Abstract
The availability of label-free data derived from yeast cells (based on the summed intensity of the three strongest, isoform-specific peptides) permitted a preliminary assessment of protein abundances for glycolytic proteins. Following this analysis, we demonstrate successful application of the QconCAT technology, which uses recombinant DNA techniques to generate artificial concatamers of large numbers of internal standard peptides, to the quantification of enzymes of the glycolysis pathway in the yeast Saccharomyces cerevisiae. A QconCAT of 88 kDa (59 tryptic peptides) corresponding to 27 isoenzymes was designed and built to encode two or three analyte peptides per protein, and after stable isotope labeling of the standard in vivo, protein levels were determined by LC-MS, using ultra high performance liquid chromatography-coupled mass spectrometry. We were able to determine absolute protein concentrations between 14,000 and 10 million molecules/cell. Issues such as efficiency of extraction and completeness of proteolysis are addressed, as well as generic factors such as optimal quantotypic peptide selection and expression. In addition, the same proteins were quantified by intensity-based label-free analysis, and both sets of data were compared with other quantification methods.
Pathway mapping and modeling requires knowledge of flux through individual steps in the pathway, a product of the specific activity of the enzyme at that node and the number of enzyme molecules present in the cell. The goal of systems biology is to be able to advance to a predictive biology, in which detailed knowledge of the cellular constituents and their quantities, dynamics, and interactions can be embedded in robust mathematical models that permit simulation of cellular state changes, testable by experiment, leading to a formal definition of living processes. It follows that the strength of the model is only as good as the data embedded in it and that these data must be rigorously quantitative. One of the requirements for such models is accurate baseline values for the cellular quantities of the constituent proteins.
As proteomics has become increasingly quantitative, new approaches have been developed to support the goal of measurement of the amount of a protein in a cell. Many of these approaches were developed to allow relative quantification, in which the protein quantity in one cell/physiological state was expressed relative to a second state: for example, a diseased state relative to a normal control. These data are dimensionless and expressed as ratios; thus, a protein might be defined as being “2.4-fold higher in cell state A compared with cell state B.” This is undoubtedly of value in the discovery of differentially expressed proteins, but the lack of formally quantitative data means that further interpretation and parameterization of system models is difficult or impossible. This is the major driver for absolute quantification in proteomics.
There are two fundamentally different approaches to the acquisition of absolute quantification data for cellular proteins. The first type of approach is based on direct assessment of the signal (or ion current) that is acquired by the mass spectrometer; these approaches are referred to collectively as “label-free” methods. These methods are based on the entirely reasonable expectation that when a mixture of proteins is digested to constituent peptides, the most abundant proteins are expected to yield more detectable ions with stronger signal intensities (1, 2). Label-mediated absolute protein quantification by mass spectrometry is based on isotope dilution. In proteomics, it is rare that the standard is a stable isotope-labeled intact protein (3, 4). More commonly, one or more representative peptides (usually tryptic) are used as standards. The standard peptide(s) can be synthesized chemically (AQUA peptides (5)), but this also brings several problems, including high cost per peptide, the difficulty of synthesizing some peptides, and the tendency of low concentration peptides to adhere irreversibly to vessel walls. Moreover, if many proteins are to be quantified, each AQUA peptide must be separately quantified at the point of use. To circumvent many of the difficulties inherent in AQUA-based quantification studies, we developed the QconCAT approach for multiplexed absolute quantification (6, 7). In brief, synthetic genes, optimized for heterologous expression in Escherichia coli, encode a single open reading frame that is a concatenation of tryptic peptides, each of which acts as an internal standard (a Q-peptide) for a defined protein. Each analyte protein is represented by at least one, but more preferably two (or more), Q-peptides. Here, we describe the quantification of enzymes of the glycolytic pathway in Saccharomyces cerevisiae, using the QconCAT strategy, quantifying 27 glycolytic proteins (including isoforms). This study has established baseline parameters for such confounding factors as completeness of extraction and completeness of digestion, issues that would affect all quantitative analyses, label-mediated or label-free. We discuss the quantification process and highlight the issues and challenges that will be posed by global quantification of a proteome.
EXPERIMENTAL PROCEDURES
The materials were sourced as described (7). [13C6]Arg and [13C6]Lys were obtained from Cambridge Isotope Laboratories courtesy of CK Gas Products (Hampshire, UK). General laboratory chemicals, MS calibration standards, glass beads, and chromatography grade solvents (ACN and water) were obtained from Sigma-Aldrich, unless otherwise stated. Chromatography grade formic acid (BDH Aristar grade) was obtained from VWR International (Leicestershire, UK).
