Abstract
SUMOylation is a relevant protein post-translational modification in eukaryotes. The C terminus of proteolytically activated small ubiquitin-like modifier (SUMO) is covalently linked to a lysine residue of the target protein by an isopeptide bond, through a mechanism that includes an E1-activating enzyme, an E2-conjugating enzyme, and transfer to the target, sometimes with the assistance of a ligase. The modification is reversed by a protease, also responsible for SUMO maturation. A number of proteins have been identified as SUMO targets, participating in the regulation of cell cycle progression, transcription, translation, ubiquitination, and DNA repair. In this study, we report that orthologous genes corresponding to the SUMOylation pathway are present in the etiological agent of Chagas disease, Trypanosoma cruzi. Furthermore, the SUMOylation system is functionally active in this protozoan parasite, having the requirements for SUMO maturation and conjugation. Immunofluorescence analysis showed that T. cruzi SUMO (TcSUMO) is predominantly found in the nucleus. To identify SUMOylation targets and get an insight into their physiological roles we generated transfectant T. cruzi epimastigote lines expressing a double-tagged T. cruzi SUMO, and SUMOylated proteins were enriched by tandem affinity chromatography. By two-dimensional liquid chromatography-mass spectrometry a total of 236 proteins with diverse biological functions were identified as potential T. cruzi SUMO targets. Of these, metacaspase-3 was biochemically validated as a bona fide SUMOylation substrate. Proteomic studies in other organisms have reported that orthologs of putative T. cruzi SUMOylated proteins are similarly modified, indicating conserved functions for protein SUMOylation in this early divergent eukaryote.
Post-translational modification of proteins with ubiquitin (Ub)1 or ubiquitin-like modifiers (Ubls) has emerged as an important intracellular signaling event (1). SUMOylation involves the covalent attachment of the Small Ubiquitin-like Modifier (SUMO) to target proteins. SUMO is initially synthesized as a precursor that is proteolytically processed at the C terminus exposing a glycine residue which is subsequently linked to an internal lysine residue of the substrate protein through the concerted action of E1-activating and E2-conjugating enzymes, and eventually an E3-ligase enzyme (2). SUMO has been shown to modulate activity, stability and/or localization of the modified protein, likely by masking or adding interaction surfaces or by inducing conformational changes that result in altered protein-protein interactions (3).
In yeast and mammalian cells a prominent role for SUMO in several important cellular processes, such as DNA replication and repair, chromosome segregation, gene expression, and nuclear trafficking was described (4, 5). Moreover, results from large-scale proteomic studies provided a great number of potential SUMO targets, suggesting that a broader spectrum of biological processes could be influenced by this post-translational modification (6–17).
Trypanosomatids are flagellated protozoan organisms representing some of the most primitive eukaryotes. Members of this group include human parasites that cause a variety of diseases: Trypanosoma cruzi is the causative agent of Chagas disease, a chronic debilitating illness widespread in Central and South America; T. brucei causes sleeping sickness in sub-Saharian Africa; and multiple species of Leishmania cause cutaneous, mucocutaneous or visceral leishmaniasis in South America, East Africa, Asia, and the Mediterranean region (http://www.who.int).
In T. cruzi, it has been demonstrated that Ub and Ubls are important for parasite survival playing a significant role in the adaptation to the different hosts. Ub controls protein fate by triggering degradation through the 26S proteasome and the Ubl Atg8 is involved in autophagosome biogenesis that recycles cellular material. In the case of SUMO studies performed in T. brucei have shown that SUMO is essential in cell cycle regulation in procyclic and bloodstream forms (18, 19). However, the SUMOylation system has not been characterized yet at the molecular level nor have the targets of SUMO been identified in any Trypanosomatid.
In this work, we present the identification of the major components of the SUMO pathway in T. cruzi, as well as demonstrate the functionality of the system in this parasite. We designed a purification scheme involving two affinity chromatography steps and obtained an enriched fraction of SUMOylated proteins from transgenic parasites expressing a tagged version of T. cruzi SUMO (TcSUMO). We identified a number of proteins present in this sample by liquid chromatography-tandem mass spectrometry (LC-MS/MS). These results suggest the involvement of SUMOylation in a broad spectrum of biological processes, either well conserved among eukaryotes or distinctive of these Trypanosomatids. Finally, we confirmed that T. cruzi metacaspase-3 is SUMOylated in vivo, thus being a novel bona fide target.
EXPERIMENTAL PROCEDURES
Parasites
The different life-cycle forms (epimastigote, metacyclic trypomastigote, cell-derived trypomastigote, and amastigote) of T. cruzi CL Brener strain were cultured in vitro. Briefly, epimastigotes were grown axenically in a medium containing brain-heart infusion-tryptose medium and fetal calf serum (BHT) as described (20). Metacyclic trypomastigotes were obtained by spontaneous differentiation of epimastigotes at 28 °C, followed by purification by DEAE-cellulose chromatography (21). Amastigotes and trypomastigotes were obtained by infection of Vero cell monolayers with trypomastigotes (22). SUMO-transfectant epimastigotes were maintained in BHT containing 0.1 mg/ml G418.
In Silico Analysis
BLAST searches were performed against T. cruzi genome database (http://www.genedb.org) using the main components of the SUMO pathway from S. cerevisiae and human as queries. The assignment of orthology was based on the number of the E-values in the BLAST output (<10−3) and in the presence of key amino acid residues or domains. Sequences were aligned with ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and occasionally refined manually.
Recombinant TcSUMO Protein Expression and Antibody Production
The sequence corresponding to Tc00.1047053511661.50 (hereafter referred to as TcSUMO) was amplified by PCR using as template T. cruzi CL Brener genomic DNA, with primers flanked by NheI and BamHI restriction sites. The resulting DNA fragment was gel-purified by Qiaquick columns (Qiagen, Valencia, CA), cloned into pGEM-T Easy vector (Promega, Madison, WI), and completely sequenced (Macrogen, Seoul, Korea). Inserts were liberated with the appropriate restriction enzymes (New England Biolabs, Ipswich, MA) and cloned into the same sites of pET-28a(+) bacterial expression vector (Novagen, EMD Chemicals, San Diego, CA) generating pET28-His6-TcSUMO-His6 for the expression of double (N- and C-terminal) His-tagged protein. The primers used in this work are listed at the end of Experimental Procedures section.
Plasmid construct pET28-His6-TcSUMO-His6 was used to transform Escherichia coli Bl21 - Codon Plus (Stratagene, La Jolla, CA). Cultures were induced with 0.5 mm isopropyl β-d-thiogalactoside for 4 h at 37 °C, harvested by centrifugation at 3000 × g for 10 min and pellets were frozen. Cells were thawed at 4 °C and lysed with Tris-buffered saline (TBS) (50 mm Tris-HCl pH 7.6, 150 mm NaCl) containing 0.1% Triton X-100 and 0.1 mg/ml lysozyme. After sonication, cell debris was removed by centrifugation at 20,000 × g for 25 min at 4 °C and the supernatant was applied to a fast flow Ni-NTA column (Amersham Biosciences, GE Healthcare), pre-equilibrated with binding buffer (50 mm Tris-HCl pH 7.6, 500 mm NaCl). The column was first washed with binding buffer, then with the same buffer supplemented with 30 mm imidazole, and finally proteins were eluted with binding buffer containing 300 mm imidazole. Eluate containing the recombinant protein was pooled and buffer was exchanged to phosphate-buffered saline (PBS) (10 mm Na2HPO4, 150 mm NaCl, pH 7.2) using a PD-10 column (Amersham Biosciences, GE Healthcare).
Polyclonal rabbit and mouse antibodies were raised by immunization with purified recombinant TcSUMO in PBS using standard protocols (23). The first immunization was performed using 200 μg (for rabbits) or 20 μg (for mice) of the protein with complete Freund's adjuvant, followed by three boosters using one quarter of the amount of protein with incomplete adjuvant. The boosters were given every 21 days for rabbits and every 14 days for mice. Ten days after the last booster the animals were bled and the antisera were obtained.
Generation of TcSUMO Transfectant Epimastigotes
A full-length version of TcSUMO (construct 1) was tagged at the N terminus with a 6 × His-HA tag and at the C terminus with a FLAG (DYKDHDGD) epitope by performing two consecutive rounds of PCR. In a first PCR, TcSUMO was amplified from the clone in pGEM-T Easy vector using a forward primer that introduced the HA tag upstream the first triplet within the open reading frame (HA sense primer) and a reverse primer that introduced the FLAG epitope and XhoI restriction site (FLAG reverse primer). The second PCR was performed using the product of the first reaction as a template, the same reverse primer and a forward primer encoding six histidine residues after the artificially added ATG start codon and a SmaI site (His×6 sense primer). Full-length version of TcSUMO with alanine replacing the scissile glycine (construct 2) was generated by site-directed mutagenesis using the Quick Change site directed mutagenesis kit (Stratagene) and the mutation was verified by automatic DNA sequencing. TcSUMO mature form (construct 3), as well as TcSUMO nonconjugatable form (construct 4), were amplified as explained for construct 1 except that no tag was added at the C terminus (using reverse primers GG and ΔGG for construct 3 and 4, respectively). The corresponding DNA fragments were completely sequenced and cloned into the pTex plasmid (24). These four constructs were used to transfect T. cruzi epimastigotes as previously described (25).
