Abstract
For many years pathologists have used Hematoxylin and Eosin (H&E), single marker immunohistochemistry (IHC) and in situ hybridization with manual analysis by microscopy or at best simple digital imaging. There is a growing trend to update pathology to a digital workflow to improve objectivity and productivity, as has been done in radiology. There is also a need for tissue-based multivariate biomarker assays to improve the accuracy of diagnostic, prognostic, and predictive testing. Multivariate tests are not compatible with the traditional single marker, manual analysis pathology methods but instead require a digital platform with brightfield and fluorescence imaging, quantitative image analysis, and informatics. Here we describe the use of the Hamamatsu NanoZoomer Digital Pathology slide scanner with HCImage software for combined brightfield and multiplexed fluorescence biomarker analysis and highlight its applications in biomarker research and pathology testing. This combined approach will be an important aid to pathologists in making critical diagnoses.
Keywords: Digital imaging pathology, multiplexed immunofluorescence, quantitative image analysis, tissue microarrays, whole-slide imaging
INTRODUCTION
Traditional pathology analyses are subjective and prone to inter- and intraindividual variation and error, which are significant causes of misdiagnosis, slow turnaround times, delays in proper treatment, and malpractice claims.[1–4] New technologies being applied to improve pathology focus on two areas: adding digital imaging and informatics to the traditionally visual approach and extracting molecular information via the use of biomarkers. The traditional pathology workflow with paper-based tracking and reporting of results and catalog storage of glass slides is inefficient and also prone to error. Pathology has been slower than other clinical areas to adopt digital technology but in recent years there has been a growing appreciation of the need to update pathology to a digital workflow, as has been done in radiology.[5–7] Digital pathology slide scanners have been increasing in adoption in medical laboratories in recent years. However, a significant barrier to full adoption is the availability of cost-effective, high-speed, high-resolution slide scanners that produce digital slide images of sufficient quality for pathology analyses at a speed that is compatible with the need for short turnaround times in pathology. In diagnostics, it has become clear that there are no “single bullet” biomarkers that can diagnose, predict clinical outcome, and response to therapies for all patients with a particular disease due to patient variability and the complexity of tissues, particularly tumors.[8,9] There is a need to assess multiple key biomarkers in order to accurately stratify patients into diagnostic, prognostic, and predictive subgroups. Multiplexed biomarker tests coupled with informatics to process the multivariate data into decision-making indices are required to improve pathology testing and personalized patient care. However, the traditional pathology methods of single marker IHC with manual analysis cannot easily be applied to multivariate biomarker testing. There is a need for high-speed, combined brightfield, and fluorescence digital slide imaging for objective quantification of multiple fluorescence biomarkers on a single slide coupled to brightfield tissue imaging in serial slides for “reference” morphological analyses. The NanoZoomer Digital Pathology system (Hamamatsu Photonics, K.K., Japan) with HCImage (Hamamatsu Corporation, Pittsburgh, PA, USA) is a high-speed, high-resolution slide scanner with both brightfield and 3-color fluorescence imaging and quantitative image analysis capabilities. The goals of this study were to investigate the potential of the NanoZoomer Digital Pathology slide scanning system and HCImage (Hamamatsu Corporation, Pittsburgh, PA, USA) for combined brightfield and fluorescence digital slide imaging in research and clinical pathology applications and to demonstrate methods to quantify multiple biomarkers per slide in tissues and cell models using this system.
MATERIALS AND METHODS
Tissue Microarray Labeling
A tissue microarray (TMA) block containing cores of formalin-fixed paraffin-embedded (FFPE) tissues and cell lines was used to demonstrate the NanoZoomer Digital Pathology slide scanner and HCImage software. Five micrometer sections of the TMA were stained with H&E by standard histology methods. Additional sections were stained by multiplexed immunofluorescence to label NF-κB p65 and CD44. Briefly, slides were deparaffinized and rehydrated according to standard histology methods followed by antigen retrieval in 1 mM EDTA pH 8.0 at 100°C for 20 minutes with subsequent washing in Tris-buffered saline 0.025% Tween 20. Slides were incubated with blocking buffer containing 5% donkey serum prior to primary labeling with rabbit anti-NF-κB p65 antibody and rat anti-CD44 antibody (both from Santa Cruz Biotechnology, CA, USA) for 1 hour at room temperature, followed by washing in Tris-buffered saline. Slides were then labeled with Alexa Fluor 488-conjugated donkey antirat secondary antibody and Alexa Fluor 568-conjugated donkey antirabbit secondary antibody (Invitrogen by Life Technologies, CA, USA) for 1 hour at room temperature, followed by washing in Tris-buffered saline, labeling with Hoechst 33342 (Invitrogen, CA, USA) and mounting with Prolong Gold Antifade Reagent (Invitrogen).
