) at the 17th International Meeting on HCV and Related Viruses, Sep. 10–14, 2010, Yokohama, Japan) reported results from the SYREN trial that included patients infected with HCV genotype 1 who did not respond to previous SOC. They compared early viral kinetic (at weeks 1, 2 and 4) and found that patients with the CT (rs12979860) allele had significantly higher viral declines than patients with the TT allele (only 3% of their cohort were patients with CC allele thus were removed from the analysis). Only 5 patients achieved SVR and all had the CT allele. This finding suggests that patients with the CT allele had a faster second phase slope of viral decline under SOC than patients with the TT allele.
IL28B polymorphisms and HCV genotypes
A higher proportion of the favorable allele was found in patients infected with HCV-genotype 2/3 than in patients infected with HCV-genotype 1 for both HIV/HCV-coinfected [80•, 90], and HCV-monoinfected individuals [91]. The reason for this is not known and might be partly related to HCV evolution driven by immune selection [92]. This may partly explain why patients infected with genotype 2/3 respond better to SOC than patients with genotype 1. Interestingly, using Eq. 2 we have previously shown that patients infected with HCV genotype 2 had significantly higher sensitivity to IFN, ε, a higher viral clearance rate, c, and a higher infected loss/death rate, δ, than patients infected with genotype 1 [13]. Similar differences were found in HIV/HCV coinfected patients infected with genotype 3 compared to patients infected with genotype 1 [18•].
Race/ethnicity, viral kinetics/dynamics and IL28 polymorphisms
Studies of HCV mono-infected patients conducted in the United States reported a higher proportion of risk alleles in African Americans than in non-Hispanic Caucasians [65, 77]. Interestingly, we did not identify a significant (p=0.5) difference in IL28B allele frequencies between African Americans and Caucasian HIV/HCV coinfected patients from Brazil [80•]. The lack of an association between race and IL28B genotype in this South American patient population might partly explain the lack of association between race/ethnicity and viral kinetic parameters or viral response patterns in our recent reports [18•, 81]. A larger retrospective analysis in 500 HCV mono-infected individuals from Brazil indicates that about 80% had risk alleles (TC or TT; rs12979860) with a similar distribution of CC/TC/TT genotypes between African Americans and Caucasians [93]. Why patient ancestry may differ in Brazil from that in the United States is not known. The results may explain why SVR rates under SOC are lower in Brazil than in North America. Very recently, it was reported that 80% of patients from Puerto Rico, Florida or Texas had risk alleles versus 60% in San Francisco and Seattle, indicating that IL28B genotype distribution may vary geographically within the United States [94].
IL28B polymorphism and DAA-based therapies
While it is anticipated that IL28B genotype will play a role in determining treatment outcome with SOC+DAAs it is less clear under DAAs alone (i.e., IFN free regimens). Akuta et al. [95] investigated the predictive factors of SVR to a 12-week or 24-week regimen of telaprevir+PEG-IFN-α-2b+ribavirin (TPR) therapy in 72 Japanese adults (treatment-experienced and treatment-naïve) infected with HCV genotype 1b (only one patient was infected with genotype 1a). SVR rates were 45% and 67% in the TPR12 or TPR24 regimens, respectively. Patients with the favorable alleles (TT (rs8099917) and CC (rs12979860)) had significantly higher SVR rates (84%) than in patients with the risk alleles (28% and 32%, respectively). The authors found high sensitivity (80%), specificity (78%), PPV (84%) and NPV (72%) values for SVR according to genotype TT (rs8099917). Increased predictive values (e.g., NPV=88%) were found with TT genotype in combination with amino acid substitutions at position 70 in the HCV core region. Muir et al. [96] reported that among subjects with the CC (rs12979860) allele (HCV genotype-1), the addition of ANA598, a non-nucleoside inhibitor of HCV polymerase, to SOC improved the percent of individuals who reached undetectable levels compared with patients treated with SOC plus placebo (40% vs. 0% and 70% vs. 18% at weeks 1 and 2, respectively); however, the difference was significantly weakened by week 12 (90% vs. 82%, respectively). Their small study suggests that ANA598 accelerates the rate of achieving undetectable levels of HCV RNA (~20% increase by week 12) in patients with T alleles.