Tandem-affinity Purification of SUMO Conjugates
For standard tandem-affinity purification (TAP), a 1-liter epimastigote T. cruzi culture was grown to a density of 5 × 107 cells per ml, which corresponded approximately to 4 g of parasites. Cells were washed twice in PBS and harvested by centrifugation at 1500 × g for 5 min. Pellets were frozen immediately in liquid nitrogen and kept at −80 °C until use. This material was resuspended in 20 ml lysis buffer (50 mm Tris-HCl pH 7.6, 0.5 m NaCl, 0.5% Nonidet P-40, 50 μm TLCK, 2 mm Iodoacetamide, 2 mm, phenylmethylsulfonyl fluoride) and incubated for 15 min on ice. When necessary, DNA present in the sample was removed by the addition of 10 μm DNase. Cell-free extracts were obtained by centrifugation at 20,000 × g for 30 min. The supernatant was incubated with 3 ml slurry of Ni-NTA resin (Amersham Biosciences, GE Healthcare) pre-equilibrated with binding buffer (50 mm Tris-HCl pH 7.6, 500 mm NaCl) for 2 h at 4 °C. The resin was washed 3 times with 25 ml binding buffer and eluted with 5 ml binding buffer containing 500 mm imidazole. Fractions containing protein were combined and diluted twofold with buffer TBS containing 1% Nonidet P-40. The sample was then mixed with 1 ml of anti-HA Affinity Matrix (Roche) and incubated for 2 h at 4 °C. The matrix was washed with 10 ml of TBS containing 0.5% Nonidet P-40, packed in a column, and the HA-tagged proteins were eluted with 1.5 ml of 50 mm Tris-HCl pH 7.6, 1% SDS. Finally proteins were precipitated with acetone 80% during 24 h at −20 °C. The control preparation for this experiment was prepared in the same manner, except that wild-type (nontransfected) epimastigotes were used.
Liquid Chromatography-Tandem Mass Spectrometry Analysis (LC-MS/MS)
Protein pellets were dissolved in 50 μl 0.4 m NH4HCO3 containing 8 m urea and reduced with 5 mm dithiothreitol for 15 min at 50 °C. Reduced thiol groups were alkylated with 10 mm iodoacetamide for 30 min at room temperature, and diluted 8 fold with HPLC-grade water. Samples were digested overnight with 6 μg sequencing-grade trypsin (Sigma-Aldrich), as described (26). After quenching the reaction with 10 μl formic acid, samples were desalted with C18 cartridges (100 mg, 1 ml, Supelco) and dried in a vacuum centrifuge. Peptides were redissolved in 50 μl 0.1% formic acid and subjected (20 μl) to two-dimensional LC-MS/MS analysis, using an Eksigent 1D-plus nanoLC coupled to a LTQ XL/ETD-MS (Thermo Fisher Scientific), equipped at the front end with a TriVersa NanoMate electrospray ionization nano-source (Advion). In this setup, peptides are loaded onto a first-dimension strong cation exchange column (5 μl, Optimized Technologies) and the elution is achieved by sequential injection (20 μl each time) of increasing salt concentrations (0, 25, 50, 100, 200, and 500 mm NaCl in 5% acetonitrile (ACN)/0.5% formic acid) through the autosampler. Eluting peptides are then captured into one of the two C18-reverse phase trap columns (1 cm, 75 μm, Phenomenex Luna C18, 5 μm) of the system and washed extensively (130 min with 5% ACN/0.5% formic acid, at 1.5 μl/min) to remove the salt. In the next round, reverse-phase (RP) chromatography is performed connected online to the mass spectrometer; whereas a second step of strong cation exchange elution is performed simultaneously through the second C18-RP column, thus avoiding time gaps between runs and increasing the capability and throughput of the system (ES Nakayasu, et al. unpublished data). The separation in the second C18-reverse phase capillary column (20 cm, 75 μm, Phenomenex Luna C18, 5 μm) was carried out using the following linear gradient: Solvent A: 5% ACN/0.1% formic acid; Solvent B: 80% ACN/0.1% formic acid, 5–40% B in 200 min, 50–90% B in 1 min, 5 min in B, 90–5% B in 1 min, and 20 min in 5% B. The MS system was set to perform one full scan (400–2000 m/z range) followed by MS/MS of the 10 most abundant parent ions (isolation width = 3.0 m/z; 35% normalized collision energy). The dynamic exclusion was set to collect each parent ion twice and subsequently excluding them for 30 s.
Protein Identification and Quantitative Analysis
The resulting MS/MS spectra (800–3500 Da, minimum of 10 counts and 15 ions) were converted to DTA files submitted to database search using SEQUEST (Available in Bioworks 3.3.1, Thermo Fisher Scientific). The database was comprised of T. cruzi, Bos taurus, human keratin, and porcine trypsin sequences (all in correct and reverse orientations, totaling 191,242 searched sequences), downloaded from GenBank on September 29, 2009. Bos taurus contaminant proteins arise from the bovine serum present in the culture medium. Epimastigote forms of T. cruzi not only adsorb these proteins to their surfaces but also take up large amounts of serum proteins and other nutrients to store them in organelles called reservosomes (27). Bovine proteins, human keratin, and porcine trypsin were filtered out before assembling the last version of the identification tables (supplemental Tables S1 and S2).
The parameters for database search were: (1) 2.0 and 1.0 Da for peptide- and fragment-mass tolerance; (2) trypsin cleavage at both termini and one missed cleavage allowed; and (3) methionine oxidation and cysteine carbamidomethylation as variable and fixed modifications, respectively. The Sequest results were filtered using Bioworks 3.3.1 with delta correlation (dCN) ≥ 0.05; peptide probability ≤ 0.05; and cross correlation (Xcorr) ≥ 1.5, 2.0, and 2.5 for singly, doubly and triply charged peptides, respectively. After filtering, peptide sequences were assembled into protein sequences and the redundant proteins into protein groups, using an in-house Perl script (28). The results were further filtered with a sum of peptide Xcorr ≥ 6, at least two unique peptides and protein probability ≤ 1E-3, which provided a false-discovery rate for protein identification of 0.7%. Differential protein expression analysis spectral count data were analyzed by QSpec, a software for differential protein expression (29). QSpec employs Poisson distribution to model the spectral count data and computes Bayes factor, equivalent to the likelihood ratio of differential expression for each protein. Large Bayes factors indicate stronger evidence of differential expression. Following the likelihood ratio computation, QSpec estimates the false discovery rates at various thresholds using an empirical Bayes approach (29). Spectra of peptides shared by more than one protein were also included in our analysis. We only considered as a SUMOylated candidate proteins with a QSpec false-discovery rate ≤ 0.01 and at least twice more abundance.
Metacaspase-3 Pull-Down Assay
For inducible expression of metacaspase-3 in the parasite, we first generated a cell line expressing T7 RNA polymerase and Tet repressor genes by transfecting epimastigotes with the plasmid pLew13 using the aforementioned electroporation method. Stable transfectants were selected and routinely grown in brain-heart-tryptose (BHT) medium supplemented with 10% inactivated fetal calf serum and 100 μg/ml G418 (Invitrogen). This parental cell line was then transfected with pTcINDEX construct (30) carrying the metacaspase-3 gene fused to a triple-FLAG epitope, and transgenic parasites were obtained after selection with 100 μg/ml G418 and 200 μg/ml hygromycin B (Calbiochem). Epimastigote cultures were grown to reach a cell density of 5–10 × 106 parasites/ml and protein expression was induced by the addition of 10 μg/ml tetracycline for 3 days. Epimastigotes (1 × 109 cells) were harvested by centrifugation at 1000 × g for 5 min, washed in PBS and lysed on ice by incubation in buffer TBS supplemented with 1% Nonidet P-40 and protease inhibitors (50 μm TLCK, 2 mm iodoacetamide) (TBS/Nonidet P-40/PI). The insoluble material was removed by centrifugation at 20,000 × g for 15 min, and the supernatant was incubated with 50 μl anti-FLAG M2 Agarose Affinity Gel (Sigma-Aldrich) pre-equilibrated with TBS/Nonidet P-40/PI. The beads were washed five times with TBS/Nonidet P-40/PI and proteins were eluted with Laemmli's sample buffer.