Label-free Identification and Quantification
For the preliminary label-free analyses, S. cerevisiae (EUROSCARF accession number Y11335 BY4742; MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YJL088w::KanMX4) was grown in C-limiting F1 medium (see supplemental information) using 10 g·l−1 of glucose as the sole carbon source. The F1 medium was supplemented with 0.5 mm arginine and 1 mm lysine to meet the auxotrophic requirements of the strain. Cultures were grown in chemostat mode at a dilution rate of 0.1 h−1, and aliquots (15 ml) of the culture were centrifuged (4000 rpm; 4 °C; 10 min). The supernatant was discarded, and the pellet was flash frozen in liquid nitrogen and stored at −80 °C for subsequent protein extraction. Proteins were extracted by resuspending the biomass pellets in 250 μl of 50 mm ammonium bicarbonate (filter sterilized) containing one tablet of Roche complete-mini protease inhibitors (with EDTA) (Roche Diagnostics) per 10 ml of ammonium bicarbonate. Acid-washed glass beads (200 μl) were then added. The pellet was subjected to repeated bead beating for 15 bursts of 30 s with a 1-min cool down in between each cycle. The biomass was centrifuged for 10 min at 13,000 rpm at 4 °C; the supernatant was removed and stored in low bind tubes on ice. Fresh ammonium bicarbonate (250 μl) with protease inhibitors was added, and the pellet was resuspended by vortexing. The bottom of the extraction vial was pierced with a hot needle, the vial placed on a fresh Eppendorf tube and quickly spun down (5 min at 4000 rpm at 4 °C). The flow through and the supernatant fraction were combined, the exact volume was measured, and the amount of protein was determined by standard assay (Bio-Rad). Protein extracts were aliquoted and stored at −80 °C prior to subsequent digestion.
An amount of lysate representing the protein from 21.5 million cells was dispensed into low protein-binding microcentrifuge tubes (Sarstedt, Leicester, UK) and made up to 160 μl by addition of 25 mm ammonium bicarbonate. The proteins were denatured using 10 μl of 1% (w/v) RapiGestTM (Waters MS Technologies, Manchester, UK) in 25 mm ammonium bicarbonate followed by incubation at 80 °C for 10 min. The sample was then reduced (addition of 10 μl of 60 mm DTT and incubation at 65 °C for 10 min) and alkylated (addition of 10 μl of 180 mm iodoacetamide and incubation at room temperature for 30 min in the dark). Trypsin (Roche Diagnostics) was reconstituted in 50 mm acetic acid to a concentration of 0.2 μg/μl. Digestion was performed by the addition of 10 μl of trypsin to the sample followed by incubation at 37 °C. After 4.5 h an additional 10 μl of trypsin was added, and the digestion was left to proceed overnight. The RapiGestTM was removed from the sample by acidification (3 μl of trifluoroacetic acid and incubation at 37 °C for 45 min) and centrifugation (15,000 × g for 15 min).
Label-free analysis was performed using a “Hi3” methodology (8). A portion of each yeast digest (100,000 cells/μl) was mixed with an equal volume of standard protein (50 fmol/μl of glycogen phosphorylase MassPREPTM digestion standard (Waters MS Technologies)). The resulting spiked digests were analyzed by LC-MSE using a nanoAcquity UPLCTM system (Waters MS Technologies) coupled to a Synapt G2 mass spectrometer (Waters MS Technologies). The sample (2 μl corresponding to 100,000 cells and 50 fmol of glycogen phosphorylase) was loaded onto the trapping column (Waters MS Technologies; C18, 180 μm × 20 mm), using partial loop injection, for 3 min at a flow rate of 5 μl/min with 0.1% (v/v) trifluoroacetic acid. The sample was resolved on an analytical column (nanoACQUITY UPLCTM BEH C18 75 μm × 150 mm 1.7-μm column) using a gradient of 97% A (0.1% formic acid) 3% B (99.9% ACN, 0.1% formic acid) to 60% A, 40% B over 90 min at a flow rate of 300 nl/min. The mass spectrometer acquired data using an MSE program with 1-s scan times and a collision energy ramp of 15–40 eV for elevated energy scans (8). The mass spectrometer was calibrated before use against the fragment ions of glufibrinopeptide and throughout the analytical run at 1-min intervals using the NanoLockSprayTM source with glufibrinopeptide. Following data processing, the database was searched using the ProteinLynx Global Server v2.5 (Waters MS Technologies). The data were processed using a low energy threshold of 100 and an elevated energy threshold of 20, and the processed spectra were searched against the complete proteome set of S. cerevisiae from Uniprot (6560 proteins) with the sequence of rabbit glycogen phosphorylase (UniProt: P00489) added. A fixed carbamidomethyl modification for cysteine and a variable oxidation modification for methionine were specified, one trypsin miscleavage was allowed, and the default settings in ProteinLynx Global Server for the precursor ion and fragment ion mass tolerance were used. The search thresholds used were: minimum fragment ion matches per peptide, 3; minimum fragment ion matches per protein, 7; minimum peptides per protein, 1; and false positive value, 4. The threshold score/expectation value for accepting individual spectra was the default value in the program, such that the false positive value was 4. Protein quantification was calculated by the software using Hi3 methodology based on the 50-fmol loading of glycogen phosphorylase. Biological variability was addressed by analyzing five yeast cultures and technical variability by digesting and analyzing each culture three times. The quantification values were averaged over technical replicates, and the resulting values were then averaged over biological replicates. The quoted standard deviations and errors refer to differences between biological replicates (supplemental Table I).
