Abstract
Introduction:
The increasing availability of whole slide imaging (WSI) data sets (digital slides) from glass slides offers new opportunities for the development of computer-aided diagnostic (CAD) algorithms. With the all-digital pathology workflow that these data sets will enable in the near future, literally millions of digital slides will be generated and stored. Consequently, the field in general and pathologists, specifically, will need tools to help extract actionable information from this new and vast collective repository.
Methods:
To address this limitation, we designed and implemented a tool (dCORE) to enable the systematic capture of image tiles with constrained size and resolution that contain desired histopathologic features.
Results:
In this communication, we describe a user-friendly tool that will enable pathologists to mine digital slides archives to create image microarrays (IMAs). IMAs are to digital slides as tissue microarrays (TMAs) are to cell blocks. Thus, a single digital slide could be transformed into an array of hundreds to thousands of high quality digital images, with each containing key diagnostic morphologies and appropriate controls. Current manual digital image cut-and-paste methods that allow for the creation of a grid of images (such as an IMA) of matching resolutions are tedious.
Conclusion:
The ability to create IMAs representing hundreds to thousands of vetted morphologic features has numerous applications in education, proficiency testing, consensus case review, and research. Lastly, in a manner analogous to the way conventional TMA technology has significantly accelerated in situ studies of tissue specimens use of IMAs has similar potential to significantly accelerate CAD algorithm development.
Keywords: IMA, SIVQ, TMA, WSI
INTRODUCTION
Tissue microarrays (TMA) are a high-throughput technology enabling the analysis (histochemistry, immunohistochemistry (IHC) and in situ hybridization (ISH)) of hundreds of tissue samples on a single slide.[1] Using this technology, tiny tissue cylinders are acquired from hundreds of different primary tissue blocks and arrayed into a single “recipient” paraffin block at a very high density.[2]
The first description of a TMA was initially described in 1986 as a “multitumor (sausage) tissue block” and developed as a novel method for IHC antibody testing.[3] One millimeter thick “rods” of various tissue types were wrapped in sheets of small intestine and then embedded in paraffin blocks allowing for the simultaneous examination of various tissues in a single experiment under identical conditions.[3,4] This technology was later improved with the introduction of an instrument for the placement of precise cores in a defined grid, improving accuracy and reproducibility of the tissue specimens within the block.[4,5] Presently, numerous experiments can be performed, considering that 200 sections can be made from one tissue block, and with contemporary workflow models utilizing as many as 500-1000 different tissue samples from routinely formalin-fixed paraffin blocks for placement on one microscope glass slide.[5,6]
The advantages of using TMAs are that they provide a judicious use of precious tissue, give experimental uniformity, and allow for analysis of a large number of samples improving statistical precision in addition to enhanced speed and quality of analysis.[1] However, there are limitations, such as: the small size of the TMAs may not provide an adequate representation of the entire tissue, the high cost of TMA production, the loss of tissue cores during processing and the complexity of IHC staining.[1,4]
TMAs can be constructed in various formats, depending on the application. Some of the more common types are multitumor TMAs, normal tissue TMAs, progression TMAs (different stages of one particular disease), prognosis TMAs (samples with available clinical follow-up data), experimental tissue arrays (cell lines - CMAs, xenografts - XMAs), and frozen tissue TMAs.[6]
The recent availability of digital whole slide imaging (WSI) data sets from glass slides creates new opportunities for possible deployment of computer aided diagnostic (CAD) technologies.[7–14] The use of CAD has the potential to improve the practice of pathology in various ways by helping the pathologist in (1) the screening of slides; (2) in the provision of real time clinical decision support tools; (3) the instantiation of additional automated layers of quality assurance and diagnostic consistency; and (4) imparting a quantitative component to the practice of diagnostic pathology.[9] In the next few years, pathology departments will be adopting all digital workflows, thus potentially generating millions of data sets (and shortly after will be able to integrate with prognostic information) to be mined along with the accessible prognostic information.
In this context, applying CAD algorithms that can quantify histopathologic and morphologic features visible via H&E, IHC, and ISH can offer new insights into disease and potentially provide a new class of biomarkers. Thus, generating tools that will leverage the tremendous value contained within these WSI data sets will be of extreme benefit to the field of pathology.
In the past, machine vision computational approaches such as CAD have been developed in an attempt to interpret this information but, with the notable exception of Pap test screening in Cytopathology, such efforts have usually fallen short of expectations. Notable obstacles have been lack of algorithmic specificity, computational power, limited data storage, and other operational issues. The application of CAD to surgical pathology specimens is incrementally challenging compared to Pap tests in that the former require the assessment of a number of features such as tissue architecture and anatomic frame of reference in addition to cellular and nuclear morphology, as well as admixed inflammatory cells and background material, to make a final diagnosis. Screening and validating CAD algorithms to account for all of these variables requires access to cohorts of such cases, while it is also computationally expensive to query and store such WSI data sets. Furthermore, to quantitatively evaluate the performance of a CAD algorithm, receiver operator characteristic (ROC) curves and area under the curve (AUC) values are a useful but time consuming process that requires a pathologist to manually annotate, pixel by pixel, the ground truth regions of interest and then exclude unwanted regions from the ground truth for each digital slide.[15] Finally, WSI data sets are computationally expensive to query and store.
To address these problems, we have developed easy-to-use tools for pathologists to enable the creation of image microarrays (IMAs) that would facilitate the creation of a single digital slide containing only the key diagnostic features and the appropriate controls (benign mimics and background features). Thus, hundreds to thousands of adjudicated features (representing diverse sets of morphologic variants) can be presented in a single image, making it more efficient and computationally less expensive for image analysis operations to generate quantitative results such as ROC curves and AUC measures.