Electrophoresis and Western blotting
Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane for probing. TcSUMO was detected using the polyclonal antibodies raised in rabbits against the recombinant purified protein diluted 1:300. The HA tag was detected using a high-affinity rat monoclonal antibody (Roche) diluted 1:1000. The FLAG epitope was detected using anti-FLAG M2 mouse monoclonal antibody (Sigma-Aldrich) diluted 1:1000. Horseradish peroxidase-conjugated goat anti-rabbit (Sigma), goat anti-rat (Calbiochem, San Diego, CA), or goat anti-mouse (Calbiochem) secondary antibody (1:5000 dilution) was detected by chemiluminescence using SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL).
Immunofluorescence Studies
Parasites were fixed with 4% paraformaldehyde in PBS for 15 min and adhered to glass slides as described (25). Coverslides were saturated in the blocking buffer (2% bovine serum albumin, 0.1% saponin in PBS) containing 3% goat serum, for 30 min and incubated for 1 h with the primary antibody diluted in the blocking buffer. Parasites were washed with PBS and incubated with the appropriate secondary antibody diluted in the blocking buffer for 1 h. After extensive washing with PBS, coverslides were mounted using FluorSaveTM reagent (Calbiochem) containing 5 μg/ml DAPI. The primary antibodies used were mouse anti-TcSUMO polyclonal antibodies (1:100 dilution) and rat anti-HA high affinity monoclonal antibodies (Roche) (1:1000 dilution). The secondary antibodies used were AlexaFluor 546-conjugated goat anti-mouse or anti-rat immunoglobulins (Molecular Probes) diluted 1:1000. Slides were examined on a Nikon Eclipse E600 fluorescence microscope and image capture was performed by a Spot RT Slider Model No.2.3.1 digital camera (Diagnostic Instruments).
Oligonucleotides
The following primers were used: for pET28-His6-TcSUMO-His6 construct, sense primer GCTAGCATGGAGGAGAATCATGCAAATG and antisense primer GGATCCCAAAACGTGTTCCCGCCTGTC; for pTex constructs HA sense primer TACCCATACGATGTTCCAGATTACGCTATGGAGGAGATTCATGCAAATG and Hisx6 sense primer CCCGGGATGCACCACCACCACCACCACTACCCATACGATGTTCC, FLAG reverse primer CTCGAGTCAATCACCGTCATGGTCTTTGTAGTCGAGTGCGGCCGCAACCTTG, GG reverse primer CTCGAGTCATGTCTGCTCAACCATGG and ΔGG reverse primer CTCGAGTCACCCGCCTGTCTGCTC. For site directed mutagenesis of the Gly residue, sense primer GGTTGAGCAGACAGGCGcGAACACGTTTTGGGATC and antisense primer GATCCCAAAACGTGTTCgCGCCTGTCTGCTCAACC.
RESULTS
Identification of SUMO Pathway Components in the T. cruzi Genome
In order to identify the core components of the SUMOylation pathway in T. cruzi we performed BLAST searches against the genome database (http://www.genedb.org) using the corresponding proteins from Saccharomyces cerevisiae and Homo sapiens as queries. The predicted gene orthologs are shown in Table I. T. cruzi possesses a single SUMO paralog which displays 41% and 39–42% sequence identity to S. cerevisiae Smt3p and the three human SUMOs (designated SUMO-1, SUMO-2, and SUMO-3), respectively. The amino acid sequence alignment (Fig. 1) shows that, although having highly variable N-terminal extensions, these proteins exhibit an overall sequence similarity including a conserved diglycine motif, critical for SUMO maturation and conjugation (31). Furthermore, TcSUMO shares with Smt3p, SUMO-2 and SUMO-3 a Lys residue lying within a SUMOylation consensus site (32) meaning that TcSUMO has the potential to form poly-SUMO chains.
Table I. Genes encoding components of the SUMOylation pathway in T. cruzi.
| Protein name | GeneDB accesion number of haplotypes | |
|---|---|---|
| SUMO | Tc00.1047053507809.70 | Tc00.1047053511661.50 |
| E1 subunit a | Tc00.1047053508177.100 | Tc00.1047053511691.30 |
| E1 subunit b | Tc00.1047053509777.100 | Tc00.1047053511655.69/Tc00.1047053509117.10 |
| E2 | Tc00.1047053508741.280 | Tc00.1047053503515.14 |
| SUMO peptidase | Tc00.1047053503407.20 | Tc00.1047053505193.40 |
| SP-RING-type E3 | Tc00.1047053509099.100 | Tc00.1047053509319.30 |
| Tc00.1047053508461.300 | - | |
| Tc00.1047053511537.16 | Tc00.104705311021.60 | |
| Tc00.1047053510445.30 | - | |
Fig. 1.
Alignment of SUMO proteins from S. cerevisiae and human with the deduced amino acid sequence of T. cruzi SUMO haplotypes. Identical and conserved amino acids are shaded in black and gray, respectively. Shade of gray depends upon number of residues conserved. The carboxyl-terminal glycine cleaved by the Ulps/SENPs is marked with an arrowhead and the internal SUMOylation site is underlined. Accession numbers or GeneDB systematic names are: T. cruzi, Tc00.1047053507809.70 and Tc00.1047053511661.50; S. cerevisiae, NP_010798; Human - SUMO-1, NP_001005781.1; SUMO-2, NP_008868.3; SUMO-3, NP_008867.2.
SUMO is synthesized as a precursor form that is post-translationally cleaved to expose the C-terminal diglycine motif. Mature SUMO becomes covalently conjugated to other proteins through an isopeptide linkage between its C terminus and the ε-amino group of a targeted lysine residue in the substrate protein. Ubiquitin like protein proteases/Sentrin specific proteases (Ulps/SENPs) mediate both processing and deconjugation of SUMO (33–35). Ulps/SENPs share a C-terminal cysteine protease domain of ∼250 amino acids containing the catalytic triad His-Asp-Cys, whereas the N-terminal domain is not well conserved, diverging in primary sequence and size (36, 37). Based on local sequence alignment of the catalytic domain T. cruzi Ulp/SENP can be assigned to Ulp group 1 (supplemental Fig. S1).
SUMO is activated by a single heterodimeric E1 enzyme. The putative E1a and E1b subunits of T. cruzi have, as described for yeast and mammals (38, 39), a multidomain architecture that includes an adenylation domain that binds ATP-Mg+2 and SUMO, a C-terminal ubiquitin-fold domain that recruits E2 for thioester transfer, and a catalytic domain that contains the active site cysteine in the E1b subunit (supplemental Fig. S2). A single SUMO-conjugating enzyme was also detected in the genome of T. cruzi and a highly conserved cysteine residue could be inferred as responsible to form the E2∼SUMO thioester adduct (supplemental Fig. S3). Finally, based on sequence homology analysis a number of potential SUMO ligases belonging only to the SP-RING-type family were identified.
Analysis of SUMOylation Pattern in Life-cycle Stages of T. cruzi
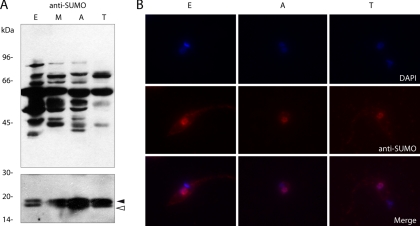
After bioinformatic analysis revealed that T. cruzi genome bears the genes encoding all components of the SUMOylation system, we decided to study its functionality in the parasite. We developed rabbit polyclonal anti-TcSUMO antibodies against the recombinant protein (see Experimental Procedures) and used them to determine SUMO expression throughout the parasite's life-cycle by Western blot analysis. Fig. 2A shows a typical SUMOylation pattern with several reactive bands consistent with a range of proteins being conjugated to SUMO. The faster-migrating double band, with apparent molecular masses of 16 and 18 kDa, possibly corresponds to the mature and precursor forms of free SUMO, respectively (Fig. 2A, bottom panel). Although their apparent molecular masses do not match with their theoretical molecular masses (11.4 and 11.8 kDa, respectively), an anomalous electrophoretic behavior has been reported for all SUMO proteins (31, 40). The high relative molecular mass bands observed at the 40–100 kDa range likely correspond to covalent adducts between SUMO and a diverse set of target proteins. Therefore, SUMOylation seems to be functional in the four major life cycle stages of the parasite (Fig. 2A, top panel). Immunofluorescence analysis performed with the same antibody on different forms of the parasite showed that the subcellular distribution of endogenous TcSUMO (free and conjugated) was predominantly nuclear, although a less intense cytoplasmic signal could also be detected (Fig. 2B). The results obtained with anti-TcSUMO polyclonal antibodies on wild-type epimastigotes were further confirmed by performing a similar analysis in transgenic epimastigotes expressing an ectopic HA-tagged version of TcSUMO using anti-HA monoclonal antibodies (see below).
Fig. 2.