Imaging Methods Using the NanoZoomer Digital Pathology System
Slides were imaged in brightfield and fluorescence using the NanoZoomer Digital Pathology-HT (C9600-03) with Fluorescence Illumination Optics (L10387) (Hamamatsu Photonics, K.K., Japan), which is a high-speed and high-resolution digital slide scanner system that acquires whole-slide digital images using TDI (time delayed integration) technology. TDI technology enables accumulation of charges on a TDI-CCD sensor (4,096 × 64 pixels) by synchronizing charge transfer with movement of a sample object. This technology makes it possible to scan the large area of pathology slides without sacrificing scanning speed, image resolution, or quality. The NanoZoomer can also scan in 3-color fluorescence using 3 CCD sensors and a full multiband triple filter set, which covers red (628 nm), green (530 nm), and blue (457 nm) fluorescence. The speed of scanning is approximately 3 minutes per slide (20 mm × 20 mm area) in the standard 20 magnification mode with high-resolution images (0.46 micrometer/pixel in 20× mode, 0.23 micrometer/pixel in 40× mode), which is a total of approximately 2 gigapixels. The acquired images can be stored as 36-bit uncompressed data as well as JPEG compressed format. The NanoZoomer Digital Pathology is also a high-throughput scanner; it can automatically process 210 slides per scanning batch using slide cassette holders and precision loading technology. Whole-slide images of the TMA slides were generated using 8× exposure (35 ms effective photon collection) and 20× magnification.
TMA Segmentation Using HCImage
A TMA extraction method was developed to automatically identify, isolate, and match TMA cores to their respective naming convention [Figure 1]. The naming convention was supplied as a matrix in a XLS or CSV file and loaded into the HCImage software. The first step allows the image to be processed to remove noise for an improved intensity threshold. An idealized box is placed around all identified cores. Unwanted artifacts are automatically removed where possible. Manual editing tools allow artifacts to be removed and undetected cores to be added. The software attempts to match the cores to their respective name, based on a regular grid pattern. Cores that fall out of a regular pattern are assessed for deviation from the grid by the amount of overlap. The software attempts to correctly match the cores based on the percentage overlap and any final matching of cores with names can easily be done manually.
Figure 1.
Extraction of individual cores from tissue microarrays. TMA sections stained with H and E (a) and multiplexed fluorescence (b) were scanned using the NanoZoomer. The whole-slide digital TMA slide images were segmented into individual cores using HCImage. Each core was labeled with the core ID, e.g., tissue type, cell line, coded patient ID, and organized into the TMA map function for review (c) and into the HCImage database for analysis
Extraction of Biomarker Data Using HCImage
Cell identification and analysis was applied to each image using an HCImage macro with the protocol described in [Figure 2]. Examples of the cell identification and analysis are shown in [Figure 3]. The resulting masks were used to make multiple measurements including (i) area: area of all pixels under the mask, not including any holes (i.e., nuclei); (ii) filled area: area of all pixels under the mask including any holes (nuclei); (iii) hole area %: area of holes/filled area; (iv) mean red: average intensity in the red channel of all pixels under the mask (cytoplasm and plasma membrane), not including any holes (nuclei); (v) mean green: average intensity in the green channel of all pixels under the mask, not including any holes; (vi) total red: the sum of all intensity in the red channel of all pixels under the mask, not including any holes; (vii) total green: the sum of all intensity in the green channel of all pixels under the mask, not including any holes; (viii) hole mean red: average hole intensity in the red channel; (ix) hole mean green: average hole intensity in the green channel; (x) red nucleus to cytoplasm ratio: mean hole intensity in the red channel/mean intensity in the red channel of all pixels under the mask, not including any holes.