QconCAT Design and Expression
A key stage in the design of a QconCAT is the selection of the appropriate proteotypic tryptic peptides to act as quantification standards. The peptides were thus selected by manual analysis of those physicochemical properties deemed to promote detectability of limit peptides following in-solution digestion, reversed phase chromatography, and electrospray ionization. Because of the anticipated molecular weight of the recombinant QconCAT, a restriction site was incorporated midway through the construct and translated to a small linker peptide, thus different peptides for each of the target proteins were separated between the two halves, and the order within each was half-randomized. This would facilitate subcloning if expression failed. The QconCAT DNA construct was synthesized de novo and cloned into pET21a by PolyQuant GmbH (Regensburg, Germany) as described (6). E. coli strain BL21(λ)DE3 (E. coli B F− dcm ompT hsdSB(rB− mB−) gal, λ(DE3) was transformed with the vector and cultured in minimal medium (1× M9 salts (7), 1 mm MgSO4, 0.1 mm CaCl2, 0.00005% (w/v) thiamine, 0.2% (w/v) glucose, unlabeled amino acids at 0.1 or 0.2 mg/ml His, Tyr, Phe, Pro, and Trp), supplemented either with unlabeled or [13C6]arginine and [13C6]lysine at 0.1 mg/ml. The cells were grown to mid log phase (A600 = 0.6–0.8), at which point expression was induced by adding 1 mm isopropyl β-d-1-thiogalactopyranoside. The cells were lysed with the BugBuster Protein Extraction Reagent (Merck). Inclusion bodies were recovered by low speed centrifugation and redissolved, and the recombinant QconCATs in labeled and unlabeled form were purified by affinity chromatography (for a detailed protocol see (7)).
Preparation of Yeast Cell Extract for QconCAT Quantification
The S. cerevisiae strain used for QconCAT quantification was YDL227C, a heterozygous deletion derivative of the diploid BY4743: MATa/MATα; his3Δ1/his3Δ1; ho::KanMX4/HO; leu2Δ0/leu2Δ0; LYS2/lys2Δ0; met15Δ0/MET15; ura3Δ0/ura3Δ0. The cultures were grown aerobically under turbidostat conditions (9) in a 3-liter fermenter (Applikon Biotechnology, Schiedam, The Netherlands) at a dilution rate of 0.198 h−1, in synthetic “footprinting” medium as described (10) or in batch culture in F1 (N-limiting) medium (see supplemental information for media details and (11)). For preparation of lysates, 40–50-ml samples were removed, and the cell numbers were determined using a hemocytometer. Biological variability of turbidostat cultures was assessed by four distinct cultures representing two cultures grown at each of two independent sites; M1 and M2 denote the two biological repeats at one site, and C1 and C2 denote the alternative site. The culture samples were centrifuged to sediment the cells, and the resulting cell pellets were mixed with 75 μl of extraction buffer (50 mm Tris-HCl, pH 7.5, 750 mm NaCl, 4 mm MgCl2, 5 mm DTT, and 10% (v/v) glycerol) and an equivalent volume of glass beads. Mechanical extraction of protein was carried out using a mini bead-beater (Biospec Products, Inc., Bartlesville, OK). For each turbidostat sample collected, five rounds of extraction were carried out until protein could no longer be detected in the supernatant fractions by a standard assay (QuantiProTM BCA assay; Sigma-Aldrich) (12, 13), and individual peptides could not be detected following LC-MS by comparing the ratio of analyte to stable isotope-labeled standard Q-peptides derived from QconCAT. For all quantification analyses, each extract was analyzed independently by co-digestion with the isotopic QconCAT standard.
For trypsin proteolysis, known amounts of the recombinant isotopically labeled analog QconCAT protein were mixed with the lysates. The samples were reduced, alkylated, and digested with sequencing grade modified trypsin (Promega, Southampton, UK) using standard procedures (7). Briefly, the samples were reduced by the addition of DTT to a final concentration of 20 mm (from a 1 m stock prepared in 50 mm ammonium bicarbonate) at 56 °C for 1 h, followed by incubation with iodoacetamide at a final concentration of 10 mm from a 1 m stock prepared in 50 mm ammonium bicarbonate for 30 min at room temperature with light exclusion (a reduced level of iodoacetamide was used to limit over-alkylation of peptide N termini). After the addition of trypsin (approximate ratio 1:50 trypsin:total cell protein), proteolysis was monitored until no residual undigested protein could be detected as assessed by SDS-PAGE, and this was further confirmed mass spectrometrically where the end point was defined as the time point where the ion intensity ratios of analyte to standard had stabilized.