While there are numerous tools and tool suites that enable image aggregation, many of them are difficult to use, with them having a much lower image size limit than currently required, as stipulated by the upper single-frame storage limit of the intrinsic DICOM 3.0 standard, upon which many of these applications were designed to operate (recognizing that 10,000 × 10,000 pixels is much less than the 500,000 × 500,000 pixel limit now possible with WSI imagers). Also, smaller, subsampled images are not amenable to a precise grid format and do not allow for the labeling of each row and column. More importantly, current software often works with standardized size and resolution images, such as the images typically obtained from a digital camera. While current WSI viewers enable the cropping and cutting out of features, this is typically done by imprecisely dragging open a cropping window, thereby making the collection of images of the same size and resolution an extremely challenging and tedious process. Here, we designed a tool that enables the viewing of WSI data sets that allows for simple systematic capturing of image features at the same size and resolution. By default, the selected areas are captured from the highest available resolution layer of the original image, preserving the greatest degree of detail.
Here we apply this tool to three use cases:
The first use case involves a recently described spatially invariant vector quantization - laser capture microdissection (SIVQ-LCM) workflow.[16] We recently integrated spatially invariant vector quantization (SIVQ), a method for image segmentation and feature extraction of histopathological images,[17] into the LCM workflow (SIVQ-LCM).[16,18] SIVQ-LCM facilitates an automated, large-scale procurement of morphologically defined cell populations for molecular analysis.[16] In this case, we utilized the creation of IMAs to screen ring vectors (predicate image features used for searching of the image) for a cytology malignant pleural effusion SIVQ-LCM workflow.
The second use case utilized a WSI data set library to create an IMA of various cancers and demonstrate its potential in educational applications.
The third use case is a prostate cancer whole mount WSI data set that was used to create an IMA of prostate cancer and its benign mimics, which can be used as a reference image that has numerous potential applications to the field of pathology (discussed below).
MATERIALS AND METHODS
Image Microarray
The Image Microarray (IMA) software application was developed with VB.net™ 2010 (Microsoft Corporation, Redmond, WA) with components from the TIFFComp ActiveX control (.OCX) (Aperio Technologies, Vista, CA) and the Aperio Viewport ActiveX control (.OCX). A user interface was similarly written in VB.net. Through the interface, a Data Grid View control was populated with image filenames in the positions that they were intended to occupy in the composite image. Two additional Data Grid View controls were utilized to hold text descriptions. Jpeg images were created to correspond to each text description along with images consisting of white space to be used as separators. The text descriptions were centered in the images. An aperio composite image (ACI) file was programmatically generated, combining each image filename with the filenames of the text images and white spaces. The magnification level and the resolution value in microns per pixel for every image were also added to the ACI file. Finally, the ACI file was converted to an Aperio SVS file using the Aperio TIFFComp ActiveX control.
The automatic loading feature was created by setting the number of rows and columns within the data grid view control to a value equal to the ceiling of the square root of the total number of image filenames and filling the cells with all the available images in the directory afterward.
A CSV export function was added to create a CSV file containing the filenames (with associated magnification and resolution values) in their corresponding locations in the array. This file then served as a key to go along with the IMA (IMA-key).
dCORE
This software application was similarly developed with VB.net™ 2010 with components from the Aperio TIFFComp Active-X control (.OCX). Its user interface was also written in VB.net, with it allowing for interactive navigation with digital images, again through the Aperio Viewport ActiveX control. This enabled the execution of image-based operations such as cropping, dynamic resizing, and dynamic arbitrary adjustment of magnification, as required. The MouseDown, MouseMove, and MouseUp events were used to trigger the execution of the SetViewXY method within the ViewPort control to change the active image. The magnification of the image was updated upon alteration of the Mouse Wheel state variable, as triggered by user actuation of the wheel situated between the left and right mouse buttons. To save the exact region being viewed by the control, the ViewPort image was transferred to an image bitmap in 24-bit RGB format using the BitBlt (pronounced “bit-blit”) operator. Finally, the resultant bitmap was saved as an uncompressed TIFF file. When captured from a digital slide, the current viewed magnification of the image was included in the image filename such that it could be later added to the description of the ACI.
Use of the IMA Software
IMA uses a graphical user interface that displays a matrix of spreadsheet-type cells [Figure 1]. Image files are selected from the plurality of choices available within these cells, with user-click-events triggering the selected image to be included in the assembled image montage. This selection process is carried out in iterative fashion. The rows and columns are labeled via the text boxes at the top and to the right of the matrix of cells. Once the matrix has been loaded with the filenames corresponding to their respective images, either an ACI or SVS file is created and viewed.
Figure 1.
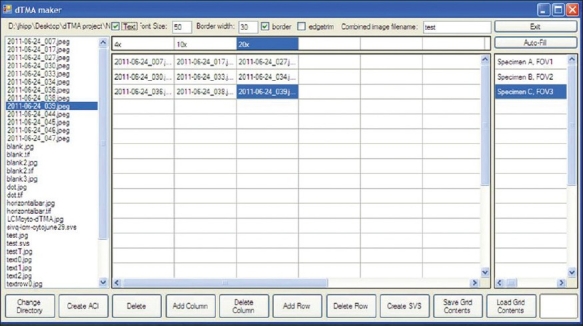
Screen capture of the IMA tool: Depicted is a representative screen capture of the user interface of the IMA tool. Files are loaded in the directory in the left hand panel. The matrix of cells in the middle represents the array. Images are loaded into the array by clicking on the file name in the directory, followed by clicking on the desired cell. The top and right-hand columns are where labels can be included.
With a graphical user interface, the image is loaded and viewed in dCORE [Figure 2]. The user navigates the image with a mouse and when a feature of interest is identified, the viewing window is adjusted to crop and capture the image. While keeping the viewing window dimensions, the user can navigate through the entire image and capture additional areas of the same dimension and resolution, as long as the viewport window has not been changed.