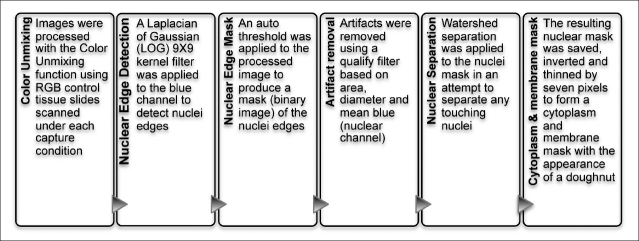
Figure 2.
Cell identification and analysis protocol using HCImage software
Figure 3.

Segmentation of fluorescence digital tissue images. Slides were scanned at 20× magnification. TMA cores were analyzed using HCImage. (a) H and E kidney core; (b) Hoechst, NF-κ B (red), CD44 (green)-stained kidney core; (c) magnified view of kidney core; (d) detection of individual nuclei edges in kidney; (e) detection of cell edges in kidney; (f) cytoplasm and membrane mask in kidney; (g) H and E MiaPaca-2 core; (h) Hoechst, NF-κ B, CD44-stained MiaPaca-2 core; (i) magnified view of MiaPaca-2 core; (j) detection of individual MiaPaca-2 nuclei edges; (k) detection of MiaPaca-2 cell edges; (l) cytoplasm and membrane mask in MiaPaca-2 core. a-b and g-h are from same cores in different serial sections of the TMA
RESULTS
Generation of Brightfield and Fluorescence Tissue Images
A TMA containing formalin-fixed paraffin-embedded (FFPE) tissues and cancer cell models was used to demonstrate the slide scanning and biomarker analysis system. CD44 was selected as a biomarker to demonstrate measurement of a plasma membrane protein. NF-κB p65 was selected as a biomarker to demonstrate both quantification of protein expression in the cytoplasmic and nuclear compartments and measurement of the ratio of a biomarker between these two subcellular compartments. Both CD44 and NF-κB p65 have been described to have diagnostic and prognostic significance in multiple cancer types.[10–14] Sections of the TMA were stained with H&E in order to assess tissue morphology in brightfield since pathologists have a long history of interpretation of morphology and by immunofluorescence for CD44 and NF-κB p65 with Hoechst staining of nuclei in order to analyze multiplexed biomarkers in fluorescence in comparison to any morphological interpretation from the H&E-stained images. The TMA slides were imaged on the NanoZoomer Digital Pathology in brightfield and 36-bit fluorescence mode; examples are shown in [Figure 3a–c, g–i].
Segmentation of Tissue and Cell Images
Cores of FFPE kidney tissue and MiaPaca-2 cells were selected as examples to demonstrate the tissue segmentation and biomarker feature data extraction using HCImage. The TMA images were segmented into individual cores [Figure 1]. Single cores were then analyzed to segment nuclei as individual objects and to create an estimated mask of the cytoplasm and plasma membrane using the process described in [Figure 2]. Examples of the segmentation procedure and masks are shown in [Figure 3d–f] and [Figure 3j–l]. In the 5 μm tissue sections used in this study, there are situations where nuclei “pile up” in 3D. In these situations a small cluster of nuclei becomes impossible to accurately segment since 3D information is not available after acquiring a 2D image. Even if the nuclear segmentation problem could be solved or approximated, extracting biomarker feature data from such clusters of nuclei would be erroneous since it will be impossible to accurately assign biomarker expression to individual cells. The 5 μ m section thickness is an established compromise that is thick enough to include an optimal number of whole nuclei, yet thin enough to avoid too many 3D overlaps. The cell density and nuclear proximity varies between tissue types. For some tissue types, e.g., kidney and breast, these clusters are rare and can be excluded from further measurements without biasing the results. Other tissues such as tonsil and colon present a significant image segmentation challenge due to the density of cells within these tissues.