Analysis of SUMOylation pattern in life-cycle stages of T. cruzi. A, Total cell lysates of 2 × 107 epimastigotes (E), metacyclic trypomastigotes (M), amastigotes (A), and cell-derived trypomastigotes (T) were electrophoresed on SDS-PAGE 7.5% (upper panel) and 15% (lower panel), transferred to nitrocellulose membranes and probed with anti-TcSUMO polyclonal antibodies. Precursor and mature forms of SUMO are depicted with a filled or empty arrowhead, respectively. B, Indirect immunofluorescence study of the different forms of the parasite using anti-TcSUMO polyclonal antibodies and AlexaFluor 546-conjugated goat anti-mouse secondary antibody. Nuclear and kinetoplast DNA were visualized by DAPI staining. The SUMO fluorescence image (red) has been merged with the corresponding DAPI staining (blue), and the resulting merged images are shown.
Requirements for TcSUMO Processing and Conjugation
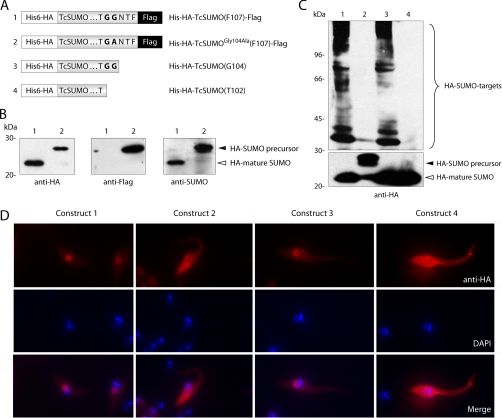
As we mentioned above, several authors have demonstrated the major role that the conserved diglycine motif (GG) plays in both, proteolytic maturation and conjugation of SUMO. To test whether SUMO has an analogous requirement in T. cruzi, we generated four stable epimastigote lines that ectopically expressed different forms of TcSUMO. Constructs are schematically summarized in Fig. 3A. In order to determine the requirements for SUMO processing in the parasite, we first analyzed whole cell lysates from epimastigotes expressing full-length His6-HA-TcSUMO-FLAG (construct 1) and the corresponding Gly104 to Ala mutant (construct 2) by Western blot using monoclonal antibodies against both, HA and FLAG tags. TcSUMO monomer in construct 1 was detected as a single band reactive only with anti-HA but not with anti-FLAG antibodies (Fig. 3B), suggesting a proteolytic event at the C terminus of the protein that removes the FLAG epitope. In construct 2, a single slower migrating band was detected with both antibodies, meaning that the proteolytic processing was inhibited when Gly104 was changed to Ala (Fig. 3B). Therefore, we confirmed that the diglycine motif is essential for maturation and found that almost all newly synthesized TcSUMO was proteolytically processed to generate the mature form. Most of the activatable precursor (construction 1) is likely activated by SENP, and does not accumulate. The mutated precursor (construction 2), on the other hand, accumulates. The double band of the mutated SUMO precursor cannot be caused by the specific action of SENP, and might be because of unspecific cleavage by SENP itself, or other parasite proteinase.
Fig. 3.
A, Schematic representation of SUMO constructs in the T. cruzi expression vector pTEX. Construct 1 corresponds to TcSUMO precursor protein, containing the conserved diglycine motif, tagged at the N- and C termini with His6-HA and FLAG, respectively. Construct 2 is identical to construct 1 except that the scissile glycine was mutated to alanine. Construct 3 represents TcSUMO mature form tagged only at the N terminus with His6-HA and construct 4 is similar to construct 3 but lacks the “GG” motif. B, Western blot analysis of in vivo SUMO maturation. Lysates from stable cell lines (5 × 106 parasites/lane) expressing constructs 1 and 2 were electrophoresed on SDS-PAGE 15% and transferred to nitrocellulose membranes. Blots were probed with anti-HA, anti-FLAG or anti-SUMO antibodies and proteins were visualized by chemiluminescence. SUMO unprocessed and processed forms are marked with a black and a white arrowhead, respectively. C, Western blot analysis of in vivo SUMO conjugation. Lysates from stable cell lines (2 × 107 parasites/lane) expressing all four constructs were electrophoresed on SDS-PAGE 7.5% (up) and 15% (down) and transferred to nitrocellulose membranes. Blots were probed with anti-HA antibodies and proteins were visualized by chemiluminescence. A number of high apparent molecular mass protein bands likely corresponding to SUMO conjugates are shown in the upper blot. SUMO unprocessed and processed forms are marked with a black and a white arrowhead, respectively in the lower blot. D, Epimastigotes transfected with the different HA-tagged TcSUMO constructs were analyzed by immunofluorescence using rat anti-HA monoclonal antibody and AlexaFluor 546-conjugated goat anti-rat secondary antibody. Nuclear and kinetoplast DNA were visualized by DAPI staining. The HA fluorescence image (red) has been merged with the corresponding DAPI staining (blue), and the resulting merged images are shown.
With respect to conjugation, a Western blot analysis of epimastigote lysates with anti-HA antibody was carried out, looking for targets of SUMOylation (Fig. 3C). Using full-length TcSUMO (already known to be processed) and mature TcSUMO (constructs 1 and 3, respectively), the appearance of high apparent molecular mass bands corresponding to TcSUMO conjugated to targeted substrates was observed. When mutated and truncated forms of TcSUMO (constructs 2 and 4, respectively), were tested, the formation of high apparent molecular mass adducts was abolished. These results suggested that TcSUMO was conjugated through its carboxy-end diglycine motif.
The subcellular localization of SUMO in cell lines expressing the constructs was studied by immunofluorescence. Cells expressing the conjugable forms of TcSUMO (constructs 1 and 3) displayed a predominantly nuclear and less intense cytoplasmic labeling, in contrast to epimastigotes expressing nonconjugatable forms of TcSUMO (constructs 2 and 4) where labeling was uniform throughout the cell. Parasites ectopically expressing precursor or mature TcSUMO (construct 1 and 3 in Fig. 3D) showed a similar distribution to endogenous TcSUMO (Fig. 2B).
Tandem-affinity Purification and Identification of Potential SUMO Targets
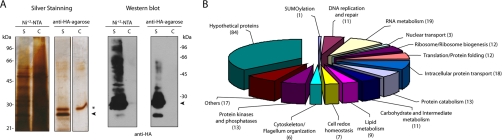
The above described cell line ectopically expressing a tagged version of full-length TcSUMO (construct 1) was used as starting material for the isolation and identification of SUMOylation targets in a large-scale proteomic study. SUMOylated proteins were enriched after two TAP steps, a Ni-NTA affinity column followed by an anti-HA affinity column. A mock purification from a wild-type cell line was also performed as control. Test and mock eluates were analyzed by silver staining and they both showed an identical pattern of bands except for the free TcSUMO. (Fig. 4A, left panel). Although few bands were detected by this technique, the sample was clearly enriched in SUMOylated proteins that were visualized by Western blot using anti-HA antibodies (Fig. 4A, right panel). Each protein mixture was prepared in triplicates, digested with trypsin and analyzed by online two-dimensional liquid chromatography-tandem mass spectrometry (2D LC-MS/MS). Peptide sequences and their corresponding proteins were identified by searching the spectra against T. cruzi and possible contaminant (non-T. cruzi) protein databases using the SEQUEST algorithm (see Experimental Procedures section).
Fig. 4.
A, Tandem affinity purification of SUMO targets. Starting material correspond to transgenic epimastigotes (S, sample) expressing the SUMO precursor form tagged at the N terminus with His6-HA (construct 1, see Fig. 3) or mock T. cruzi wild-type epimastigotes (C, control). Samples collected at the two different purification stages (Ni+2-NTA and anti-HA-agarose) were resolved in 10% SDS-PAGE and silver stained (left) or blotted and probed with anti-HA antibodies (right). The arrowhead denotes free SUMO and the asterisk indicates the immunoglobulin light chain. B, Pie chart summarizing the functional classification of the proteins identified by the proteomic analysis of the purified proteins.
The large number of background proteins (nonspecific binders) that is cocaptured during tandem-affinity purification of SUMOylated proteins is a well-documented phenomenon (12, 41, 42). Thus, the identification of potential SUMOylated candidates needs to be done by quantitative analysis comparing test with mock affinity purification. In our analysis, over two thousand (2686) nonredundant proteins were identified in both test and mock eluates out of a total of 7178 protein hits (supplementary Tables S1 and S2). Next, to state the proteins that were enriched in the test tandem-affinity purifications, we performed a quantification using the spectral count approach. The enriched protein hits were determined by statistical analysis using QSpec (supplementary Table S3). After carrying out the comparison between test and mock control samples, a group of 236 proteins that were more abundant in the test sample was identified. The complete list of proteins is given in supplementary Table S4, including annotation ID of gene alleles. Our triplicate data allowed us to determine the reproducibility, i.e. interaction specificity in test and mock tandem-affinity purifications (supplementary Table S5). For example, all our selection of SUMOylation targets demonstrated the replication in two or more test affinity purifications, but only few were present in mock experiments. By contrast, in the next 236 potential targets that we did not consider as significant evidence, most targets showed replication in two or three test affinity purifications, but the majority of them were also present in all three mock purifications. The remainder of data showed relatively worse replication rates even in test purifications, let alone the sporadic patterns in mock experiments. This clearly shows that our selection reflects the most representative fraction of replicated SUMOylation targets in the current data.