Conclusion
Mathematical modeling of HCV RNA kinetics during treatment with various direct anti-HCV agents (DAA) pose new challenges and has the potential to reveal new insights about the HCV lifecycle and HCV-host dynamics. Further studies on the role of IL28B polymorphisms, viral kinetics, baseline characteristics and other new markers such as IP-10 in predicting response are needed to optimize therapeutic regimens with SOC alone and in combination with DAAs. The immediate challenge will likely be to optimize therapeutic regimens that include a DAA (such as telaprevir and boceprevir) in order to avoid unnecessary treatments and the emergence of DAA resistance strains in patients that have poor chance to achieve SVR. This may be very relevant for thousands of non-SVR patients (and their physicians) who are waiting to be treated with new drugs.
Acknowledgments
Portions of this work were done under the auspices of the U. S. Department of Energy under contract DE-AC52-06NA25396 and supported by NIH grants AI28433, RR06555, P20-RR18754 and R56-AI078881 and by the University of Illinois Walter Payton Liver Center GUILD.
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1. [Accessed July 13, 2009];Hepatitis C fact sheet. Available from: [ http://www.who.int/mediacentre/factsheets/fs164/en/]
- 2.Davis GL, Alter MJ, El-Serag H, et al. Aging of hepatitis C virus (HCV)-infected persons in the United States: a multiple cohort model of HCV prevalence and disease progression. Gastroenterology. 2009;138(2):513–521. 521 e511–516. doi: 10.1053/j.gastro.2009.09.067. [DOI] [PubMed] [Google Scholar]
- 3.TenCate V, Sainz B, Jr, Cotler SJ, Uprichard SL. Potential treatment options and future research to increase hepatitis C virus treatment response rate. Hepatic Medicine: Evidence and Research. 2010 doi: 10.2147/HMER.S7193. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawlotsky JM, Dahari H, Neumann AU, et al. Antiviral action of ribavirin in chronic hepatitis C. Gastroenterology. 2004;126(3):703–714. doi: 10.1053/j.gastro.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen TT, Sedghi-Vaziri A, Wilkes LB, et al. Fluctuations in viral load (HCV RNA) are relatively insignificant in untreated patients with chronic HCV infection. J Viral Hepat. 1996;3(2):75–78. doi: 10.1111/j.1365-2893.1996.tb00084.x. [DOI] [PubMed] [Google Scholar]
- 6.Nainan OV, Alter MJ, Kruszon-Moran D, et al. Hepatitis C virus genotypes and viral concentrations in participants of a general population survey in the United States. Gastroenterology. 2006;131(2):478–484. doi: 10.1053/j.gastro.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347(13):975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 8.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358(9286):958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 9.Zeuzem S, Fried MW, Reddy KR, et al. Improving the clinical relevance of pre-treatment viral load as a predictor of sustained virological response (SVR) in patients infected with hepatitis C genotype 1 treated with peginterferon alfa-2a (40KD) (PEGASYS®) plus ribavirin (COPEGUS®) Hepatology. 2006;44(Suppl 1):267A–268A. [Google Scholar]
- 10.Zehnter E, Mauss S, John C, et al. Better prediction of SVR in patients with HCV genotype 1 (G1) with peginterferon alfa-2a (PEGASYS) plus ribavirin: Improving differentiation between low (LVL) and high baseline viral load (HVL) Hepatology. 2006;44(Suppl 1):328A. [Google Scholar]
- 11.Neumann AU, Lam NP, Dahari H, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282(5386):103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 12.Dahari H, Sainz B, Jr, Perelson AS, Uprichard SL. Modeling subgenomic hepatitis C virus RNA kinetics during treatment with alpha interferon. J Virol. 2009;83(13):6383–6390. doi: 10.1128/JVI.02612-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neumann AU, Lam NP, Dahari H, et al. Differences in viral dynamics between genotypes 1 and 2 of hepatitis C virus. J Infec Dis. 2000;182(1):28–35. doi: 10.1086/315661. [DOI] [PubMed] [Google Scholar]