Extraction and Analysis of Multiplexed Biomarker Feature Data
A variety of biomarker feature measurements were made in the nuclear, cytoplasmic, and membrane masks. The nuclear mask enables cells to be included/excluded from further analysis based on size [Figure 4a–b]. Small objects that are unlikely to be cells or large objects that may represent aggregates of multiple cells that are beyond segmentation can be excluded from biomarker feature measurements. Red blood cells, which are a common source of autofluorescence in tissues due in part to the presence of lipofuscin, are excluded by the image analysis script since they lack a nucleus. The extracted biomarker feature measurements included mean red and green intensity and total red and green intensity in “holes” (i.e., nuclei) and in “donuts” (i.e., cytoplasm and plasma membrane) and the hole:donut ratio of mean red intensity [Figure 4c–d]. High outliers were removed from further analysis by reviewing the images to determine the signal intensity of artifacts and debris, e.g., > 1500 in mean green signal or red nucleus:cytoplasm ratio > 4. In the green channel (CD44), a threshold of 250 was set based on the mean signal in a negative control cell line contained on the TMA (T47D breast cancer cell line that does not express CD44), signal resulting from staining the TMAs with the secondary antibodies only and by visual assessment of signal in cells expressing green signal. Cells with signal above the threshold were considered positive for CD44 expression [Figure 4e–f]. The image analysis showed the expected plasma membrane staining pattern for CD44 in the majority of MiaPaca-2 cells and in a small fraction of kidney cells. This is consistent with the published literature demonstrating plasma membrane expression of CD44 in MiaPaca-2 cells and low frequency of CD44-expressing interstitial immune cells in normal kidney tissue.[15–17] A threshold was not applied to the red channel (NF-κB p65) since all cells express this protein and the ratio of nuclear:cytoplasmic NF-κB p65 is relevant to its measurement and interpretation rather than overall intensity [Figure 4c–d]. NF-κB p65 is located in the cytoplasm in its resting state and translocates to the nucleus upon activation. The image analysis showed the expected staining pattern for NF-κB p65 in both the MiaPaca-2 cells and kidney tissue, with the majority of cells showing low nuclear:cytoplasmic ratios of NF-κB p65 and subpopulations with high nuclear:cytoplasmic ratios indicating activation of NF-κB in these cells.[18] Normal kidney tissue produces low levels of chemokines that activate NF-κB, which explains the staining pattern detected by the image analysis software in this study. Bivariate analysis of CD44 and NF-κB p65 was also performed; the cells of the kidney and MiaPaca-2 cell cores could be divided into 4 subpopulations based on varying expression of these two biomarkers [Figure 4g–h].
Figure 4.
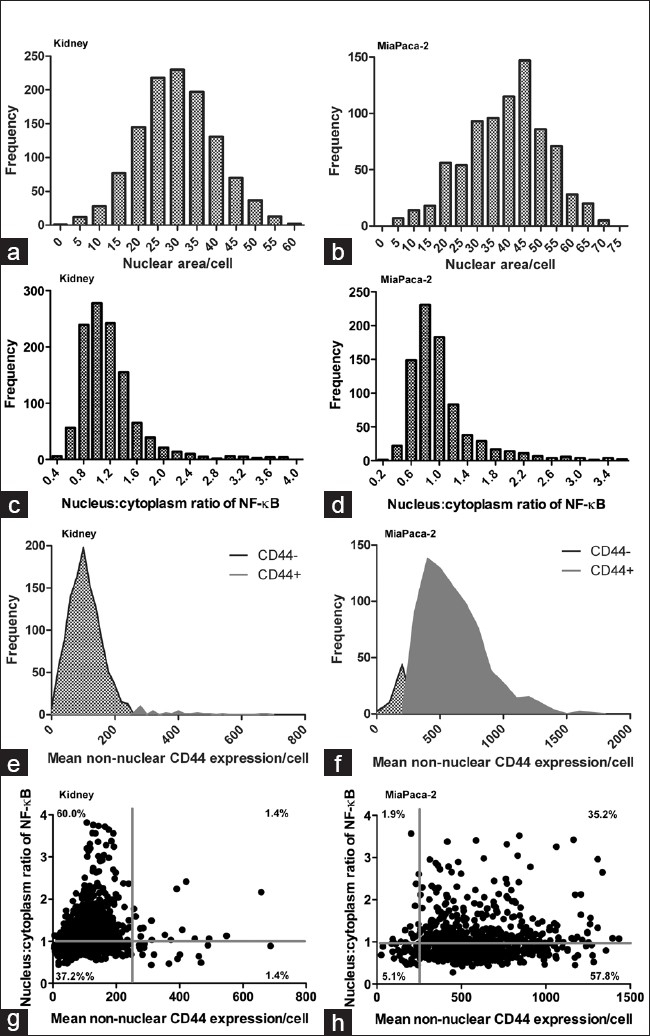
Multiplexed biomarker expression in example tissue and cell line. Multiple measurements were made in the segmented fluorescence digital tissue images. Kidney tissue and MiaPaca-2 cell line were used as example tissue and cancer cell model, respectively. Extracted data were analyzed in Prism 5 (GraphPad Software, Inc., CA, USA) using frequency distributions to assess the nuclear area (a and b), nuclear:cytoplasm NF-κB p65 ratio (c and d), CD44+/- cells (e and f) and using bivariate analysis to assess expression of CD44 and the ratio nuclear:cytoplasmic NF-κB expression in single cells (g and h)
DISCUSSION