Manual inspection of the data allowed us to provide functional annotation for 152 proteins, whereas the rest of them remained as hypothetical proteins. In the group of proteins with functional annotation we found that almost 40% had at least one typical consensus site of SUMOylation (SUMOsp 2.0) (43) and 35 proteins have been previously identified in proteomic studies of SUMOylation in other organisms. Moreover, some of them have been clearly demonstrated to be SUMOylated by biochemical approaches as discussed below. With respect to annotation, proteins were categorized according to the biological processes in which they participate and a representative distribution is shown in Fig. 4B and Table II. We found proteins involved in classical nuclear processes that have been described to be regulated by SUMO, such as DNA replication and repair, chromatin remodeling, mRNA metabolism, ribosome biogenesis, and nuclear transport. In addition, we found well represented cytoplasmic and mitochondrial proteins involved in biological processes, such as lipid metabolism, carbohydrate metabolism, RNA editing, and intracellular protein transport. Recently, these processes have also been proposed to be associated with SUMOylation (44), providing further confidence to our proteomic results.
Table II. Functional categorization of identified SUMOylated protein candidates.
| Biological Process | Protein Name |
|---|---|
| Sumoylation | SUMO |
| DNA Replication and Repair | |
| DNA Replication and Repair | PCNA, Rep Fac C, RAD51, DNA Pol B, DNA Pol Y |
| DNA Biosynthesis | HGPRTase |
| DNA Catabolism | SSE-1 |
| Chromatin Remodeling | |
| Chromatin Organization | NAP, acetyltransferase |
| DNA Modification | JBP-1 |
| Chromosome Segregation | SMC3 |
| mRNA Metabolism | |
| RNA helicase | DEAD/H RNA helicase, RNA helicase |
| mRNA metabolism | TRRM1, PUF6, RNAse, RBP, PABP1, hnRNPH, U5 snRNP-specific 40 kDa protein |
| RNA editing | RNA editing endoribonuclease and ligase, oligoU binding protein |
| tRNA Metabolism | PseudoU Synthase I, Tyr, Val, Trp and Queuine tRNA Synthetases |
| Nuclear Transport | Nup54/57, Nup53a and b |
| Ribosome/Ribosome Biogenesis | |
| Structural Constituent of Ribosome | Ribosomal proteins 60S L2, L4, L13, and P0; Ribosomal proteins 40S S6, 8 and 27, Ub/ribosomal protein S27a |
| rRNA processing | PUF7 |
| Translation | |
| Translation Initiation | IF-2, eIF4a, SUI1 |
| Translation Elongation | EF-2 |
| Translation Termination | eRF1 and eRF3 |
| Protein Folding | α-Glucosidase II, DNAJ protein and Cyclophilin |
| Intracellular Protein Transport | |
| Vesicle Mediated Transport | Sec1, Sec23A, Rab7, Rab-GAP, coatomer-ϵ, μ-adaptin 3 |
| Vacuolar Transport | Vps26, Vps16, |
| Mitochondrial Transport | Mitochondrial carrier |
| Golgi Organization | COG2 |
| Microtubule Motor Activity | Kinesin, MCAK-like kinesin |
| Protein Targeting to Membrane | SRP, SRPR |
| Protein Catabolism | |
| Proteases | Peptidase M1, M16, M41and M67, metacaspase, cruzipain, oligopeptidase B, calpain-like |
| Ubiquitination Cycle | Ub hydrolase and Ub-protein ligase |
| Carbohydrate Metabolism | UDPGP, DAK1, Enolase, TKT, PGK, 6PF2K |
| Intermediate Metabolism | Pyr dh E1, 2-oxoacid dh (E1, E2 and E3), Iso dh |
| Lipids Metabolism | |
| Fatty Acid Metabolism | Thiolase, 2,4-dienoyl-Coa reductase, 3HCDH, fatty acid desaturase, Ac-CoA Synthetase |
| Coenzyme A Biosynthesis | PPC Synthetase, Pantothenate Kinase |
| Glycolipid Synthesis | Ppyr Decarboxylase, LPG1L |
| Cell Redox Homeostasis | |
| Oxidoreductase Activity | Adrenodoxin reductase, monooxygenase, quinone-oxidoreductase, Cyt P450, Cyt b5 reductase, G3P dh |
| Glutathion Metabolism | 5-Oxoprolinase |
| Cytoskeleton | Tubulin-Tyr ligase, γ-tubulin, MAP Gb4 |
| Flagellum Organization | Paraflagellar rod protein, flagellar transport protein, dynein heavy chain |
| Kinases and Phosphatases | |
| Kinases | Rac Ser/Thr kinase, Ser/Thr-protein kinase, protein kinase, Ser/Arg-rich protein kinase, cdc2-related kinase |
| Phosphatases | dual specificity protein phosphatase, TFIIF-stimulated CTD phosphatase, Ser/Thr-protein phosphatase 2A |
| Other | vATPase subc, TM9SF, GTP-binding protein, Tc40 Antigen, RHS protein, IFT81, kinesin divergent sequence, P-glycoprotein, WD40 repeat protein, Zn Finger proteins, calmodulin, AcOrn deacetylase, TAT |
Metacaspase-3 is a Substrate of SUMOylation
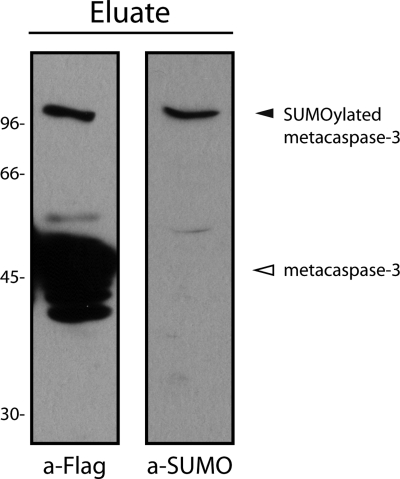
In the list of potential targets of SUMOylation, metacaspase-3 drew our attention because it has been the subject of studies in our laboratory (25). Therefore, we decided to perform additional experiments to validate it as a SUMO target. To achieve this goal, we have generated trypanosome cell lines expressing a version of metacaspase-3 tagged with a triple FLAG epitope at the C terminus, under the control of a tetracycline-regulated promoter using pTcINDEX vector (30) (see Experimental Procedures section). We confirmed the expression of epitope-tagged metacaspase-3 by Western blot after inducing with tetracycline (data not shown). Then, we performed a purification of tagged-metacaspase-3 using anti-FLAG affinity column and eluted the bound proteins with Laemmli's sample buffer. Western blot analysis of the eluate with anti-FLAG antibodies showed a strong band at the expected apparent molecular mass for metacaspase-3 and an extra band with higher molecular weight. This band was also reactive when the eluate was analyzed with anti-TcSUMO antibodies. These results showed that a minor fraction of metacaspase-3 was SUMOylated (Fig. 5), thus validating this protein as a bona fide SUMOylation target.
Fig. 5.
Validation of metacaspase-3 as a SUMOylation target in vivo. A stable cell line of epimastigotes expressing T. cruzi metacaspase-3 tagged with a 3 × FLAG (TcMCA3–3×FLAG) was grown in the presence of tetracycline for 60 h before cell lysate was prepared. TcMCA3–3×FLAG protein was purified by immunoaffinity chromatography using anti-FLAG M2-agarose affinity gel. Eluate was electrophoresed on 7.5% SDS-PAGE, transferred to nitrocellulose membranes and probed with either anti-FLAG (left) or anti-SUMO (right) antibodies. Protein bands corresponding to metacaspase and SUMOylated metacaspase-3 are shown with an empty or filled arrowhead.
DISCUSSION
A bioinformatic analysis of the T. cruzi genome unambiguously confirmed the existence of all essential genes of the SUMO conjugation and deconjugation pathway. Western blot analysis of total parasite homogenates using anti-TcSUMO polyclonal antibodies showed a band pattern compatible with a classical SUMOylation pattern. The presence of several slower migrating bands in addition to the SUMO precursor and mature bands support the existence of a functional SUMOylation system and multiple SUMO targets in the four major life cycle stages of the parasite. Although a number of the bands observed are common to all stages, there are some bands which are not. This might be because of a stage-specific expression or a stage-specific SUMOylation of the target protein. The functionality of the system was further confirmed when an HA-tagged version of TcSUMO was ectopically overexpressed in the parasite. Analysis of the transfected cell line showed that although the bulk of the SUMO protein remained unconjugated, high apparent molecular mass adducts were still detected. This suggests that SUMOylation is a tightly regulated process where only a small fraction of a target protein is modified with SUMO at any given time.