Figure 2.
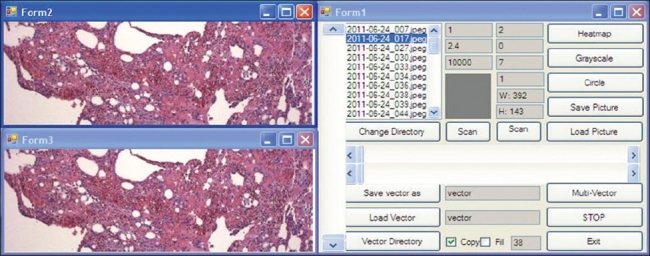
Screen capture of the dCORE tool: Depicted is a representative screen capture of the dCORE user interface. Images are loaded into the directory in the center right. Images can be panned using the upper left viewing window. Features can be cropped by altering the size of the viewing window. Clicking on the “save picture” button on the right hand panel saves the image displayed in the viewing window
Images
For the cytology use case, jpeg images were captured of an uncoverslipped (xylene served as a pseudo-coverslip),[16] H and E-stained, formalin-fixed paraffin embedded (FFPE) cell block section of a malignant pleural effusion from a lung adenocarcinoma patient, using with the camera on the Arcturus XT™ at multiple magnifications (4×, 10×, 20×) and a single z-plane. The brightness, color, and contrast of the images were “Auto Corrected” with Microsoft Office Picture Manager™.[16]
Digital slides of various cancers from the WSI database at the University of Michigan, Department of Pathology were used by dCORE. The digital slides were scanned using the Aperio XT™ whole slide scanner. Representative Tiff images were captured from our digital slide library at various magnifications.
For the prostate cancer use case, a WSI data set provided by Monaco et al.[11] was analyzed for this study; for a detailed description, the reader is directed to Monaco et al.[11] Briefly, an H and E-stained, whole-mount histological section of the prostate gland was cut into four quadrants, which were formalin fixed and paraffin embedded, and subsequently digitized at 20× magnification (0.46 um per pixel) via an Aperio Technology XT scanner, as previously described. A digital slide representing one quadrant with prostate cancer was used in this study.
The images reported here are available at the WSI repository (www.WSIrepository.org) as described by Hipp et al.[19]
Spatially Invariant Vector Quantization
The use of SIVQ image analysis has been previously described in detail by Hipp et al.[13,16]
RESULTS
IMA is a software application that allows for the simplified, interactive creation of an array of digital images, in the form of a monolithic presentation layer that allows for the comparison of preaggregated collections of digital images. dCORE is a software application that enables the user to easily capture standardized images of constrained pixel dimensions and associated resolution. While the viewer window can be adjusted to various sizes, multiple preset windows were created to enable the user to iteratively change from a large panel (often used for navigating through the image) to a series of small panels, to enable better cropping of the image feature of interest while maintaining a standardized dimension and resolution. The case number and name of the digital slide, along with the magnification at which the image was captured, is integrated into the new “image core” file name. In addition to creating the IMA digital slide, the user can also create an Excel spreadsheet of the filenames that correspond to the image locations within the IMA.
These technologies were applied to three use cases as described below:
SIVQ-LCM Use Case
Despite a number of technological advances, laser capture micro dissection remains a relatively tedious procedure, requiring visual microscopic identification of each target cell population by a trained investigator. The time consumed via the cell-by-cell selection process is often rate-limiting when many cells and/or many samples need to be reviewed and subsequently dissected. Moreover, a lengthy dissection interval can compromise the molecular integrity of the recovered biomolecules. The integration of SIVQ (SIVQ-LCM) improves the efficiency of this step;[16] however, the selection of the optimal vector then becomes the rate limiting step.
Ring vectors need to be screened for their sensitivity and specificity. For example, it is important to screen ring vectors to assess their specificity for various morphologic presentations of cells and features of interest and their sensitivity in the context of potential benign background cells and features. Screening entire slides or multiple fields of views is a time consuming and computationally expensive process. However, the efficiency of this process can be greatly improved with the creation of a single image that contains all the possible features.
Using the Arcturus XT™ machine, multiple images of different fields of view at various magnifications (4×, 10×, and 20×) were captured from a glass slide with an unconverslipped section (H and E stained) prepared from a pleural effusion specimen cell block preparation.
dCORE was used to capture the various morphologies of the tumor cells, in addition to the benign cells (including red blood cells, lymphocytes, neutrophils, histiocytes, and mesothelial cells). While SIVQ operates at multiple length scales (i.e., magnifications),[18] the vectors are selected and used at a single length scale to remain comparable; thus images that were captured at 10× were chosen and consolidated into a single image collage [Figure 3].
Figure 3.
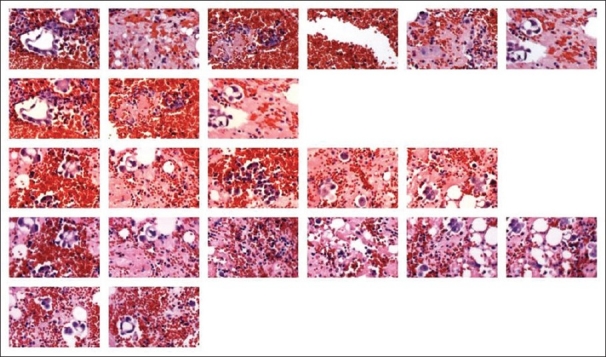
Malignant pleural effusion of a patient with lung adenocarcinoma. A cell block of a pleural effusion from a patient with metastatic lung adenocarcinoma was stained with H and E. Multiple images representative of different fields of view were taken using the camera on the Arcturus XT™ LCM stage. The slide was not coverslipped and was coated with xylene. The images were then “autocorrected” using Microsoft Picture Manager. dCORE was used to capture representative malignant and benign features, which were then loaded into IMA to create the image array shown here.