The current standard in pathology is to measure biomarkers singly by IHC and in situ hybridization with visual inspection and manual scoring or at best simple digital imaging of single biomarkers. Digital pathology slide scanners and IHC image analysis applications are increasing in adoption in medical laboratories; however, there is a need for faster slide scanning and for multiplexed approaches to quantify multiple key biomarkers in the same tissue section to improve pathology testing. The use of an H and E tissue section image is important as an aid to pathologists, since this is the standard of practice and the value of the pathologists′ interpretation of the tissue morphology, especially in conjunction with multiple biomarkers, is essential. Fluorescence digital imaging pathology with image analysis algorithms will enable objective, quantitative analysis of multiple biomarkers per slide to meet this need to incorporate novel multivariate biomarker tests into pathology testing. We investigated the potential applications of combined brightfield and fluorescence digital slide imaging using the NanoZoomer Digital Pathology slide scanner with HCImage image analysis software. This system has various potential clinical applications and advantages versus traditional pathology methods and over other digital pathology systems. The slide scanner digitizes slides at exceptional speed in both brightfield and fluorescence imaging modes, which make it an ideal slide scanning solution for both research and clinical pathology applications. There are various brightfield digital slide scanning systems available from multiple vendors; however, scanning speed is a significant barrier to clinical implementation due to the need for short turnaround times in pathology testing. The NanoZoomer used in this study is faster than any other fluorescence slide scanner on the market and Hamamatsu is further improving the scanning speed in a second-generation version of the NanoZoomer (NanoZoomer 2.0-HT (C9600-13)). Fluorescence slide scanning systems are also being introduced to meet the need for multiplexed biomarker analysis in pathology. Fluorescence slide scanners that image 4-6 fluorescence channels individually are available based on multipass single sensor approaches. The slower speed of these scanners and the lack of FDA-approved image analysis algorithms for specific clinical applications make them currently suitable only for research. Adoption of fluorescence digital imaging pathology will continue to increase as improvements are made to the scanning speed and as clinical applications are developed and validated.
The NanoZoomer Digital Pathology system combines fluorescence digital imaging of multiple biomarkers on one slide with brightfield digital imaging of tissue morphology and includes image analysis software (HCImage) to objectively extract quantitative data on multiple biomarkers per tissue section. The dual brightfield – fluorescence capabilities will be a useful aid to pathologists to assess fluorescence biomarker images in the context of the tissue morphology. In this study we demonstrated quantitative analysis of multiplexed biomarkers in example tissues and control cells lines in TMA format. The HCImage image analysis software-enabled segmentation of individual cells within tissues into subcellular compartments such that we were able to measure the mean and total intensity of biomarkers in nuclei and the cytoplasm and plasma membrane of single cells within tissues and measure the ratio of biomarker intensity between the different subcellular compartments. These measurements have various potential clinical applications since for many biomarkers the expression, tissue localization, subcellular localization, and ratio between different tissue and cellular compartments are relevant to their diagnostic, prognostic and/or predictive significance[19–22] (reviewed in[8]). The software includes TMA segmentation and extracted tissue core organization to accommodate high-throughput analysis of multiple biomarkers in hundreds to thousands of patient samples per slide for biomarker assay development and validation studies.