- 14.Guedj J, Rong L, Dahari H, Perelson AS. A perspective on modelling hepatitis C virus infection. J Viral Hepat. 2010;17:825–833. doi: 10.1111/j.1365-2893.2010.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rong L, Perelson AS. Treatment of hepatitis C virus infection with interferon and small molecule direct antivirals: viral kinetics and modeling. Crit Rev Immunol. 2010;30(2):131–148. doi: 10.1615/critrevimmunol.v30.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahari H, Layden-Almer JE, Perelson AS, Layden TJ. Hepatitis C Viral Kinetics in Special Populations. Curr Hepat Rep. 2008;7(3):97–105. doi: 10.1007/s11901-008-0022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Adiwijaya BS, Hare B, Caron PR, et al. Rapid decrease of wild-type hepatitis C virus on telaprevir treatment. Antivir Ther. 2009;14(4):591–595. Using the biphasic decline model (Eq. 1), the authors suggested that telaprevir effectiveness in blocking HCV production/release and the loss rate of infected cells are significantly higher than with IFN-based therapy. [PubMed] [Google Scholar]
- 18•.Dahari H, Araújo ES, Haagmans BL, et al. Pharmacodynamics of PEG-IFN-alpha-2a in HIV/HCV co-infected patients: Implications for treatment outcomes. J Hepatol. 2010;53(3):460–467. doi: 10.1016/j.jhep.2010.03.019. This is the first modeling paper that couples PEG-IFN-α-2a plasma concentrations with its effectiveness in blocking HCV production, and treatment outcome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahari H, Lo A, Ribeiro RM, Perelson AS. Modeling hepatitis C virus dynamics: Liver regeneration and critical drug efficacy. J Theor Biol. 2007;247(2):371–381. doi: 10.1016/j.jtbi.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahari H, Ribeiro RM, Perelson AS. Triphasic decline of hepatitis C virus RNA during antiviral therapy. Hepatology. 2007;46(1):16–21. doi: 10.1002/hep.21657. [DOI] [PubMed] [Google Scholar]
- 21.Dahari H, Major M, Zhang X, et al. Mathematical modeling of primary hepatitis C infection: Noncytolytic clearance and early blockage of virion production. Gastroenterology. 2005;128(4):1056–1066. doi: 10.1053/j.gastro.2005.01.049. [DOI] [PubMed] [Google Scholar]
- 22••.Rong L, Dahari H, Ribeiro RM, Perelson AS. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci Transl Med. 2010;2(30):30ra32. doi: 10.1126/scitranslmed.3000544. This theoretical study explains the observed rapid emergence of telaprevir resistant strains during monotherapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dahari H, Layden-Almer JE, Kallwitz E, et al. A mathematical model of hepatitis C virus dynamics in patients with high baseline viral loads or advanced liver disease. Gastroenterology. 2009;136(4):1402–1409. doi: 10.1053/j.gastro.2008.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berg T, von Wagner M, Nasser S, et al. Extended treatment duration for hepatitis C virus type 1: comparing 48 versus 72 weeks of peginterferon-alfa-2a plus ribavirin. Gastroenterology. 2006;130(4):1086–1097. doi: 10.1053/j.gastro.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez-Tapias JM, Diago M, Escartin P, et al. Peginterferon-alfa2a plus ribavirin for 48 versus 72 weeks in patients with detectable hepatitis C virus RNA at week 4 of treatment. Gastroenterology. 2006;131(2):451–460. doi: 10.1053/j.gastro.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Brouwer JT, Nevens F, Bekkering FC, et al. Reduction of relapse rates by 18-month treatment in chronic hepatitis C. A Benelux randomized trial in 300 patients. J Hepatol. 2004;40(4):689–695. doi: 10.1016/j.jhep.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 27.Ferenci P, Laferl H, Scherzer TM, et al. Peginterferon alfa-2a/ribavirin for 48 or 72 weeks in hepatitis C genotypes 1 and 4 patients with slow virologic response. Gastroenterology. 2010;138(2):503–512. 512, e501. doi: 10.1053/j.gastro.2009.10.058. [DOI] [PubMed] [Google Scholar]