Fig. 2A shows the endogenous SUMOylation pattern, in parasite extracts using a polyclonal antibody raised against a purified recombinant TcSUMO. Fig. 3C, on the other hand, shows the SUMOylation pattern in extracts of parasites expressing an ectopic SUMO containing His-HA tags in the N terminus, obtained with an anti-HA monoclonal antibody. It is likely that the greater number of bands in Fig. 3C is caused by the high affinity of the commercial anti-HA antibody, which would improve the detection of the conjugates, as compared with Fig. 2A.
Analysis by Western blotting, using anti-SUMO antibodies, of the expression of endogenous SUMO suggests that the majority of SUMO is present in conjugates and the remaining SUMO is free (precursor and mature forms). A similar analysis of epimastigotes transfected with tagged SUMO, using anti-HA antibodies, suggests that in this case a smaller proportion of SUMO is present in conjugates whereas a greater amount appears as free form. Analysis of the overexpression of ectopic SUMO using the anti-SUMO serum suggests that overexpression results in an increase of about fivefold as compared with the endogenous protein. Analysis by immunofluorescence of the endogenous SUMO, as well as the conjugatable forms of tagged TcSUMO, suggests that in both cases most of the protein is present in the nucleus. The nonconjugatable forms of TcSUMO show a uniform distribution throughout the parasite cell.
Parasites transfected with construct 1 (full TcSUMO precursor) were used for the sample because they must follow the complete SUMOylation pathway, including proteolytic processing, activation and conjugation to targets, and so they are very similar to the wild type parasites undergoing natural SUMOylation. The mutant constructs 2 and 4 were not used as mock treatments, because in both cases a small amount of conjugates was formed, despite the mutations. This was not unexpected, because it has been reported that Saccharomyces cerevisiae mutants with the SUMO gene deleted (which are not viable) can be complemented with the GxA mutant, analogous to construct 2, giving a petite phenotype (38). Moreover, Western blot analysis of the mutant proteins GxA in both, yeast and human SUMO isoforms, presented a low degree of processing and conjugation to targets (31, 38, 45). A low formation of conjugates is not important when working in a small scale, as in Fig. 3C, but it certainly would be important when scaling up in order to obtain a comparable control, thus invalidating the mock treatment. A mock purification from a wild-type cell line was performed as a negative control.
Here we developed a purification scheme with two TAP steps to reduce the number of nonspecific binding proteins. Although this allowed us to obtain a fraction enriched in SUMOylated proteins (n = 236) identified by 2D LC-MS/MS, we still found a significant number of nonspecific binders or interactors (n = 2,450). Despite TAP being the current approach of choice for reducing nonspecific interactions, thus far there is no data to set a normal level of contaminants in this type of experiment, even if two-step purification is allegedly considered “specific ” in general. To further address this issue, we processed our data using the same criteria as described by Wohlschlegel et al. (12), which validated SUMOylated targets in S. cerevisae. Accordingly, we considered as potential SUMOylated proteins only those identified with at least two peptides and not present in the mock sample. For this analysis we used only one of our replicates (mock 2 versus test 2), because the data reported by Wohlschlegel et al. (12) was based on a single experiment. With these criteria, we identified 2083 proteins, of which 851 (40.9%) are present in the test but not in the mock sample (supplemental Fig. S4). Thus, our data has similar fold of enrichment compared with the data reported by Wohlschlegel et al. (12) (271 (38.5%), SUMOylated proteins out of 704 identified proteins). Unfortunately, we were unable to use similar criteria as described by Galisson et al. (41) for SUMOylated proteins of human cells, because their approach employed a quantification based on the intensity of MS1 scans. Moreover, in a recent publication by Westman et al. (42) only 25 (4.2%) out of 590 identified proteins were validated as SUMO targets in a nucleolar fraction of human cells. Therefore, in general the data described in the present study is in fair agreement with those described in the literature (12, 42).
In good agreement with the nuclear localization of the HA-tagged SUMO construct, many of the SUMOylated proteins detected in our proteomic analysis are predicted to perform their functions within the nucleus, including DNA replication and repair, chromatin remodeling, pre-mRNA splicing, rRNA processing, ribosome biogenesis, and nuclear transport. Replication and repair of DNA is a well-studied biological process regulated by SUMOylation. In this category, we identified three putative components of the replication fork: DNA pol δ, PCNA and RFC; and two proteins likely to participate in the mechanism of DNA repair, namely, DNA pol ξ and RAD51 (46). Two of these proteins have already been validated; PCNA is a well conserved SUMO target in several organisms and RFC was recently demonstrated to be SUMOylated in Drosophila. (15). In addition, PCNA is one example of SUMO and ubiquitin crosstalk, because both molecules modify the same lysine residue but confer opposite properties (47). Other levels of SUMO and Ub crosstalk exist, involving two components of the ubiquitination system: the E3 ubiquitin protein ligases and the deubiquitinating enzymes (DUBs). In both cases the enzymes have SUMO-interacting motifs (SIM) (48, 49) stabilizing their binding to SUMO and facilitating subsequent conjugation (50). In the former case, the interaction promotes a covalent linkage of one Ubiquitin molecule to the end of the polySUMO chain of conjugated protein, driving it to degradation (51, 52). In the latter, a specific DUB named USP25 is SUMOylated at a Lys residue embedded in a region critical for Ub recognition, thus decreasing its isopeptidase activity (53). Interestingly, we also detected homologs to E3 ligases and DUBs in our proteomic study.
It has been previously reported that diverse chromatin-modifying enzymes and chromatin-associated proteins are the effectors of SUMO-dependent changes in chromatin structure and gene expression in yeast and mammalian cells (54). In our list of potential SUMO substrates we detected the thymidine-hydroxylase JBP-1, a nucleosome assembly protein and a putative GC5N histone acetyltransferase. Because RNA polymerase II transcription seems to be constitutive in Trypanosomatids, with promoters and transcription factors likely to be absent in the genome (55), the presence of proteins involved in DNA modification and chromatin organization in our SUMO targets' data set represents a very interesting finding and reflects a potential role for SUMO in determining chromatin conformation and transcriptional activity.
We also identified the poly(A)-binding protein PABP-1, as well as other putative RNA recognition motif-containing proteins such as TRRM1 and PUF6 in our proteomic analysis, meaning that SUMO could also play a role at a post-transcriptional level regulating mRNA abundance. Another unique feature of all Trypanosomatids is the fact that transcription is polycistronic and a trans-splicing event is needed in order to produce mature mRNAs (56). We believe SUMO could participate as well in this process, because several splicing factors (e.g. hnRNPH, U5 snRNP-specific 40 kDa protein, and pseudoU Synthase I) were found in our analysis.
It has been noticed in Trypanosomatids that there is a very weak correlation between mRNA abundance and protein expression (57–59), suggesting an important regulation step at the translational level and/or at the level of protein degradation. Very recently, SUMO has been implicated in a novel fundamental regulatory mechanism of protein synthesis. SUMOylation of mammalian translation initiation factor 4 (eIF4E) has been shown to promote the formation of the active translation initiation complex (60) in a conjugation reaction that depends on the E3 ligase activity of the histone deacetylase (HDAC2) (61). We were not able to detect T. cruzi eIF4E as a SUMO target in our proteomic approach; however, other components of the translation initiation complex (i.e. eIF4A (orthologous to LmeIF4A2), SUI-1 (corresponding to eIF-3 small subunit), and eIF-2/eIF-5) were present in our study.
Another distinctive aspect of Trypanosomatids is the extensive editing of the RNA that takes place in the parasite's single mitochondrion (62). We found proteins that are known to be involved in the editing mechanism like the RNA editing endoribonuclease and ligase, and the oligoU-binding protein. The fact that a mitochondrial protein could be a SUMO substrate has been investigated lately, and very recently a specific E3 ligase catalyzing specific SUMO conjugation to mitochondrial targets has been identified (4).
A large number of proteins that are structural constituents of ribosomes or are potentially involved in ribosome biogenesis were found in our proteomic experiment. This is not surprising because many proteomic studies propose ribosomal particles and trans-acting factors as potential SUMO substrates. Furthermore, it has been recently documented that preribosome assembly and nuclear export is functionally linked to the SUMO pathway in yeasts. Eighteen factors involved in ribosome biogenesis were validated as bona fide SUMO targets (63). Diverse aminoacyl-tRNA synthetases were detected in our list of potential SUMO targets. In Drosophila glutamyl-prolyl-tRNA synthetase and methionyl-tRNA synthetase were demonstrated to be conjugated with SUMO in vivo, and a potential role for this modification in increasing the traffic of tRNAs from the nucleus to the ribosomes was suggested (64).