The applications of the NanoZoomer Digital Pathology system include combined transmitted light and 3-color multiplexed fluorescence quantitative biomarker assays in research and clinical pathology. Multiple separate IHC biomarker scores have been used to stratify cancer patients according to prognosis;[23,24] however, there are difficulties scoring and interpreting multiple IHC stains in single cells within tissues and this tedious manual method of measuring many markers singly with manual scoring is neither realistic in terms of efficiency nor reproducible clinically. Digital pathology has been applied to IHC analysis to improve objectivity and reproducibility.[25,26] Even with digital analysis, the accuracy of IHC is only semiquantitative and is limited by inherent variability in staining intensity and the difficulties in multiplexing. Multiplexed fluorescence with digital imaging offers significant improvements on IHC, including: (i) more consistent, higher resolution in the labeling and imaging since fluorophores are directly conjugated to antibodies, (ii) greater dynamic detection range, (iii) the ability to quantify multiple antigens per section, including multiple antigens colocalized within the same subcellular compartment, (iv) objective, reproducible extraction of quantitative biomarker data by image analysis software. Quantitative fluorescence biomarker analysis has been shown to correlate with Western blot analysis,[27] to match or exceed the accuracy of manual IHC scoring,[28–30] and to improve standardization of tissue biomarker testing.[31]
The NanoZoomer system has a 3-color/multiplexed biomarker limit, due to the detection method. However, there are many current and potential future clinical applications that require only 2-3 biomarkers coupled to a reference H&E image test, e.g., cytokeratins and/or Ki-67 in various tumor types, estrogen receptor (ER), progesterone receptor (PgR) and HER2-/neu in breast cancer or p63, high molecular weight cytokeratin, and P504S in prostate cancer. Using three simultaneous sensors for fluorescence scanning as the NanoZoomer does is very efficient for such applications. Novel multivariate index biomarker tests are likely to be based on 6--10, or possible even higher numbers of biomarkers. Higher multiplexing can be achieved by staining and imaging biomarkers in serial sections using the NanoZoomer. The ideal system for tissue-based multivariate index assays would accommodate all the relevant biomarkers on one slide. There is a challenge for next-generation systems to provide the optimal trade-off between scanning speed and number of fluorescence channels. In order to overcome the single exposure time for each channel with the NanoZoomer, it is critical to optimize the immunofluorescence biomarker staining to improve signal:noise in each fluorescence channel.
In summary, the NanoZoomer Digital Pathology system with HCImage image analysis software enables high-throughput brightfield imaging of tissue morphology and fluorescence imaging of multiple biomarkers per slide with quantitative, objective image analysis at the subcellular compartments. The system has potential applications in 3-color fluorescence biomarker testing to improve the objectivity and efficiency of pathology testing and in high-throughput analysis of biomarkers in research pathology.
ACKNOWLEDGEMENT
Cernostics would like to acknowledge the valuable collaboration with Hamamatsu Photonics, K.K., on the development of the tools to complete this study.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2011/2/1/50/89849
REFERENCES
- 1.Raab SS, Grzybicki DM, Janosky JE, Zarbo RJ, Meier FA, Jensen C, et al. Clinical impact and frequency of anatomic pathology errors in cancer diagnoses. Cancer. 2005;104:2205–13. doi: 10.1002/cncr.21431. [DOI] [PubMed] [Google Scholar]