- 28.Buti M, Lurie Y, Zakharova NG, et al. Randomized trial of peginterferon alfa-2b and ribavirin for 48 or 72 weeks in patients with hepatitis C virus genotype 1 and slow virologic response. Hepatology. 2010;52(4):1201–1207. doi: 10.1002/hep.23816. [DOI] [PubMed] [Google Scholar]
- 29.McHutchison JG, Manns MP, Muir AJ, et al. Telaprevir for previously treated chronic HCV infection. N Engl J Med. 2010;362(14):1292–1303. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 30•.Snoeck E, Chanu P, Lavielle M, et al. A comprehensive hepatitis C viral kinetic model explaining cure. Clin Pharmacol Ther. 2010;87(6):706–713. doi: 10.1038/clpt.2010.35. This study models various observed HCV kinetics (including late viral rebounds such as viral breakthrough) during SOC and predicts SVR. Some features of the model are discussed in [31] [DOI] [PubMed] [Google Scholar]
- 31.Dahari H, Rong L, Layden TJ, Cotler SJ. Hepatocyte proliferation and hepatitis C virus (HCV) kinetics during treatment. Clin Pharmacol Ther. 2010 doi: 10.1038/clpt.2010.238. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436(7053):933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 33.Zhong J, Gastaminza P, Cheng G, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102(26):9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wakita T, Pietschmann T, Kato T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11(7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindenbach BD, Evans MJ, Syder AJ, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 36.Einav S, Gerber D, Bryson PD, et al. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat Biotechnol. 2008;26(9):1019–1027. doi: 10.1038/nbt.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu X, Uprichard SL. Curr Protoc Microbiol. Unit 17. Chapter 17. 2010. Cell-based hepatitis C virus infection fluorescence resonance energy transfer (FRET) assay for antiviral compound screening; p. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho NJ, Dvory-Sobol H, Lee C, et al. Identification of a class of HCV inhibitors directed against the nonstructural protein NS4B. Sci Transl Med. 2010;2(15):15ra16. doi: 10.1126/scitranslmed.3000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Gao M, Nettles RE, Belema M, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465(7294):96–100. doi: 10.1038/nature08960. This study provides the first clinical validation of an inhibitor of HCV NS5A (BMS-790052). Viral kinetics after one dose of BMS-790052 was characterized by a rapid biphasic decline that persisted in some patients give a 100 mg dose for as long as 6 days. Using the biphasic decline model, Eq. 1, it has been shown that the HCV clearance rate under BMS-790052, is significantly higher than under telaprevir or IFN [59] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pawlotsky JM. Hepatitis virus resistance. In: Fong I, Drlica K, editors. Antimicrobial Resistance and Implications for the 21st Century. Spirnger; 2008. pp. 291–323. [Google Scholar]
- 41.Thompson AJ, McHutchison JG. Antiviral resistance and specifically targeted therapy for HCV (STAT-C) J Viral Hepat. 2009;16(6):377–387. doi: 10.1111/j.1365-2893.2009.01124.x. [DOI] [PubMed] [Google Scholar]
- 42.Forestier N, Reesink HW, Weegink CJ, et al. Antiviral activity of telaprevir (VX-950) and peginterferon alfa-2a in patients with hepatitis C. Hepatology. 2007;46(3):640–648. doi: 10.1002/hep.21774. [DOI] [PubMed] [Google Scholar]
- 43.Kieffer TL, Sarrazin C, Miller JS, et al. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology. 2007;46(3):631–639. doi: 10.1002/hep.21781. [DOI] [PubMed] [Google Scholar]
- 44.Jacobson IM, McHutchison JG, Dusheiko GM, et al. Telaprevir in Combination with Peginterferon and Ribavirin in Genotype 1 HCV Treatment-Naïve Patients: Final Results of Phase 3 ADVANCE Study. Hepatology. 2010;52(Suppl):427A. [Google Scholar]
- 45.Sherman KE, Flamm SL, Afdhal NH, et al. Telaprevir in Combination with Peginterferon Alfa2a and Ribavirin for 24 or 48 weeks in Treatment-Naïve Genotype 1 HCV Patients who Achieved an Extended Rapid Viral Response: Final Results of Phase 3 ILLUMINATE Study. Hepatology. 2010;52(Suppl):401A. [Google Scholar]