Following digestion, the resultant peptide mixture was analyzed in triplicate by LC-MS; each biologically independent sample was therefore analyzed over 15 separate LC-MS analyses, a total of 90 analytical runs (a total of 60 for the turbidostats M1, M2, C1, and C2 and 30 for batch cultures B1 and B2). Any residual protein remaining after the five successive extractions was also analyzed by QconCAT co-digestion and LC-MS, by subjecting the final pellet to the same reduction/alkylation/trypsinization protocol.
In-solution Protein Digest
Typically, 3 μl of analyte (corresponding to a maximum of 100 μg total cell protein) and 5.4 μg of recombinant QconCAT were digested with 2 μg of trypsin, in a final volume of 50 μl, following reduction/alkylation as described above. The tryptic digests were further diluted 50-fold in water, 0.1% (v/v) formic acid (Buffer A) prior to analysis, and 4 μl was loaded on a column, corresponding to approximately 8.6 ng/100 fmol of QconCAT and 160 ng of total cell protein. The amount of QconCAT added to the same amount of lysate was adjusted as required for low abundant proteins.
Nano-LC-MS/MS
The digested peptide mixtures were resolved by LC-MS using a nanoACQUITY chromatograph (Waters MS Technologies) coupled to either an LTQ-Orbitrap XL or a TSQ VantageTM triple quadrupole mass spectrometer (ThermoFisher Scientific, Bremen, Germany); in both cases the mass spectrometers were equipped with the manufacturer's dynamic nanospray source and fitted with a coated PicoTip Emitter 20–10 μm (New Objective, Woburn, MA), with the voltage applied at the tip.
Liquid Chromatography
The sample temperature was maintained at 10 °C, and 4 μl of each sample was injected initially onto a trapping column (C18, 180 μm × 20 mm; Waters MS Technologies), using the partial loop mode of injection, at a flow rate of 18 μl/min 99% (v/v) A, 1% (v/v) B (A as described above, and B consisting of 100% ACN, 0.1% (v/v) formic acid). The analytical column (nanoACQUITY UPLCTM BEH C18 75 μm × 150 mm, 1.7-μm column) was maintained at 35 °C and was developed at 300 nl/min by incrementing buffer B from 1% (v/v) to 50% (v/v) Buffer B over 30 min, followed by a rapid ramp to 85% buffer B over 1 min and then a return to the starting mobile phase conditions for re-equilibration prior to the next injection.
Mass Spectrometry
The LTQ-Orbitrap XL was calibrated prior to use according to the manufacturer's instructions, and the data were acquired using Xcalibur version 2.0.5/Tuneplus version 2.4SP1/configured with Waters Acquity driver (build 1.0). The Orbitrap was used for two types of analysis, depending on the extent of MS/MS acquisition required. In both cases, full scan MS spectra (m/z range, 300–1600) were acquired with the Orbitrap operating at a resolution (R) of 30,000 (as defined at m/z 400). For unbiased analyses, the top five most intense ions from the MS1 scan (full MS) were selected for tandem MS by collision-induced dissociation with helium as collision gas (hereafter referred to as data-dependent analysis), and for quantification applications, the data were acquired with a “preferred” inclusion list (i.e. most intense precursor from m/z list selected, or most intense ion in MS1 scan if no listed precursors detected), directing collision-induced dissociation. The latter approach was used to maximize the data points across the chromatographic peak while concomitantly acquiring tandem MS data for sequence verification. In both cases, a normalized collision energy of 30% was applied with an activation q of 0.25. Dynamic exclusion was enabled for 30 s with a repeat count of two, and all product ion spectra were acquired in the LTQ. The automatic gain control feature was used to control the number of ions in the linear trap and was set to 1 × 106 charges for a full MS scan, and 1 × 104 for the LTQ (MSn) i.e., higher order MS scans, with maximum injection times of 50 and 500 ms applied for the LTQ and Orbitrap, respectively. All Orbitrap scans consisted of one microscan.
Selected Reaction Monitoring Analysis
The TSQ VantageTM (ThermoFisher Scientific) was calibrated according to the manufacturer's instructions, and the data were acquired using Xcalibur version 2.0.6 SP1/Tuneplus version 2.2.0 Eng2, configured with an Acquity driver (build1.0) (Waters MS Technologies). Where possible, transitions were selected based on experimental tandem MS data obtained on the LTQ-Orbitrap XL. The y-series ions were selected as product ions, not only because this series is preferentially observed in the triple quadrupole analysis but also because the isotopic variants of the tryptic peptides labeled with [13C6]Arg and [13C6]Lys retain the label at the C terminus and therefore the mass difference (the list of transitions used are given in supplemental materials). The vendor-supplied software Pinpoint (v 1.1.12.0) (for a more detailed description see (14)) was used in parallel to predict/confirm appropriate transitions (thereby providing accurate m/z for product ions, not possible experimentally in the LTQ) and for in silico prediction of collision energies (by solution of the equation y = mx + c, where m = 0.034 and 0.044 for +2 and +3 charge states, respectively, c = 3.314 in both cases, x corresponds to mass m/z, and y corresponds to collision energy). The resolutions of both the first and third quadrupoles were set to 0.7 full width at half-maximum, and for high resolution analysis (highly selective reaction monitoring), the first quadrupole was set to 0.2 full width at half-maximum. The scan time was set to 0.005 s/transition, and the m/z width was 0.005. The collision gas used was argon according to the manufacturer's instructions. Lysates were applied (initially with 160 ng on column) with a range of QconCAT concentrations (1, 10, and 100 fmol) on column.