The resulting image was then imported and analyzed by SIVQ. A single ring vector was selected to capture the dark, hyperchromatic nuclear features of the tumor cells and was then screened against the IMA. The bottom panel of Figure 4 shows the results of the SIVQ analysis a heatmap of match quality. This can then be compared to the top image of Figure 4 (the original image) for qualitative validation.[1] The heatmap can then be reimported into the Arcturus XT™ machine and those “painted” cells could be microdissected so that molecular analysis could be performed.
Figure 4.
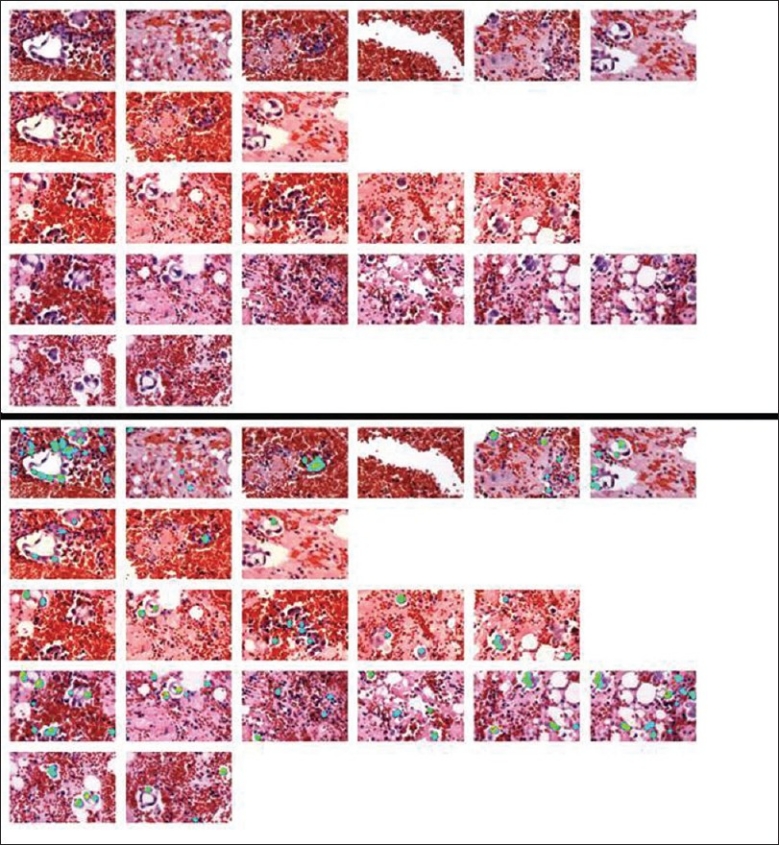
SIVQ analysis of the pleural effusion IMA. The top panel was loaded into SIVQ. A vector was selected from a tumor cell and was then used to search the image. A heatmap corresponding to the quality of matches is displayed in the lower panel.
SIVQ appears to identify all the tumor cells; however, by H and E alone, it is impossible to distinguish tumor cells from reactive mesothelial cells and immunohistochemical staining for TTF1 would be required. The heat map can then be reimported into the Arcturus XT™ machine and the corresponding cells would be microdissected and a molecular analysis performed.
IMA Created from a WSI Database
The creation of large IMAs has many educational applications. Creation of IMAs of key diagnostic features would allow pathologists, trainees, and other laboratory staff to efficiently review these image databases and be tested if required. Using the WSI database at the University of Michigan, Department of Pathology, dCORE was used to mine this image database and create 147 “image cores” (image subsets) of representative features from various cancers. These images were then assembled into a single IMA [Figure 5].
Figure 5.
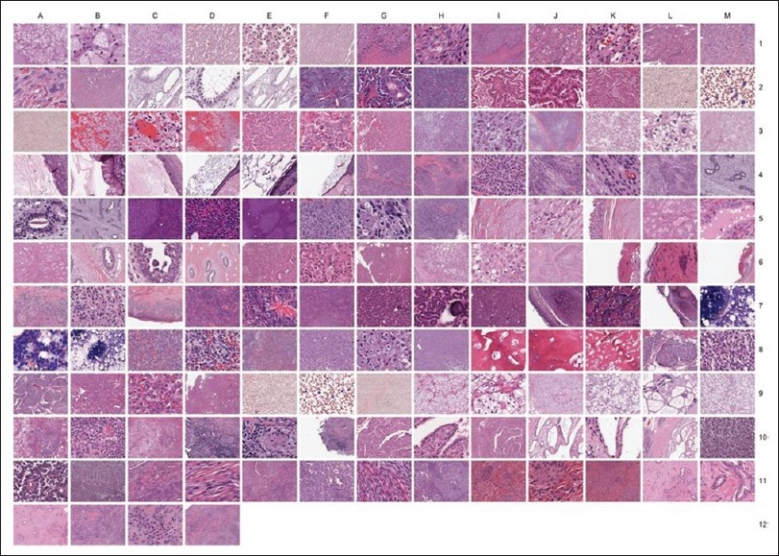
An IMA was created from the University of Michigan, Department of Pathology WSI library: Various tumors were used to create this digital slide IMA
Because we enabled the image capture and save feature to also save the original file from which it was generated, the magnification at which it was captured, and the microns per pixel recorded, into the newly generated file name, we developed an application that output this file name information into an Excel spread sheet, thus providing an annotation log corresponding to the IMA [Figure 6]. From an educational perspective, rather than studying and screening entire digital slides, users can instead more efficiently study and test themselves on key diagnostic features.
Figure 6.