- 46.Kieffer TL, Bartels DJ, Sullivan J, et al. Clinical Virology Results from Telaprevir Phase 3 Study ADVANCE. Hepatology. 2010;52(Suppl):879A. [Google Scholar]
- 47.Kwo PY, Lawitz EJ, McCone J, et al. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet. 2010;376(9742):705–716. doi: 10.1016/S0140-6736(10)60934-8. [DOI] [PubMed] [Google Scholar]
- 48.Vierling J, Poordad F, Lawitz E, et al. Once daily narlaprevir (SCH 900518) in combination with peginterferon alfa-2b/ribavirin for treatment-naive patients with genotype-1 chronic hepatitis C: interim results from the NEXT-1 study. Hepatology. 2009 [Google Scholar]
- 49.Sulkowski M, Bourliere M, Bronowicki J-P, et al. SILEN-C2: early antiviral activity and safety of BI 201335 combined with pegintergeron alfa-2a and ribavirin (PEGIFN/RBV) in chronic HCV genotype-1 patients with non-response to PEGIFN/RBV. Journal of Hepatology. 2010;52 (Suppl 1):S462–S463. [Google Scholar]
- 50.Sulkowski MS, Ferenci P, Emanoil C, et al. SILEN-C1: early antiviral activity and safety of BI 201335 combined with peginterferon alfa-2a and ribavirin in treatment-naïve patients with chronic genotype 1 HCV infection. The 60th meeting of the American Association for the Study of Liver Diseases (AASLD); Boston, MA, USA. 2009. p. Abstract LB3. [Google Scholar]
- 51.Sarrazin C, Zeuzem S. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology. 2010;138(2):447–462. doi: 10.1053/j.gastro.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 52.Reesink HW, Fanning GC, Farha KA, et al. Rapid HCV-RNA decline with once daily TMC435: a phase I study in healthy volunteers and hepatitis C patients. Gastroenterology. 2010;138(3):913–921. doi: 10.1053/j.gastro.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 53.Morcos PN, Kulkarni R, Ipe D, et al. Pharmacokinetics/pharmacodynamics (PK/PD) of combination R7227 and R7128 therapy from INFORM - 1 demonstrates similar early HCV viral dynamics when R7227 is combined with either PEG-IFN/Ribavirin (SOC) or R7128. Hepatology. 2009;50(S4):1041A. [Google Scholar]
- 54.Sarrazin C, Kieffer TL, Bartels D, et al. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology. 2007;132(5):1767–1777. doi: 10.1053/j.gastro.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 55.Herrmann E, Zeuzem S, Sarrazin C, et al. Viral kinetics in patients with chronic hepatitis C treated with the serine protease inhibitor BILN 2061. Antiviral Therapy. 2006;11(3):371–376. [PubMed] [Google Scholar]
- 56.Panorchan P, Nachbar R, Saltzman J, et al. Evaluation of the dose-response relationship to short-term monotherapy with the hcv protease inhibitor, mk-7009. 2nd American Conference on Pharmacometrics (ACoP).; 2009. [Google Scholar]
- 57.Ramratnam B, Bonhoeffer S, Binley J, et al. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet. 1999;354(9192):1782–1785. doi: 10.1016/S0140-6736(99)02035-8. [DOI] [PubMed] [Google Scholar]
- 58.Reesink HW, Fanning GC, Farha KA, et al. Rapid HCV-RNA decline with once daily TMC435: a phase I study in healthy volunteers and hepatitis C patients. Gastroenterology. 2010;138(3):913–921. doi: 10.1053/j.gastro.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 59•.Dahari H, Guedj J, Cotler SJ, et al. Higher hepatitis C virus (HCV) clearance rates during treatment with direct acting agents compared to interferon-alpha. Hepatology. 2010;52(Suppl):718A–719A. This study shows that the HCV clearance rate in serum is higher than previuosly estimated under IFN-based treatments and suggests that in the era of DAAs intracellular features of HCV dynamics are needed to be included in models in order to explain these new HCV kinetics. [Google Scholar]
- 60.Dahari H, Shudo E, Cotler SJ, et al. Modelling hepatitis C virus kinetics: the relationship between the infected cell loss rate and the final slope of viral decay. Antiviral Therapy. 2009;14(3):459–464. [PMC free article] [PubMed] [Google Scholar]