Irrespective of the analytical platform used, sample acquisitions were alternated with “buffer only” blank (defined as the starting mobile phase) injections to ensure that data analysis/quantification was not compromised by sample carryover. Data analysis was carried out using Xcalibur 2.0.6, which supports the raw files from both analytical platforms.
Peptide Identification and Quantification
Peptide sequences were verified using the search engines SequestTM (15, 16) (v.28, ©1998–2007, on license from ThermoFisher Scientific) and Mascot (v2.2.03, Matrix Science) (17), facilitated through the vendor-supplied software Proteome DiscovererTM (version 1.0 Build 43; ThermoFisher Scientific). Tandem MS data were searched using the following databases: Sequest, yeast.fasta 22.3.07 (which contains 14,580 entries and a customized version containing the QconCAT recombinant protein sequence); Mascot, Swiss-PROT (v.56.0, 6735 entries from a total of 392,667 entries). Because the tandem MS data were used for verification rather than identification, taxonomy restriction was applied. The search parameters used were: trypsin, two missed cleavages permitted, precursor mass tolerance of 50 ppm, and fragment mass tolerance set to 0.8 Da. The following modifications were included: static/fixed modifications carbamidomethyl (C), and variable modifications; oxidation (M), label ([13C6]Lys)/label ([13C6]Arg). A high confidence significance threshold of 0.01 was applied to the mascot ion score for mascot search results, and the cut-off score was set to allow 5% false positive, because the purpose of the database search was to confirm the presence of peptides rather than to identify them. The following default thresholds were applied to Sequest results: z = 2 and high confidence XCorr = 1.9, z = 3 and XCorr = 2.3, by the same rationale. Where post-translational modifications are described in the text, e.g. deamidation of Asn, conversion of Gln to pyro-Glu, they were initially assigned through the search engines described above and checked by manual inspection of the tandem MS data (see supplemental information).
For quantification of Orbitrap data, extracted ion chromatograms of the monoisotopic peaks were used to compare the ratios of analyte to standard (following verification of tandem MS data from one or both of the heavy and light peptides), and the peak area was determined using the default interactive chemical integration system algorithm peak detection settings (baseline window = 40, area noise factor = 5, and peak noise factor = 10) in the Qual Browser (version 2) component of Xcalibur (version 2.0.6). For quantification of data from the TSQ Vantage, TICs (i.e., summation of signal from the transitions) of the heavy and light were used to determine ratios. The ratios were converted to molecules/cell and then parts per million for comparison between different quantitative approaches.
RESULTS
Label-free Preliminary Profiling of Glycolytic Enzymes
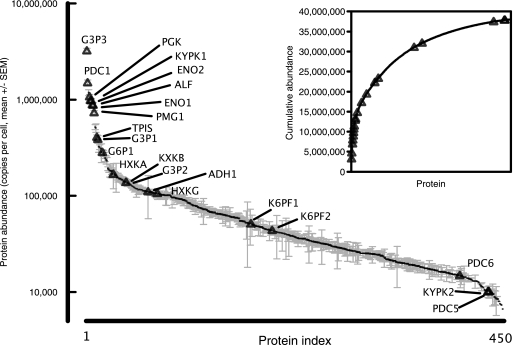
The availability of data-independent label-free quantitative values derived from haploid yeast cells (based on the summed intensity of the three strongest, isoform-specific peptides) (8) permitted a preliminary assessment of protein abundances for the glycolytic proteins (Fig. 1). Label-free quantification, based on 100,000 cells equivalent digest on column, permitted quantification of over 450 proteins between 5000 and 3,200,000 copies/cell, a dynamic range of between two and three logs. Assuming 4–5 pg of protein from a typical haploid yeast cell and an average protein molecular weight of 50 kDa, this gives a total constituency of ∼50 million protein molecules. The label-free analysis revealed a cumulative constituency of 38 million molecules over 450 proteins. There are several sources of error in these calculations, but the numbers are substantially in agreement, and this implies that the remaining, undetected yeast proteins are present at low levels. As is evident from the figure, the largest contribution to the cumulative protein content is derived from relatively few high abundance proteins; 50% of the total molecules determined by label-free quantification are derived from the top 40 protein molecules, including 14 of the glycolytic enzymes in this study. In total, 21 of the 27 enzymes were detectable at levels above 5000 copies/cell. This suggested that quantification of most of the high abundance members of the pathway should be possible, based on comparative intensities of the standard and analyte peptides, by analysis of precursor ions in an accurate mass/retention time (AMRT)1 experimental workflow. However, of the 27 proteins specified in this study, six were not detectable by label-free analysis at a relatively modest protein load on column (100,000 cells), suggesting expression levels below 5,000 copies/cell. Moreover, the extent to which label-free approaches are comparable with other quantification approaches (tagging) remains unclear, because the overall correlations are poor (18). Accordingly, we quantified the same proteins using a QconCAT.