A representative “key” to the IMA in Figure 5: The IMA tool has a feature that will export the file names of the images from the corresponding IMA, thus creating an Excel file which serves as a key to the IMA.
Prostate WSI Use Case
An important step in the validation and screening of image analysis/pattern recognition algorithms is in assessing their sensitivity and specificity. This task often requires the analysis of numerous digital slides to ensure adequate screening to capture the various heterogeneous morphologic presentations and to encompass all the potential background features. All tasks taken together, this effort is computationally expensive and time consuming. Creating a single IMA that captures these key visual features of pathology would improve algorithm screening efficiency. Such benchmark data sets of small images have existed outside of pathology in the field of computer vision for quite some time.[20–23]
For example, there are many morphologic and architectural features within prostate tissue sections. Using a single H and E prostate tissue section, we captured the key architectural/morphologic features of cancer, benign mimics, and related morphologic features (e.g., prostate intraepithelial neoplasia, various hyperplasias) at different magnifications using dCORE. Using the IMA application, these image subsets were then assembled into a single digital slide collage and categorized based on their magnification and cellular morphology [Figure 7]. This approach can be easily applied to other types of cancers and diseases.
Figure 7.
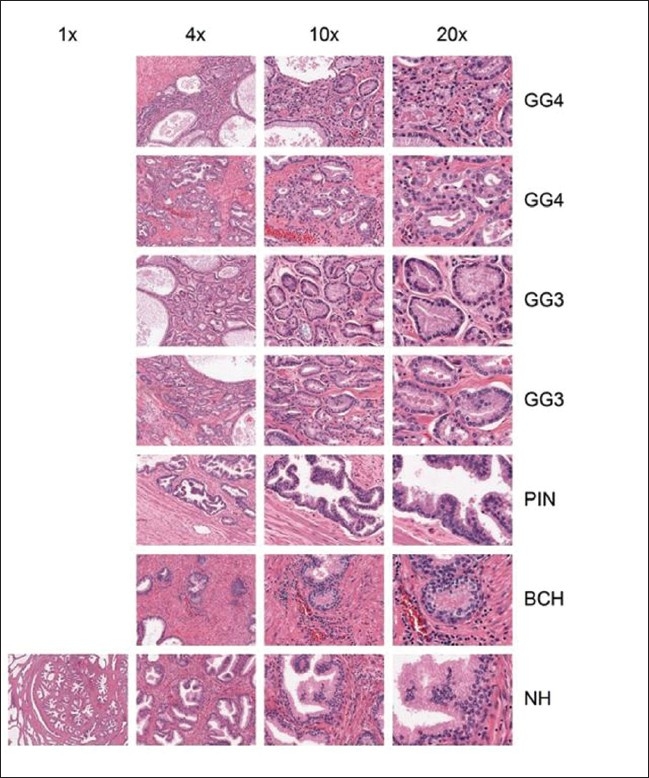
Prostate IMA: A digital slide of a prostate cancer tissue section was used by dCORE to capture various malignant and benign morphologic features, as described in the right hand column of the text. Various magnifications were taken of these features, displayed in the top row of the text
DISCUSSION
We developed a digital construct referred to as IMA that can easily create a collage of virtual image data sets in a grid-based format, analogous to a conventional TMA. Like conventional TMAs that require creating cores out of tissue blocks with standardized sized needle cores, IMAs also require a similar tool to collect images that are all of the same size and resolution. We therefore created dCORE, which performs the digital equivalent of making cores out of a conventional tissue sample, but with the resultant construct being an “image core.” The availability of IMAs allows for the comparison of preaggregated collections of digital images, thus facilitating the development of new image analysis algorithms.
With the ability to easily create and compare high-resolution subsets of WSI data, there are numerous applications of IMA in pathology such as the following:
Algorithm Development
Having a curated set of various tumor and benign lesions in a single IMA would make algorithm screening more efficient, with this innovation enabling researchers to assess performance across hundreds or thousands of samples. It would also improve the efficiency of algorithm development in as much as pathologists could concentrate their annotation efforts on higher order tasks. This would improve the efficiency of computer scientists in collaborating with diagnostic pathologists. Lastly, distributing a single IMA that contains both experimental and control images would standardize and “democratize” algorithm development to include researchers (nationally and internationally) that may not have access to digital slide archives. Standardized reference IMA data sets would provide an opportunity to enable algorithm validation where ROC curves and AUC values can easily be generated from a single IMA construct to determine potential clinical utility.
Education
Many tumors types have several subclassifications and variants. One can create an IMA atlas to represent all these variants, even within different subcategories, in support of education and training of other pathologists.
Clinical Classification and Grading of Pathology Cases
Creating an IMA of different morphologic features and allowing them to be scored by leading panel of experts can help facilitate the development of new classification and grading systems. Trainees could compare their results on scoring or grading with those of a recognized expert.[24] IMAs may also facilitate the introduction of quantitative image metrics into new diagnostic classification and grading systems.
Regulatory Affairs
IMAs can be generated using different equipment (i.e., different slide scanners) to better enable image quality comparison. Designing studies to determine intra- and interobserver variability are challenging because pathologists often remember key features unique to a slide/case. However, with the ability to create IMAs, creating large collages of image subsets and manipulating them (rotating, capturing their inverse, cropping the fields of view differently) might overcome this limitation in designing such studies.
Proficiency Testing and Quality Reviews
Creating IMAs may allow for even better standardization of image materials used in interlab proficiency testing. Similarly, daily quality reviews, including cross-modality comparisons, would be facilitated by IMAs. This may be particularly important for disciplines such as cytopathology and hematopathology, which require triaged review of the daily work of lab professional screeners.