An outstanding finding in our proteomic analysis is the occurrence of metacaspase-3 among the potential SUMO targets. We have confirmed that at least one metacaspase-3 isoform is conjugated to SUMO in vivo, even under conditions when SUMO is expressed at endogenous levels. The band with the lower mobility shows an apparent molecular mass, which suggests that two molecules of TcSUMO have been covalently linked, but we cannot tell if the second is linked to the first, or if both are covalently bound to different sites of the target. To our knowledge, this is the first metacaspase being post-translationally modified with SUMO. Metacaspases are distant caspase relatives present in plant, fungi, and protozoa (65). This novel family of peptidases shares with caspases a common fold but displays radically different substrate specificity and it is not clear whether their members play an analogous role in programmed cell death in nonmetazoan organisms (66). Caspase-2 and -7 have been reported to be SUMOylated (67, 68), and a detailed study on caspase-8 elegantly demonstrated that SUMOylation of this protein is determinant for its nuclear localization (69). Thus, modulation of metacaspase subcellular localization by SUMO conjugation is an intriguing possibility. However we cannot rule out that SUMOylation could be affecting the proteolytic activity of metacaspase, as it was shown to be the case for another peptidase, calpain-2 (70).
The fact SUMO resulted essential for T. brucei viability (18, 19) reflects that the SUMOylation system is also important in these early divergent eukaryotes, and emphasizes the relevance of studying the proteins modified by SUMO. According to the conditions in which the purification of SUMOylated proteins is performed, a different kind of information can be extracted from a proteomic study. If strong denaturating conditions are used, a more refined information about covalent target is obtained. However, if it is performed under native conditions, as in the present work, the data may include proteins that directly or indirectly interact with SUMOylated targets (15, 44). This is the first large-scale proteomic study aimed to identify SUMO interactors in trypanosomatids performed under native conditions. We found a broad spectrum of biological processes potentially influenced by SUMO, some of them representing general cellular processes whereas some others are distinctive of trypanosomatids. Unraveling the role of SUMOylation in these processes requires a more detailed study on individual targets and on how SUMO could affect their subcellular localization, activity or interactions.
Acknowledgments
We thank Omar Harb (University of Pennsylvania) for helping connecting the GenBank and TriTrypDB/GeneDB accession numbers.
Footnotes
* This study was supported by the National Institutes of Health (NIH) grants 5S06GM08012-37, 1R01AI070655-04, and 3R01AI070655-04S1 (to ICA), and 2G12RR008124-16A1 and 2G12RR008124-16A1S1 (to BBRC/UTEP). We are thankful to the Biomolecule Analysis Core Facility/BBRC/UTEP, supported by NIH grants 2G12RR008124-16A1 and 2G12RR008124-16A1S1 for the access to the LC-MS instrument and other proteomic resources. JJC and VEA are members of the research career of the Argentinean National Research Council (CONICET); JCB had fellowships from CONICET and from the grant “Parasitic Diseases Research at the IIB” (Grant # 1D43TW007888-01, Sub-award # RR374-041/350037, from FIRCA, NIH); and ML had fellowships from AMSUD-Pasteur and CONICET. The work performed in Argentina was financed by PICT 2006 02381 from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT, MinCyT), Argentina.
 This article contains supplemental Figs. S1 to S4 and Tables S1 to S5.
This article contains supplemental Figs. S1 to S4 and Tables S1 to S5.
Data Availability: The raw LC-MS/MS files and the protein sequence data base are available free of charge at Tranche database (proteomecommons.org) at hash: cYL4uA12vLUKO9z0Dmrj0Gh3NtO+02j6HKFon4NNL3HcBwNwVRywx4Ii0wsCelb8HlBpMVVmijlY4yFJq283aa9MgEsAAAAAAAACfQ==. The identified proteins were deposited at the TriTrypDB (tritrypdb.org).
1 The abbreviations used are:
- Ub
- ubiquitin
- ATG
- autophagy-related gene
- DAPI
- 4′,6-diamidino-2-phenylindole
- G418
- Geneticin
- HA
- hemagglutinin
- IPTG
- isopropyl β-D-1-thiogalactopyranoside
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- NP-40
- Nonidet-P40 (octyl phenoxylpolyethoxylethanol)
- NTA
- nitrilotriacetic acid
- PBS
- phosphate-buffered saline
- PMSF
- phenylmethylsulfonyl fluoride
- SUMO
- small ubiquitin-like modifier
- TAP
- tandem- affinity purification
- TBS
- Tris-buffered saline
- TLCK
- tosyl-lysyl chloromethyl ketone
- Ubl
- ubiquitin-like modifiers.
REFERENCES
- 1. Kerscher O., Felberbaum R., Hochstrasser M. (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 2. Johnson E. S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 3. Meulmeester E., Melchior F. (2008) Cell biology: SUMO. Nature 452, 709–711 [DOI] [PubMed] [Google Scholar]
- 4. Braschi E., Zunino R., McBride H. M. (2009) MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 10, 748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 6. Li T., Evdokimov E., Shen R. F., Chao C. C., Tekle E., Wang T., Stadtman E. R., Yang D. C., Chock P. B. (2004) Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc. Natl. Acad. Sci. U.S.A. 101, 8551–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Denison C., Rudner A. D., Gerber S. A., Bakalarski C. E., Moazed D., Gygi S. P. (2005) A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell Proteomics 4, 246–254 [DOI] [PubMed] [Google Scholar]
- 8. Issar N., Roux E., Mattei D., Scherf A. (2008) Identification of a novel post-translational modification in Plasmodium falciparum: protein sumoylation in different cellular compartments. Cell Microbiol. 10, 1999–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vertegaal A. C., Ogg S. C., Jaffray E., Rodriguez M. S., Hay R. T., Andersen J. S., Mann M., Lamond A. I. (2004) A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 279, 33791–33798 [DOI] [PubMed] [Google Scholar]
- 10. Rosas-Acosta G., Russell W. K., Deyrieux A., Russell D. H., Wilson V. G. (2005) A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell Proteomics 4, 56–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou W., Ryan J. J., Zhou H. (2004) Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 279, 32262–32268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wohlschlegel J. A., Johnson E. S., Reed S. I., Yates J. R., 3rd (2004) Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279, 45662–45668 [DOI] [PubMed] [Google Scholar]
- 13. Panse V. G., Hardeland U., Werner T., Kuster B., Hurt E. (2004) A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 279, 41346–41351 [DOI] [PubMed] [Google Scholar]
- 14. Hannich J. T., Lewis A., Kroetz M. B., Li S. J., Heide H., Emili A., Hochstrasser M. (2005) Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280, 4102–4110 [DOI] [PubMed] [Google Scholar]
- 15. Nie M., Xie Y., Loo J. A., Courey A. J. (2009) Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS One 4, e5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller M. J., Barrett-Wilt G. A., Hua Z., Vierstra R. D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 16512–16517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Braun L., Cannella D., Pinheiro A. M., Kieffer S., Belrhali H., Garin J., Hakimi M. A. (2009) The small ubiquitin-like modifier (SUMO)-conjugating system of Toxoplasma gondii. Int. J. Parasitol. 39, 81–90 [DOI] [PubMed] [Google Scholar]
- 18. Obado S. O., Bot C., Echeverry M. C., Bayona J. C., Alvarez V. E., Taylor M. C., Kelly J. M. (2010) Centromere-associated topoisomerase activity in bloodstream form Trypanosoma brucei. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liao S., Wang T., Fan K., Tu X. (2010) The small ubiquitin-like modifier (SUMO) is essential in cell cycle regulation in Trypanosoma brucei. Exp. Cell Res. 316, 704–715 [DOI] [PubMed] [Google Scholar]
- 20. Cazzulo J. J., Franke de Cazzulo B. M., Engel J. C., Cannata J. J. (1985) End products and enzyme levels of aerobic glucose fermentation in trypanosomatids. Mol. Biochem. Parasitol. 16, 329–343 [DOI] [PubMed] [Google Scholar]
- 21. de Sousa M. A. (1983) A simple method to purify biologically and antigenically preserved bloodstream trypomastigotes of Trypanosoma cruzi using DEAE-cellulose columns. Mem. Inst. Oswaldo Cruz. 78, 317–333 [DOI] [PubMed] [Google Scholar]
- 22. Andrews N. W., Colli W. (1982) Adhesion and interiorization of Trypanosoma cruzi in mammalian cells. J. Protozool. 29, 264–269 [DOI] [PubMed] [Google Scholar]