Fig. 1.
Label-free quantification of glycolytic enzymes in S. cerevisiae. Samples of chemostat-grown S. cerevisiae equivalent to 100,000 cells were analyzed by Hi3 label free quantification on a QToF (Quadrupole Time of Flight) mass spectrometer. The expression data, calculated as copies/cell, were obtained for five biological replicates (the data are expressed as the means ± S.E., n = 5). The glycolytic enzymes that were quantified by this methodology are highlighted on the expression diagram (triangles). The inset shows the cumulative abundance of yeast proteins (expressed as copies/cell), and the same glycolytic enzymes are identified with triangles.
QconCAT Design and Expression and Technical Deployment
To determine the glycolytic enzyme abundances by a method independent to and for comparison with label-free methodology, we adopted the QconCAT approach. A QconCAT was designed to quantify each of the 27 glycolytic enzymes with at least two peptides per protein (supplemental Table II).
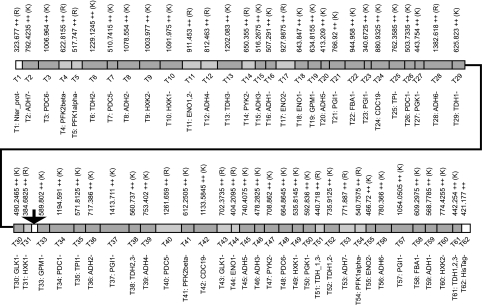
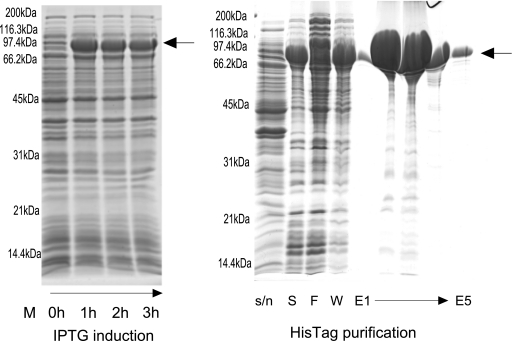
The design process led to a final QconCAT of 804 amino acids (average mass, 87.8 kDa), including a sacrificial N-terminal segment to protect the true peptide standards and a C-terminal hexahistidine tag to aid purification of the QconCAT (Fig. 2). After synthesis of the gene, insertion into a suitable vector, and transformation into bacterial cells, induction of expression led to production of a recombinant protein band that was the most abundant protein in a whole bacterial cellular extract. This protein migrated on SDS-PAGE with a mobility consistent with an approximate molecular mass of ∼85 kDa, implying that the correct QconCAT had been expressed (Fig. 3). The putative QconCAT protein band from the gel of total bacterial protein extract was digested with trypsin, and on MALDI-TOF mass spectrometric analysis, multiple peptides of masses commensurate with those predicted by the QconCAT were observed, confirming the identity of the major band on the gel and thus successful expression of the QconCAT (MALDI-TOF data; supplemental Fig. 1 and supplemental Table III). Fresh cultures were established to express the QconCAT in unlabeled and labeled forms. After purification using the hexahistidine tag, the QconCAT was essentially pure and was used without further purification. A typical 200-ml bacterial culture, grown to a cell density of A600 = 0.6–0.8, yielded 8 mg (approximately 90 nmol) of the QconCAT. The identity and chromatographic retention time of the Q-peptides from the unlabeled and labeled QconCAT recombinant proteins were established by preliminary tandem MS analyses of pure QconCATs. The labeling efficiency was high, and the QconCAT peptides were labeled to > 99%, reflecting the quality of the starting isotopes [13C6]Arg and [13C6]Lys. Moreover, a minor peak at (+5.02)/z, resulting from incomplete labeling of the carbon atoms, was insignificant. For expression of the recombinant labeled QconCAT protein, it is particularly important that E. coli cells are grown in the presence of excess unlabeled proline to limit the conversion of isotopically labeled arginine to [13C5]proline via the ornithine cycle as occurs during proline synthesis, because some of the Q-peptides selected contain proline residues (a potential problem that could also be circumvented in future by expression in a proline auxotroph host). The absence of labeled proline through this conversion in the QconCAT recombinant protein was confirmed experimentally, by data-dependent acquisition of tandem MS data and inspection of the MS1 data to check for the increase in m/z where appropriate.
Fig. 2.