Histology Stain Digital Workflow
Histology laboratories that routinely perform special stains, including immunohistochemistry, routinely prepare control slides associated with each distinct epitope class. The workflow for producing these control studies can be logistically complex, as some protocols stipulate on-slide controls (with actual control tissue juxtaposed to the specimen in question) or conversely, batch-level controls where one separate slide suffices for validating the batch of slides being run in the immunostainer. As a consequence of this plurality of control slide rendering strategies, combined with commonly encountered geographic constraints of complex and distributed pathology practices, it may be difficult to provide all pathologists, on a daily basis, with access to all the controls associated with their ordered studies. Already, it is not an uncommon (but perhaps unfortunate) practice to observe larger academic departments where the control slides themselves are replaced by a proxy document, which stipulates that all controls were reviewed centrally, and were found to be in compliance with acceptance criteria. While acceptable, from a compliance perspective, this practice is undesirable, as it potentially removes subtle visual cues to the interpreting pathologist about absolute stain intensity and qualitative interbatch variations, which might be helpful in establishing grading thresholds. Thus, the assemblage of an IMA construct specifically tailored to house all daily controls (whether intraslide or interslide) would offer a cogent and thorough mechanism for their simplified distribution, by proxy, and subsequent review. Such digital image constructs could be easily created and distributed along with the primary data of the actual specimen WSI files (or similarly, stained slides), thus avoiding the unnecessary and wasteful step of shuttling the control slides themselves. Certainly, the control slide/IMA constructs could and should be archived, in support of possible consultation, rereview or as otherwise required, such as in this case of audits. Finally, in contrast to the need for managing distinct static images, the IMA construct exhibits would exhibit the added benefit of allowing for accelerated and simplified slide-to-slide registration across multiple semantically and spatially coupled IMA planes, such that a pathologist can rapidly switch between all control and specimen sections of a given case, on a per-stain basis.
Cytology and Hematology
In cytopathology, automated Pap test screening workflow relies on imaging systems that detect and record cytological abnormalities present on glass slides. To review these selected abnormal areas, users (i.e., cytotechnologists) rely on stored coordinates to find the exact location on the glass slide (so-called Pap map).[25]
In hematology, automated digital imaging technology has been utilized to review peripheral blood smears and perform differentials instead of relying upon manual microscopy, which is time consuming and inconsistent.[26] With this digital cell morphology system, digital images captured during review of a peripheral blood smear can be preclassified and displayed for analysis in real-time or via remote teleconsultation. In both instances, key diagnostic morphologies from cases are captured and stored in a single file, making them easier to display, share and to identify, in terms of diagnostic cell. These image subsets can be built into one IMA, which would generate a much smaller size file, allowing for easier review and simplified distribution via wireless and distant networks. In addition, use of IMAs could provide a digital archive for multiple uses (e.g., presentation at tumor boards, simplified incorporation into reports, transmission into the patient's chart within the electronic medical record, and enabling of simplified image analysis).
Gross Images
The addition of gross images to the internal IMA representational data model adds compelling opportunities for anatomic frame-of-reference spatial fiduciary tracking, where the location and extent of every microscopic field could be coregistered with its parent gross image. Additionally, the presence of gross images would allow for multimodality evaluation of IMA-encoded cases, where the following could be conveniently considered all together: gross images, H and E stained images, special-stained images, and IHC stained images. Similarly, viewers could easily be extended to allow for side by side comparisons of large cohorts of IMA-encoded cases; a potential aid to both clinical consensus activities and collation of ground truth common morphologic features, during histopathologic discovery.
Research
IMAs can be used to better understand the statistical sampling problem in conventional TMAs. For example, when using a TMA to screen for tumor cell expression of a protein, it is generally assumed that using three cores is sufficient to allow for coverage of biologic heterogeneity of the protein in the disease class being studied. This is true for proteins with a certain degree of homogeneity but a rigorous image-based statistical approach has not been performed to understand the true number of cores needed for a protein under nonhomogenous expression conditions. One can therefore create IMAs on IHC slides where the protein in question varies in expression for 0-10% expression (low expresser), to intermediate expresser (20-50% expression), to high expresser (>50% cells express). IMAs could then be used to define the number of virtual cores required to statistically represent the starting population, thereby allowing optimum TMA construction. In addition, IMAs could be used to investigate quantitative and qualitative drift of IHC staining by comparing control tissues for an antibody run over time.
IMAs would allow for more efficient evaluation of tumor heterogeneity in research projects by capturing all of a tumor's morphologic variation on a single slide. Because of the spatial heterogeneity of the target molecular expression in cancer tissues, Halama et al.[27] demonstrated that for personalized medicine applications, WSI is irreplaceable when compared to TMAs. This reality is true during the identification of prognostic markers, as well as in their subsequent application because spatial orientation of marker expression is preserved in evaluating the whole tissue section rather than stochastically sampled smaller regions (i.e., cores).[5,27] Therefore, taking multiple cores from the same paraffin block is not only required but is also laborious and doing so increases the risk of destroying the block. Thus, the use of WSI as an alternative to TMAs has the following advantages: (1) the laborious work of repeated TMA sampling is avoided; (2) the statistical uncertainty of number of cores needed is avoided because the whole tissue is scanned; (3) the spatial context of each area imaged is preserved.[27] Using the WSI, Halama et al. were able to create several 1 mm2 fields of view and constitute individual “tumor maps” [Figure 4].[27]
Potentially, the creation of “tumor maps” with dCORE and IMA synergistically combines the advantages of using conventional TMAs with the advantages of using WSI, allowing pathologists to easily create IMAs containing cohorts of “tumor maps.” Thus pathologists can ensure that adequate and representative areas of the digital tissue section are sampled and yet their location in the array can ensure their spatial preservation.