- 61•.Guedj J, Neumann AU. Understanding hepatitis C viral dynamics with direct-acting antiviral agents due to the interplay between intracellular replication and cellular infection dynamics. J Theor Biol. 2010;267(3):330–340. doi: 10.1016/j.jtbi.2010.08.036. This theoretical paper explores the possibility that the new patterns of viral kinetics observed with DAAs (rapid decline of wild type virus and drug resistance related viral breakthrough) may be attributed to the dynamics of intracellular HCV RNA. [DOI] [PubMed] [Google Scholar]
- 62.Adiwijaya BS, Herrmann E, Hare B, et al. A multi-variant, viral dynamic model of genotype 1 HCV to assess the in vivo evolution of protease-inhibitor resistant variants. PLoS Comput Biol. 2010;6(4):e1000745. doi: 10.1371/journal.pcbi.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hezode C, Forestier N, Dusheiko G, et al. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N Engl J Med. 2009;360(18):1839–1850. doi: 10.1056/NEJMoa0807650. [DOI] [PubMed] [Google Scholar]
- 64.McHutchison JG, Everson GT, Gordon SC, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360(18):1827–1838. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- 65.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41(10):1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 67.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41(10):1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 68.Rauch A, Kutalik Z, Descombes P, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138(4):1338–1345. 1345 e1331–1337. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 69.Rauch A, Rohrbach J, Bochud PY. REVIEW: The recent breakthroughs in the understanding of host genomics in hepatitis C. Eur J Clin Invest. 2010;40(10):950–959. doi: 10.1111/j.1365-2362.2010.02337.x. [DOI] [PubMed] [Google Scholar]
- 70.Ahlenstiel G, Booth DR, George J. IL28B in hepatitis C virus infection: translating pharmacogenomics into clinical practice. J Gastroenterol. 2010;45(9):903–910. doi: 10.1007/s00535-010-0287-4. [DOI] [PubMed] [Google Scholar]
- 71.Imazeki F, Yokosuka O, Omata M. Impact of IL-28B SNPs on control of hepatitis C virus infection: a genome-wide association study. Expert Rev Anti Infect Ther. 2010;8(5):497–499. doi: 10.1586/eri.10.30. [DOI] [PubMed] [Google Scholar]
- 72.Akkarathamrongsin S, Sugiyama M, Matsuura K, et al. High sensitivity assay using serum sample for IL28B genotyping to predict treatment response in chronic hepatitis C patients. Hepatol Res. 2010;40(10):956–962. doi: 10.1111/j.1872-034X.2010.00702.x. [DOI] [PubMed] [Google Scholar]
- 73.McCarthy JJ, Li JH, Thompson A, et al. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138(7):2307–2314. doi: 10.1053/j.gastro.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stattermayer AF, Stauber R, Hofer H, et al. Impact of IL28B genotype on the early and sustained virologic response in treatment-naive patients with chronic hepatitis C. Clin Gastroenterol Hepatol. 2010 doi: 10.1016/j.cgh.2010.07.019. In press. [DOI] [PubMed] [Google Scholar]
- 75.Askarieh G, Alsio A, Pugnale P, et al. Systemic and intrahepatic interferon-gamma-inducible protein 10 kDa predicts the first-phase decline in hepatitis C virus RNA and overall viral response to therapy in chronic hepatitis C. Hepatology. 2010;51(5):1523–1530. doi: 10.1002/hep.23509. [DOI] [PubMed] [Google Scholar]
- 76.Darling JM, Aerssens J, Fanning GC, et al. Quantitation of pretreatment serum IP-10 improves the predictive value of an IL28B gene polymorphism for hepatitis C treatment response. Hepatology. 2010;52(Suppl):382A–383A. doi: 10.1002/hep.24056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Howell CD, Thompson AJ, Ryan K, et al. IL28B genetic variation association with early viral kinetics and SVR in HCV henotype 1 the VIRAHEP-C study. J Hepatol. 2010;52(Suppl 1):S451. [Google Scholar]