Organization of a S. cerevisiae glycolysis QconCAT. A QconCAT was designed to quantify the yeast glycolytic pathway. This is illustrated diagrammatically, and the peptides and expected masses (expressed as [M+2H]2+) are highlighted. Dark and light grey blocks indicate Q-peptides terminated with lysine and arginine residues, respectively. Three additional peptides were present in the QconCAT. The peptide indicated by the arrow in the center of the QconCAT did not report on a yeast protein but was a small in-frame linker peptide, encoded to contain a unique restriction site, permitting expression of the QconCAT in two halves if desired. Also included was a short sacrificial peptide at the N terminus, and a His tag (n = 6) at the C terminus to facilitate purification. Further information on the target proteins and the peptides is provided in supplemental Table S2.
Fig. 3.
Expression of the yeast glycolytic QconCAT. The yeast glycolytic QconCAT protein was expressed heterologously in E. coli by induction with 1 mm isopropyl β-d-1-thiogalactopyranoside (IPTG). Throughout growth and induction, samples of the culture were withdrawn and used for A600 determination and analysis by SDS-PAGE. An equivalent of 0.1 A600 units were loaded onto the gel at each time point. The culture was grown for 5 h post-induction. The QconCAT (expressed as inclusion bodies that were recovered by centrifugation) was dissolved in binding buffer (20 mm sodium phosphate, 500 mm NaCl, 20 mm imidazole, pH 7.4) containing 6 m guanidine HCl before purification by nickel-nitrilotriacetic acid affinity chromatography. Lanes S, F, and W correspond to starting material, flow through, and wash, respectively, and E1–E5 refer to successive elution fractions from the column.
From the label-free data, many of the glycolytic proteins were expected to be at high abundance in yeast, and we therefore adopted an AMRT strategy for the majority of quantification analyses. In brief, extracted ion chromatograms were used to generate peak areas for the analyte and standard peptides. The linearity of the response was established prior to these analyses using labeled and unlabeled QconCAT mixed at different ratios (supplemental Fig. 2). For the lower abundance proteins, we supplemented the AMRT strategy with an SRM-based method.
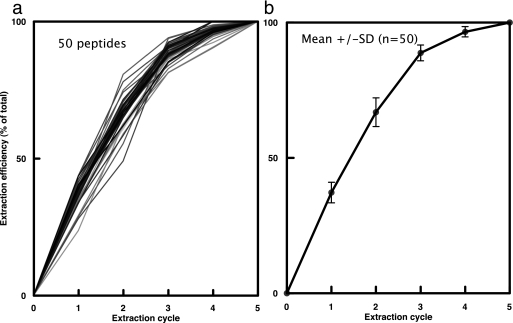
With any quantification procedure in proteomics, the extraction and digestion efficiencies are critical. Incomplete cell breakage or recovery of analyte will give erroneous measures of quantities; even before the proteomic analysis is commenced. To ensure that the protein extract contained all of the proteins to be quantified, the processes of cell breakage and recovery were monitored and optimized. Yeast cells were broken using glass beads for five successive disruption cycles, the supernatant fraction from each round of extraction was combined with the heavy labeled QconCAT, and the samples were reduced, alkylated, digested, resolved, and quantified independently through separate LC-MS analyses (supplemental Fig. 3a). Surprisingly, two rounds of extraction could only recover 50–68% of the proteins, and it was necessary to repeat the extraction for three further cell breakage cycles to recover all 99% (±1%) of the proteins. Fig. 4 shows the extraction efficiency for all proteins over the sequence of extractions. Analysis of the residual pellet showed that less that 1% of the glycolytic enzymes remained, consistent with 99% extraction, although the SDS-PAGE analysis indicated that some proteins were, as expected, still in the pellet. The five combined soluble extracts therefore contained >99% of each of the enzymes.
Fig. 4.
Efficiency of protein extraction for quantification. Cells were subjected to successive rounds of disruption using “bead beating.” After each cycle of disruption, the supernatant fraction was recovered, and the pellet (comprising cell debris and undisrupted cells) was resuspended to the same volume prior to another round of disruption. After five rounds of disruption, proteins were supplemented with labeled QconCAT and digested to completion, prior to LC-MS. Panel a) indicates data for 50 peptides at each extraction, expressed as percentage of total. Panel b) shows the accumulated statistics expressed as mean +/− SD.