CONCLUSION
Akin to how TMAs revolutionized biomarker development by enabling the efficient and rapid screening of IHC and in situ molecular probes, we envision IMAs serving similar functions for CAD algorithm development. While one can argue that creating digital slides of TMAs could serve a similar function,[28] the advantages of creating IMAs are that they are much simpler, quicker and less expensive to produce since access to the tissue block is not necessary; key morphologic features can be easily and definitively captured instead of blindly punching into the tissue block with a needle). Similarly, compliance issues may be reduced since only image data are being used. Furthermore, with the adoption of an all-digital workflow, we envision that there will be archives of WSI data sets representing virtually every morphologic variant of entire disease classes readily available to be mined. In addition, these tools were created with the end user (the pathologist) in mind; thus the worldwide pathology community can contribute and share their IMAs, thus readily ensuring that all disease entities are eventually captured. The use of these tools will also lessen the divide between pathologists and computer vision experts, providing the latter with access to high quality referenced data sets to more efficiently facilitate algorithm development.
Currently, there exist tools such as CellaVision™, which automatically analyze slides and categorize individual images of cells based on their morphology.[26,29] This technology locates the erythrocyte monolayer and leukocytes at low magnification and captures images at higher magnification.[30] In comparison, reviewing surgical pathology slides is often a different and more complex process because it often requires assessment at different length scales (zooming in and out at different magnifications) for both single cellular morphologies and architectural features. Thus, WSI data sets are needed to capture these dynamic length scales. Although use of IMA construct does notafford automation as is present with the use of platfroms such as CellaVistaTM (which makes it more time consuming and user dependent), IMA use is none the less more applicable to all types of pathology use cases. Moreover, the IMA construct is easy to use and leverages the domain expertise intrinsic to the pathalogist. In addition to being easy to use, and most importantly, it leverages the domain expertise of the pathologist. Since IMA use is manually controlled, and thus customizable, it allows the pathologist to control the capture of the pertinent diagnostic features at the necessary length scales (i.e., architectural features, small groups of cells, and/or specific cytoplasmic/nuclear morphologies) pertinent to the diagnostic work-up of the case. This enables of the gathered collection as a single image (even when sourced from multiple cases) for the numerous applications discussed above.
The key benefit of using IMAs (for research or clinical use) is the ability to perform rapid analysis over a panel of images obtained from different original samples collected independently, with this attribute being particularly compelling where pair-wise and multi-image comparisons (or annotations) are an integral component of the analytical process. Additionally, the smaller absolute data size, as represented by the highly concentrated IMA format, represents an opportunity for expedited exchange of many encapsulated images in a single convenient and compact electronic construct, which opens a plurality of possibilities for simplified collaboration, tele-health, and indeed global health applications.
Lastly, the field of computer aided diagnostics is limited due to the domain expertise divide between pathology, computer science, and electrical engineering experts. Typical algorithm development and validation requires the pathologist to identify key features, send these to an imaging scientist, who develops a potential algorithm, analyzes the images and sends them back to the pathologist for evaluation. To an active pathologist performing clinical service, this iterative loop is time consuming and inefficient. The tools described here, IMA and dCORE, will better enable the pathologist to help computer scientists/engineers by allowing them to easily capture the key diagnostic and benign features of interest and efficiently organize the images (i.e., benign and malignant) at different length scales, with this accelerating the screening and discovery process.
A key area for future development are software packages that enables color normalization to correct for differences in slide staining and tissue processing. One can envision in the near future that a user might select textual metadata and then be presented with an IMA for this search. Alternately, the user might start with a ring vector as a search predicate and then be driven to a dynamically constructed IMA of cases with similar ring vectors.
In conclusion, in a manner analogous to the way that conventional TMA technology has significantly accelerated in situ studies of tissue specimens, use of IMA construct has similar potential to significantly accelerate CAD development.
Footnotes
1SIVQ identifies all the tumor cells. However, by H and E alone, it is nearly impossible to distinguish tumor cells from reactive mesothelial cells and immunohistochemical staining for TTF1 would be required. Therefore, further confirmation with use of SIVQ with the corresponding TTF-1 immunohistochemical stained slide would be required for definitive confirmation.
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2011/2/1/47/86829
REFERENCES
- 1.Avninder S, Ylaya K, Hewitt SM. Tissue microarray: A simple technology that has revolutionized research in pathology. J Postgrad Med. 2008;54:158–62. doi: 10.4103/0022-3859.40790. [DOI] [PubMed] [Google Scholar]
- 2.Nocito A, Kononen J, Kallioniemi OP, Sauter G. Tissue microarrays (TMAs) for high-throughput molecular pathology research. Int J Cancer. 2001;94:1–5. doi: 10.1002/ijc.1385. [DOI] [PubMed] [Google Scholar]
- 3.Battifora H. The multitumor (sausage) tissue block: Novel method for immunohistochemical antibody testing. Lab Invest. 1986;55:244–8. [PubMed] [Google Scholar]