- 23. Harlow E., Lane D. eds. (1988) Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 24. Kelly J. M., Ward H. M., Miles M. A., Kendall G. (1992) A shuttle vector which facilitates the expression of transfected genes in Trypanosoma cruzi and Leishmania. Nucleic Acids Res. 20, 3963–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kosec G., Alvarez V. E., Aguero F., Sanchez D., Dolinar M., Turk B., Turk V., Cazzulo J. J. (2006) Metacaspases of Trypanosoma cruzi: possible candidates for programmed cell death mediators. Mol. Biochem. Parasitol. 145, 18–28 [DOI] [PubMed] [Google Scholar]
- 26. Stone K. L., Willians K. R. (1996) Enzymatic digestion of proteins in solution and in SDS polyacrylamide gel. In: Walker J. M. ed. The protein protocol handbook, pp. 415–421, Humana Press Inc., Totowa, N.J [Google Scholar]
- 27. Sant'Anna C., Nakayasu E. S., Pereira M. G., Lourenço D., de Souza W., Almeida I. C., Cunha-E-Silva N. L. (2009) Subcellular proteomics of Trypanosoma cruzi reservosomes. Proteomics 9, 1782–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emmer B. T., Nakayasu E. S., Souther C., Choi H., Sobreira T. J., Epting C. L., Nesvizhskii A. I., Almeida I. C., Engman D. M. (2011) Global analysis of protein palmitoylation in African trypanosomes. Eukaryot. Cell 10, 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choi H., Fermin D., Nesvizhskii A. I. (2008) Significance analysis of spectral count data in label-free shotgun proteomics. Mol. Cell Proteomics 7, 2373–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taylor M. C., Kelly J. M. (2006) pTcINDEX: a stable tetracycline-regulated expression vector for Trypanosoma cruzi. BMC Biotechnol. 6, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kamitani T., Nguyen H. P., Yeh E. T. (1997) Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J. Biol. Chem. 272, 14001–14004 [DOI] [PubMed] [Google Scholar]
- 32. Rodriguez M. S., Dargemont C., Hay R. T. (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654–12659 [DOI] [PubMed] [Google Scholar]
- 33. Li S. J., Hochstrasser M. (2000) The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell Biol. 20, 2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li S. J., Hochstrasser M. (1999) A new protease required for cell-cycle progression in yeast. Nature 398, 246–251 [DOI] [PubMed] [Google Scholar]
- 35. Mukhopadhyay D., Dasso M. (2007) Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32, 286–295 [DOI] [PubMed] [Google Scholar]
- 36. Yeh E. T., Gong L., Kamitani T. (2000) Ubiquitin-like proteins: new wines in new bottles. Gene 248, 1–14 [DOI] [PubMed] [Google Scholar]
- 37. Bailey D., O'Hare P. (2004) Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J. Biol. Chem. 279, 692–703 [DOI] [PubMed] [Google Scholar]
- 38. Johnson E. S., Schwienhorst I., Dohmen R. J., Blobel G. (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16, 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lois L. M., Lima C. D. (2005) Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Desterro J. M., Thomson J., Hay R. T. (1997) Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417, 297–300 [DOI] [PubMed] [Google Scholar]
- 41. Galisson F., Mahrouche L., Courcelles M., Bonneil E., Meloche S., Chelbi-Alix M. K., Thibault P. (2011) A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol. Cell Proteomics 10, M110.004796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Westman B. J., Verheggen C., Hutten S., Lam Y. W., Bertrand E., Lamond A. I. (2010) A proteomic screen for nucleolar SUMO targets shows SUMOylation modulates the function of Nop5/Nop58. Mol. Cell 39, 618–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ren J., Gao X., Jin C., Zhu M., Wang X., Shaw A., Wen L., Yao X., Xue Y. (2009) Systematic study of protein sumoylation: Development of a site-specific predictor of SUMOsp 2.0. Proteomics 9, 3409–3412 [DOI] [PubMed] [Google Scholar]
- 44. Makhnevych T., Sydorskyy Y., Xin X., Srikumar T., Vizeacoumar F. J., Jeram S. M., Li Z., Bahr S., Andrews B. J., Boone C., Raught B. (2009) Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol. Cell 33, 124–135 [DOI] [PubMed] [Google Scholar]
- 45. Kamitani T., Kito K., Nguyen H. P., Fukuda-Kamitani T., Yeh E. T. (1998) Characterization of a second member of the sentrin family of ubiquitin-like proteins. J. Biol. Chem. 273, 11349–11353 [DOI] [PubMed] [Google Scholar]
- 46. Moldovan G. L., Pfander B., Jentsch S. (2007) PCNA, the maestro of the replication fork. Cell 129, 665–679 [DOI] [PubMed] [Google Scholar]
- 47. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 48. Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hecker C. M., Rabiller M., Haglund K., Bayer P., Dikic I. (2006) Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281, 16117–16127 [DOI] [PubMed] [Google Scholar]
- 50. Kerscher O. (2007) SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sun H., Leverson J. D., Hunter T. (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Uzunova K., Göttsche K., Miteva M., Weisshaar S. R., Glanemann C., Schnellhardt M., Niessen M., Scheel H., Hofmann K., Johnson E. S., Praefcke G. J., Dohmen R. J. (2007) Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282, 34167–34175 [DOI] [PubMed] [Google Scholar]
- 53. Meulmeester E., Kunze M., Hsiao H. H., Urlaub H., Melchior F. (2008) Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell 30, 610–619 [DOI] [PubMed] [Google Scholar]
- 54. Ouyang J., Gill G. (2009) SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics 4, 440–444 [DOI] [PubMed] [Google Scholar]
- 55. Campbell D. A., Thomas S., Sturm N. R. (2003) Transcription in kinetoplastid protozoa: why be normal? Microbes Infect 5, 1231–1240 [DOI] [PubMed] [Google Scholar]
- 56. Liang X. H., Haritan A., Uliel S., Michaeli S. (2003) trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2, 830–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Minning T. A., Weatherly D. B., Atwood J., 3rd, Orlando R., Tarleton R. L. (2009) The steady-state transcriptome of the four major life-cycle stages of Trypanosoma cruzi. BMC Genomics 10, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cuervo P., Domont G. B., De Jesus J. B. Proteomics of trypanosomatids of human medical importance. J. Proteomics 73, 845–867 [DOI] [PubMed] [Google Scholar]
- 59. Atwood J. A., 3rd, Weatherly D. B., Minning T. A., Bundy B., Cavola C., Opperdoes F. R., Orlando R., Tarleton R. L. (2005) The Trypanosoma cruzi proteome. Science 309, 473–476 [DOI] [PubMed] [Google Scholar]
- 60. Xu X., Vatsyayan J., Gao C., Bakkenist C. J., Hu J. (2010) Sumoylation of eIF4E activates mRNA translation. EMBO Rep. 11, 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu X., Vatsyayan J., Gao C., Bakkenist C. J., Hu J. (2010) HDAC2 promotes eIF4E sumoylation and activates mRNA translation gene specifically. J. Biol. Chem. 285, 18139–18143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stuart K. D., Schnaufer A., Ernst N. L., Panigrahi A. K. (2005) Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 30, 97–105 [DOI] [PubMed] [Google Scholar]
- 63. Panse V. G., Kressler D., Pauli A., Petfalski E., Gnädig M., Tollervey D., Hurt E. (2006) Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic 7, 1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Smith M., Bhaskar V., Fernandez J., Courey A. J. (2004) Drosophila Ulp1, a nuclear pore-associated SUMO protease, prevents accumulation of cytoplasmic SUMO conjugates. J. Biol. Chem. 279, 43805–43814 [DOI] [PubMed] [Google Scholar]
- 65. Uren A. G., O'Rourke K., Aravind L. A., Pisabarro M. T., Seshagiri S., Koonin E. V., Dixit V. M. (2000) Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 6, 961–967 [DOI] [PubMed] [Google Scholar]
- 66. Vercammen D., Declercq W., Vandenabeele P., Van Breusegem F. (2007) Are metacaspases caspases? J. Cell Biol. 179, 375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shirakura H., Hayashi N., Ogino S., Tsuruma K., Uehara T., Nomura Y. (2005) Caspase recruitment domain of procaspase-2 could be a target for SUMO-1 modification through Ubc9. Biochem. Biophys. Res. Commun. 331, 1007–1015 [DOI] [PubMed] [Google Scholar]
- 68. Hayashi N., Shirakura H., Uehara T., Nomura Y. (2006) Relationship between SUMO-1 modification of caspase-7 and its nuclear localization in human neuronal cells. Neurosci. Lett 397, 5–9 [DOI] [PubMed] [Google Scholar]
- 69. Besnault-Mascard L., Leprince C., Auffredou M. T., Meunier B., Bourgeade M. F., Camonis J., Lorenzo H. K., Vazquez A. (2005) Caspase-8 sumoylation is associated with nuclear localization. Oncogene 24, 3268–3273 [DOI] [PubMed] [Google Scholar]
- 70. Wang H. C., Huang Y. S., Ho C. C., Jeng J. C., Shih H. M. (2009) SUMO modification modulates the activity of calpain-2. Biochem. Biophys. Res. Commun. 384, 444–449 [DOI] [PubMed] [Google Scholar]