- 2.Raab SS, Meier FA, Zarbo RJ, Jensen DC, Geisinger KR, Booth CN, et al. The “Big Dog” effect: variability assessing the causes of error in diagnoses of patients with lung cancer. J Clin Oncol. 2006;24:2808–14. doi: 10.1200/JCO.2005.04.3661. [DOI] [PubMed] [Google Scholar]
- 3.Frable WJ. Surgical pathology-second reviews, institutional reviews, audits, and correlations: What's out there. Error or diagnostic variation? Arch Pathol Lab Med. 2006;130:620–5. doi: 10.5858/2006-130-620-SPRIRA. [DOI] [PubMed] [Google Scholar]
- 4.Singh H, Sethi S, Raber M, Petersen LA. Errors in cancer diagnosis: current understanding and future directions. J Clin Oncol. 2007;25:5009–18. doi: 10.1200/JCO.2007.13.2142. [DOI] [PubMed] [Google Scholar]
- 5.Soenksen D. Advances in digital pathology drive continued momentum and globalization. MLO Med Lab Obs. 2009;41:31. [PubMed] [Google Scholar]
- 6.Reiner BI, Siegel EL, Siddiqui K. Evolution of the digital revolution: A radiologist perspective. J Digit Imaging. 2003;16:324–30. doi: 10.1007/s10278-003-1743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montalto MC. Pathology RE-imagined: The history of digital radiology and the future of anatomic pathology. Arch Pathol Lab Med. 2008;132:764–5. doi: 10.5858/2008-132-764-PRTHOD. [DOI] [PubMed] [Google Scholar]
- 8.Critchley-Thorne RJ, Miller SM, Taylor DL, Lingle WL. Applications of Cellular Systems Biology in Breast Cancer Patient Stratification and Diagnostics. Comb Chem High Throughput Screen. 2009;12:860–9. doi: 10.2174/138620709789383222. [DOI] [PubMed] [Google Scholar]
- 9.Critchley-Thorne RJ, Yu HX, Lee PP. New York: Springer; 2010. Immune Signatures Associated with the Cancer Bearing State. Signatures of Rejection. [Google Scholar]
- 10.Morris SF, O’Hanlon DM, McLaughlin R, McHale T, Connolly GE, Given HF. The prognostic significance of CD44s and CD44v6 expression in stage two breast carcinoma: An immunohistochemical study. Eur J Surg Oncol. 2001;27:527–31. doi: 10.1053/ejso.2001.1167. [DOI] [PubMed] [Google Scholar]
- 11.Diaz LK, Zhou X, Wright ET, Cristofanilli M, Smith T, Yang Y, et al. CD44 expression is associated with increased survival in node-negative invasive breast carcinoma. Clin Cancer Res. 2005;11:3309–14. doi: 10.1158/1078-0432.CCR-04-2184. [DOI] [PubMed] [Google Scholar]
- 12.van Laere SJ, van der Auwera I, van den Eynden GG, van Dam P, van Marck EA, Vermeulen PB, et al. NF-kappaB activation in inflammatory breast cancer is associated with oestrogen receptor downregulation, secondary to EGFR and/or ErbB2 overexpression and MAPK hyperactivation. Br J Cancer. 2007;97:659–69. doi: 10.1038/sj.bjc.6603906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Rodriguez L, Sancho-Torres I, Mesonero C, Gibbon DG, Shih WJ, Zotalis G. The CD44 receptor is a molecular predictor of survival in ovarian cancer. Med Oncol. 2003;20:255–63. doi: 10.1385/MO:20:3:255. [DOI] [PubMed] [Google Scholar]
- 14.Al-Saad S, Al-Shibli K, Donnem T, Persson M, Bremnes RM, Busund LT. The prognostic impact of NF-kappaB p105, vimentin, E-cadherin and Par6 expression in epithelial and stromal compartment in non-small-cell lung cancer. Br J Cancer. 2008;99:1476–83. doi: 10.1038/sj.bjc.6604713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy-Chaudhury P, Khong TF, Williams JH, Haites NE, Wu B, Simpson JG, et al. CD44 in glomerulonephritis: Expression in human renal biopsies, the Thy 1.1 model, and by cultured mesangial cells. Kidney Int. 1996;50:272–81. doi: 10.1038/ki.1996.312. [DOI] [PubMed] [Google Scholar]
- 16.Stamenkovic I, Aruffo A, Amiot M, Seed B. The hematopoietic and epithelial forms of CD44 are distinct polypeptides with different adhesion potentials for hyaluronate-bearing cells. Embo J. 1991;10:343–8. doi: 10.1002/j.1460-2075.1991.tb07955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugahara KN, Hirata T, Hayasaka H, Stern R, Murai T, Miyasaka M. Tumor cells enhance their own CD44 cleavage and motility by generating hyaluronan fragments. J Biol Chem. 2006;281:5861–8. doi: 10.1074/jbc.M506740200. [DOI] [PubMed] [Google Scholar]