- 4.Singh A, Sau AK. Tissue Microarray: A powerfuland rapidly evolving tool for high-throughput analysis of clinical specimens. IJCRI. 2010;1:1–6. [Google Scholar]
- 5.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–7. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 6.Simon R, Sauter G. Tissue microarrays for miniaturized high-throughput molecular profiling of tumors. Exp Hematol. 2002;30:1365–72. doi: 10.1016/s0301-472x(02)00965-7. [DOI] [PubMed] [Google Scholar]
- 7.Madabhushi A. Digital Pathology Image Analysis: Opportunities and Challenges. Imaging Med. 2009;1:4. doi: 10.2217/IIM.09.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fatakdawala H, Xu J, Basavanhally A, Bhanot G, Ganesan S, Feldman M, et al. Expectation-maximization-driven geodesic active contour with overlap resolution (EMaGACOR): Application to lymphocyte segmentation on breast cancer histopathology. IEEE Trans Biomed Eng. 2009;57:1676–89. doi: 10.1109/TBME.2010.2041232. [DOI] [PubMed] [Google Scholar]
- 9.Basavanhally AN, Ganesan S, Agner S, Monaco JP, Feldman MD, Tomaszewski JE, et al. Computerized image-based detection and grading of lymphocytic infiltration in HER2+ breast cancer histopathology. IEEE Trans Biomed Eng. 2010;57:642–53. doi: 10.1109/TBME.2009.2035305. [DOI] [PubMed] [Google Scholar]
- 10.Lexe G, Monaco J, Doyle S, Basavanhally A, Reddy A, Seiler M, et al. Towards improved cancer diagnosis and prognosis using analysis of gene expression data and computer aided imaging. Exp Biol Med (Maywood) 2009;234:860–79. doi: 10.3181/0902-MR-89. [DOI] [PubMed] [Google Scholar]
- 11.Monaco JP, Tomaszewski JE, Feldman MD, Hagemann I, Moradi M, Mousavi P, et al. High-throughput detection of prostate cancer in histological sections using probabilistic pairwise Markov models. Med Image Anal. 2010;14:617–29. doi: 10.1016/j.media.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurcan MN, Boucheron L, Can A, Madabhushi A, Rajpoot N, Yener B. Histopathological Image Analysis: A Review. IEEE Rev Biomed Eng. 2009;2:147–71. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hipp J, Flotte T, Monaco J, Cheng J, Madabhushi A, Yagi Y, et al. Computer aided diagnostic tools aim to empower rather than replace pathologists: Lessons learned from computational chess. J Pathol Inform. 2011;2:25. doi: 10.4103/2153-3539.82050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle S, Feldman M, Tomaszewski J, Madabhushi A. A Boosted Bayesian Multi-Resolution Classifier for Prostate Cancer Detection from Digitized Needle Biopsies. IEEE Trans Biomed Eng. 2010:99. doi: 10.1109/TBME.2010.2053540. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez DC, Bhargava R, Hewitt SM, Levin IW. Infrared spectroscopic imaging for histopathologic recognition. Nat Biotechnol. 2005;23:469–74. doi: 10.1038/nbt1080. [DOI] [PubMed] [Google Scholar]
- 16.Hipp J, Cheng J, Hanson JC, Yan W, Taylor P, Hu N, et al. SIVQ-aided laser capture microdissection: A tool for high-throughput expression profiling. J Pathol Inform. 2011;2:19. doi: 10.4103/2153-3539.78500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hipp JD, Cheng JY, Toner M, Tompkins R, Balis U. Spatially Invariant Vector Quantization: A pattern matching algorithm for multiple classes of image subject matter- including Pathology. J Pathol Inform. 2011;2:13. doi: 10.4103/2153-3539.77175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, et al. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 19.Hipp JD, Lucas DR, Emmert-Buck MR, Compton CC, Balis UJ. Digital slide repositories for publications: lessons learned from the microarray community. Am J Surg Pathol. 2011;35:783–6. doi: 10.1097/PAS.0b013e31821946b6. [DOI] [PubMed] [Google Scholar]
- 20.Russell B, Torralba A, Murphy K, Freeman W. LabelMe: A Database and Web-Based Tool for Image Annotation. Int J Comp Vision. 2008;77:157–73. [Google Scholar]
- 21.Computer Vision Test Images. [Last cited on 2011 July 22]. available from: http://www.cs.cmu.edu/~cil/vimages.html .
- 22.Databases or Datasets for Computer Vision Applications and Testing. [Last cited on 2011 July 22]. available from: http://datasets.visionbib.com/index.html .
- 23.CV Datasets on the web. [Last cited on 2011 July 22]. available from: http://www.cvpapers.com/datasets.html .
- 24.Bubendorf L, Nocito A, Moch H, Sauter G. Tissue microarray (TMA) technology: Miniaturized pathology archives for high-throughput in situ studies. J Pathol. 2001;195:72–9. doi: 10.1002/path.893. [DOI] [PubMed] [Google Scholar]
- 25.Pantanowitz L, Hornish M, Goulart RA. The impact of digital imaging in the field of cytopathology. Cytojournal. 2009;6:6. doi: 10.4103/1742-6413.48606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Briggs C, Longair I, Slavik M, Thwaite K, Mills R, Thavaraja V, et al. Can automated blood film analysis replace the manual differential. An evaluation of the CellaVision DM96 automated image analysis system? Int J Lab Hematol. 2009;31:48–60. doi: 10.1111/j.1751-553X.2007.01002.x. [DOI] [PubMed] [Google Scholar]
- 27.Halama N, Zoernig I, Spille A, Michel S, Kloor M, Grauling-Halama S, et al. Quantification of prognostic immune cell markers in colorectal cancer using whole slide imaging tumor maps. Anal Quant Cytol Histol. 2010;32:333–40. [PubMed] [Google Scholar]
- 28.Krenacs T, Ficsor L, Varga SV, Angeli V, Molnar B. Digital microscopy for boosting database integration and analysis in TMA studies. Methods Mol Biol. 2010;664:163–75. doi: 10.1007/978-1-60761-806-5_16. [DOI] [PubMed] [Google Scholar]
- 29.Cornet E, Perol JP, Troussard X. Performance evaluation and relevance of the CellaVision DM96 system in routine analysis and in patients with malignant hematological diseases. Int J Lab Hematol. 2008;30:536–42. doi: 10.1111/j.1751-553X.2007.00996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ceelie H, Dinkelaar RB, van Gelder W. Examination of peripheral blood films using automated microscopy; evaluation of Diffmaster Octavia and Cellavision DM96. J Clin Pathol. 2007;60:72–9. doi: 10.1136/jcp.2005.035402. [DOI] [PMC free article] [PubMed] [Google Scholar]