- 78.Neumann AU, Bibert S, Haagmans B, et al. IL28B polymorphism is significantly correlated with IFN anti-viral effectivness already on first day of pegylated interferon-a and ribavirin therapy of chronic HCV infection. J Hepatol. 2010;52(Suppl 1):S468. [Google Scholar]
- 79.Trippler M, Schumacher S, Poggenpohi L, et al. Immediate and early antiviral responses to Peg-IFN or consensus IFN and ribavirin therapy for HCV correlate with the “upstream-of-IL28B” SNP (rs12979860) Hepatology. 2010;52(Suppl):1229A. [Google Scholar]
- 80•.Araújo ES, Dahari H, Cotler SJ, et al. Pharmacodynamics of PEG-IFN alpha-2a and HCV response as a function of IL28B polymorphism in HIV/HCV co-infected patients. J Acquir Immune Defic Syndr. 2010 doi: 10.1097/QAI.0b013e3182020596. In press. This is the first paper that explores the association among early viral kinetic and PEG-IFN alpha-2a pharmacodynamic parameters and IL28B genotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Araújo ES, Dahari H, Neumann AU, et al. Very early prediction of response to HCV treatment with peg-IFN-alfa-2a and ribavirin in HIV/HCV coinfected patients. J Viral Hepat. 2010 doi: 10.1111/j.1365-2893.2010.01358.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marcello T, Grakoui A, Barba-Spaeth G, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131(6):1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 83.Honda M, Sakai A, Yamashita T, et al. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139(2):499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 84.Thompson AJ, Muir AJ, Sulkowski MS, et al. IL28B Polymorphism Improves Viral Kinetics and Is the Strongest Pre-treatment Predictor of SVR in HCV-1 Patients. Gastroenterology. 2010:15. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 85.Zeuzem S, Herrmann E, Lee JH, et al. Viral kinetics in patients with chronic hepatitis C treated with standard or peginterferon alpha2a. Gastroenterology. 2001;120(6):1438–1447. doi: 10.1053/gast.2001.24006. [DOI] [PubMed] [Google Scholar]
- 86.Pilli M, Zerbini A, Penna A, et al. HCV-specific T-cell response in relation to viral kinetics and treatment outcome (DITTO-HCV project) Gastroenterology. 2007;133(4):1132–1143. doi: 10.1053/j.gastro.2007.06.059. [DOI] [PubMed] [Google Scholar]
- 87.Thompson AJ, Clark PJ, Zhu M, et al. Genome wide-association study identifies IL28B polymorphism to be associated with baseline ALT and hepatic necro-inflammatory activity in chronic hepatitis C patients enrolled in the IDEAL study. Hepatology. 2010;52(Suppl):1220A. [Google Scholar]
- 88.Thompson AJ, Clark PJ, Fellay J, et al. IL28B genotype is not associated with advanced hepatic fibrosis in chronic hepatitis C patients enrolled in the IDEAL study. Hepatology. 2010;52(Suppl):437A. [Google Scholar]
- 89.Barreiro P, Pineda JA, Rallon N, et al. Influence of Interleukin-28B Single Nucleotide Polymorphisms (SNP) on Progression to Liver Cirrhosis in HIV/Hepatitis C Virus Coinfected Patients. ICAAC; September 12–15 2010; Boston. 2010. [DOI] [PubMed] [Google Scholar]
- 90.Rallon NI, Naggie S, Benito JM, et al. Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS. 2010;24(8):F23–29. doi: 10.1097/QAD.0b013e3283391d6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Montes-Cano MA, Garcia-Lozano JR, Abad-Molina C, et al. Interleukin-28B genetic variants and hepatitis virus infection by different viral genotypes. Hepatology. 52(1):33–37. doi: 10.1002/hep.23624. [DOI] [PubMed] [Google Scholar]
- 92.Pang PS, Planet PJ, Glenn JS. The evolution of the major hepatitis C genotypes correlates with clinical response to interferon therapy. PLoS One. 2009;4(8):e6579. doi: 10.1371/journal.pone.0006579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Araújo ES, Melo CE, Martins LP, et al. Brazilian profile of IL28-B Single Nucleotide Polymorphism (SNP): a retrospective analysis and possible consequences for Interferon-alpha based therapies. Hepatology. 2010;52(Suppl):779A. [Google Scholar]
- 94.McHutchison JG, Goldstein DB, Shianna K, et al. IL28B SNP geographical distribution and antiviral responses in a 28-day Phase 2a trial of PSI-7977 daily dosing plus PEG-IFN/RBV. Hepatology. 2010;52(Suppl):711A. [Google Scholar]