- 18.Wang JM, Shen W, Chertov O, Damme JV, Oppenheim JJ. Totowa, NJ: Humana Press; 1999. Chemokine Modulation of Tumor Cell Physiology. Chemokines in Cancer. [Google Scholar]
- 19.Nevalainen MT, Xie J, Torhorst J, Bubendorf L, Haas P, Kononen J, et al. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J Clin Oncol. 2004;22:2053–60. doi: 10.1200/JCO.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 20.Nakopoulou L, Mylona E, Papadaki I, Kavantzas N, Giannopoulou I, Markaki S, et al. Study of phospho-beta-catenin subcellular distribution in invasive breast carcinomas in relation to their phenotype and the clinical outcome. Mod Pathol. 2006;19:556–63. doi: 10.1038/modpathol.3800562. [DOI] [PubMed] [Google Scholar]
- 21.Bell D, Chomarat P, Broyles D, Netto G, Harb GM, Lebecque S, et al. In breast carcinoma tissue, immature dendritic cells reside within the tumor, whereas mature dendritic cells are located in peritumoral areas. J Exp Med. 1999;190:1417–26. doi: 10.1084/jem.190.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wakabayashi O, Yamazaki K, Oizumi S, Hommura F, Kinoshita I, Ogura S, et al. CD4+ T cells in cancer stroma, not CD8+ T cells in cancer cell nests, are associated with favorable prognosis in human non-small cell lung cancers. Cancer Sci. 2003;94:1003–9. doi: 10.1111/j.1349-7006.2003.tb01392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyall MS, Dundas SR, Curran S, Murray GI. Profiling markers of prognosis in colorectal cancer. Clin Cancer Res. 2006;12:1184–91. doi: 10.1158/1078-0432.CCR-05-1864. [DOI] [PubMed] [Google Scholar]
- 24.Zlobec I, Minoo P, Baumhoer D, Baker K, Terracciano L, Jass JR, et al. Multimarker phenotype predicts adverse survival in patients with lymph node-negative colorectal cancer. Cancer. 2008;112:495–502. doi: 10.1002/cncr.23208. [DOI] [PubMed] [Google Scholar]
- 25.Sharangpani GM, Joshi AS, Porter K, Deshpande AS, Keyhani S, Naik GA, et al. Semi-automated imaging system to quantitate estrogen and progesterone receptor immunoreactivity in human breast cancer. J Microsc. 2007;226:244–55. doi: 10.1111/j.1365-2818.2007.01772.x. [DOI] [PubMed] [Google Scholar]
- 26.Slodkowska J, Filas V, Buszkiewicz E, Trzeciak P, Wojciechowski M, Koktysz R, et al. Study on breast carcinoma Her2/neu and hormonal receptors status assessed by automated images analysis systems: ACIS III (Dako) and ScanScope (Aperio) Folia Histochem Cytobiol. 2010;48:19–25. doi: 10.2478/v10042-010-0015-1. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Barnes RO, West NR, Olson M, Chu JE, Watson PH. Jab1 is a target of EGFR signaling in ERalpha-negative breast cancer. Breast Cancer Res. 2008;10:R51. doi: 10.1186/bcr2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–7. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 29.Giltnane JM, Molinaro A, Cheng H, Robinson A, Turbin D, Gelmon K, et al. Comparison of quantitative immunofluorescence with conventional methods for HER2/neu testing with respect to response to trastuzumab therapy in metastatic breast cancer. Arch Pathol Lab Med. 2008;132:1635–47. doi: 10.5858/2008-132-1635-COQIWC. [DOI] [PubMed] [Google Scholar]
- 30.Bloom K, Harrington D. Enhanced accuracy and reliability of HER-2/neu immunohistochemical scoring using digital microscopy. Am J Clin Pathol. 2004;121:620–30. doi: 10.1309/Y73U-8X72-B68T-MGH5. [DOI] [PubMed] [Google Scholar]
- 31.Gustavson MD, Bourke-Martin B, Reilly D, Cregger M, Williams C, Mayotte J, et al. Standardization of HER2 immunohistochemistry in breast cancer by automated quantitative analysis. Arch Pathol Lab Med. 2009;133:1413–9. doi: 10.5858/133.9.1413. [DOI] [PubMed] [Google Scholar]