- 95.Akuta N, Suzuki F, Hirakawa M, et al. Amino acid substitution in hepatitis C virus core region and genetic variation near the interleukin 28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatology. 2010;52(2):421–429. doi: 10.1002/hep.23690. [DOI] [PubMed] [Google Scholar]
- 96.Muir AJ, Lawitz E, Rodriguez-Torres M, et al. IL28B polymorphism and kinetics of antiviral activity for ANA598 in combination with pegylated interferon α2A plus ribavirin in treatment-naïve genotype-1 chronic HCV patients. Hepatology. 2010;52(Suppl):1200A. [Google Scholar]
- 97.Layden JE, Layden TJ, Reddy KR, et al. First phase viral kinetic parameters as predictors of treatment response and their influence on the second phase viral decline. J Viral Hepat. 2002;9(5):340–345. doi: 10.1046/j.1365-2893.2002.00377.x. [DOI] [PubMed] [Google Scholar]
- 98.Cotler SJ, Layden JE, Neumann AU, Jensen DM. First phase hepatitis c viral kinetics in previous nonresponders patients. J Viral Hepat. 2003;10(1):43–49. doi: 10.1046/j.1365-2893.2003.00401.x. [DOI] [PubMed] [Google Scholar]
- 99.Colombatto P, Civitano L, Oliveri F, et al. Sustained response to interferon-ribavirin combination therapy predicted by a model of hepatitis C virus dynamics using both HCV RNA and alanine aminotransferase. Antivir Ther. 2003;8(6):519–530. [PubMed] [Google Scholar]
- 100.Colombatto P, Ciccorossi P, Maina AM, et al. Early and accurate prediction of Peg-IFNs/ribavirin therapy outcome in the individual patient with chronic hepatitis C by modeling the dynamics of the infected cells. Clin Pharmacol Ther. 2008;84(2):212–215. doi: 10.1038/clpt.2008.21. [DOI] [PubMed] [Google Scholar]
- 101.Herrmann E, Lee JH, Marinos G, et al. Effect of ribavirin on hepatitis C viral kinetics in patients treated with pegylated interferon. Hepatology. 2003;37(6):1351–1358. doi: 10.1053/jhep.2003.50218. [DOI] [PubMed] [Google Scholar]
- 102.Reluga TC, Dahari H, Perelson AS. Analysis of Hepatitis C Virus Infection Models with Hepatocyte Homeostasis. SIAM J Appl Math. 2009;69(4):999–1023. doi: 10.1137/080714579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Talal AH, Ribeiro RM, Powers KA, et al. Pharmacodynamics of PEG-IFN alpha differentiate HIV/HCV coinfected sustained virological responders from nonresponders. Hepatology. 2006;43(5):943–953. doi: 10.1002/hep.21136. [DOI] [PubMed] [Google Scholar]
- 104.Rozenberg L, Haagmans BL, Neumann AU, et al. Therapeutic response to peg-IFN-alpha-2b and ribavirin in HIV/HCV co-infected African-American and Caucasian patients as a function of HCV viral kinetics and interferon pharmacodynamics. AIDS. 2009;23(18):2439–2450. doi: 10.1097/QAD.0b013e32832ff1c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dixit NM, Layden-Almer JE, Layden TJ, Perelson AS. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature. 2004;432(7019):922–924. doi: 10.1038/nature03153. [DOI] [PubMed] [Google Scholar]
- 106.Brunetto MR, Colombatto P, Bonino F. Bio-mathematical models of viral dynamics to tailor antiviral therapy in chronic viral hepatitis. World J Gastroenterology. 2009;15(5):531–537. doi: 10.3748/wjg.15.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sarrazin C, Schwendy S, Moeller B, et al. Completely individualized treatment durations with peginterferon- alfa - 2b and ribavirin in HCV genotype 1 - infected patients and importance of IL28B genotype (INDIV-2 study) Hepatology. 2010;52(Suppl):384A. [Google Scholar]
- 108.Mangia A, Thompson AJ, Santoro R, et al. Rapid virological response (RVR) vs IL28B CC genotype in HCV-1 infected patients treated with an individualized course of peginterferon and weight based ribavirin. Hepatology. 2010;52(Suppl):750A. [Google Scholar]
- 109.Poordad F, McCone J, Bacon BR, et al. Boceprevir (BOC) combined with peginterferon alfa - 2b/ribavirin(P/R) for treatment-naive patients with hepatitis C virus (HCV) genotype (G) 1: SPRINT-2 final results. Hepatology. 2010;52(Suppl):402A. [Google Scholar]