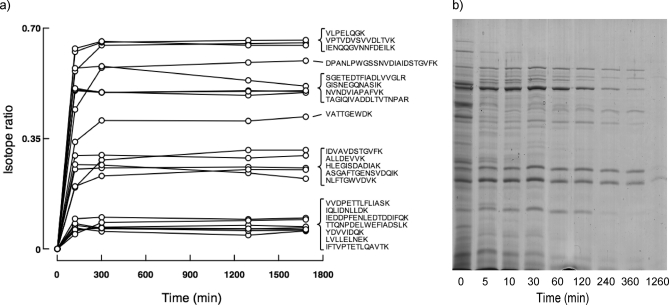
A second confounding factor in a peptide-based quantitative analysis is the impact of incomplete proteolysis. Even with synthetic peptides, it is necessary to ensure that the equivalent analyte peptide was quantitatively released from the parent protein. In addition, with QconCATs, the standard peptides must also be completely released from the concatamer (18). However, in our experience, the QconCAT, being an unstructured protein that is isolated in chaotropic buffers, is usually rapidly and fully proteolyzed (6, 19). To confirm complete proteolysis of the recombinant QconCAT, we generated a customized FASTA database including the sequence of the recombinant QconCAT protein, and searched against this with MS/MS data to facilitate easy detection of any problematic Q-peptides that might be susceptible to miscleavage. For the analyte, we explored the kinetics of release of the peptides from the proteins analyzed here. The progress of the digestion was monitored by selecting samples post-digest at the time points indicated in Fig. 5. After 300 min of digestion, the relative proportions of analyte to standard had reached a stable plateau for the majority of peptides, consistent with complete proteolysis. In all instances, the light:heavy signal increased over time, indicating the expected behavior of rapid proteolysis of the QconCAT relative to analyte. If the QconCAT recombinant protein had been more difficult to digest than the analyte proteins, the light:heavy ratio should have declined over time. We were therefore confident that the analyte mixture was fully representative of the total protein pools, that the digestion of analyte and standard was complete, and that the linearity of the signal was appropriate for our analysis. These issues, rarely overtly explored in quantitative analyses, are critical for fully quantitative studies. Also included in the supplemental materials are the sequence context for both the native and QconCAT proteins (supplemental Table IV).
Fig. 5.
Completeness of proteolytic digestion. The trypsin digestion protocol was scaled up such that 750 μg of yeast cell lysate protein was digested with trypsin (at a substrate:proteinase ratio = 50:1) together with isotopically labeled QconCAT (81 μg), and at different times as indicated, the samples were removed for analysis. The extent of digestion of the analyte was assessed (a) as the proportion of unlabeled peptide (light) relative to the total pool (light + heavy). During the digestion, the samples were removed and analyzed by SDS-PAGE (b).
Quantification of Individual Proteins
Quantification by AMRT was as described under “Experimental Procedures.” The complete set of quantification data for the enzymes of the pathway are provided in supplemental Table V. In a typical AMRT experiment, we added labeled standard such that the final amount of QconCAT, once digested and diluted, was equivalent to the application of 100 fmol of protein on column. Assuming accurate quantification at a level of 5% of the intensity of the standard, this would allow us to quantify down to 5 fmol of each peptide. A typical yeast cell contains ∼5–6 pg of protein, and the on-column load of digest (150 ng), expressed in terms of “cell equivalents” was ∼30,000 cells. An analyte signal equivalent to 5 fmol of standard would therefore be generated by a protein that was present at 100,000 copies/cell.
For example, for Hexokinase 2 (M1 extract 1), we obtained 5.4 fmol on column (heavy:light ratio, approximately 20:1), which (combined with the other extracts) gave a total of 123,000 molecules/cell. In a targeted experimental design, we also used a triple quadrupole mass spectrometer, which increased the limits of quantification by more than 1 order of magnitude to approximately 800 amol on column (M1 extract_1) and thus extends to approximately 16,000 molecules/cell. The detection limits could be improved still further by scheduling the SRM transitions over the analytical LC run time, improving the allocation of instrument duty cycle to successive transitions. Previous data for the glycolytic enzymes, based on antigen tagging, indicated a working range of between 1,200 (P52489_PYK2, YOR347C) and 1,000,000 (P14540_FBA1, YKL060C) molecules/cell (20). We therefore expected to achieve measurable heavy:light ratios for most proteins in the pathway without further refinement of the methodology. The results of SRM analysis are given in supplemental Table Vb.
The study was conducted across four biological replicates of cells grown in continuous culture. Careful control of set point (and hence, dilution rate parameters) and culture conditions is essential for exploration of reproducibility, and batch (shake flask) culture does not offer sufficient precision of control over growth conditions, nutrient utilization, and sampling strategy. Accordingly, the majority of our analyses have been conducted on cells grown in continuous culture, in aerobic turbidostat cultures grown at two sites. One pair of cultures was set up at Manchester, UK (M1 and M2), and two independent cultures were prepared at a second geographic location (Cambridge, UK; C1 and C2). The strain, media, growth rates, and sampling regimen were identical in both centers, as were the turbidostat operating conditions. This allowed exploration of the consistency of protein expression data that might be expected across the proteomics community, a prerequisite for comparative analyses. Because the four cultures were demonstrated to yield remarkably consistent data (see below), the analyses reported here are average values from all four biological replicates, each of which is, in turn, the mean of three technical replicates. The error terms (expressed as S.E., n = 4) reflect the errors in the four biological replicates (supplemental Table V).
For all of the peptides used in the QconCAT analysis, we apply a simple classification. Type A quantifications are where both standard and analyte are detected. Type B quantifications are where the standard could be detected, but the analyte is absent; this sets an upper boundary on the abundance of the protein. Finally, Type C is reserved for the rare situations where neither the standard nor analyte could be detected, usually attributable to selection of a peptide with poor chromatographic properties or weak fragmentation in the collision cell (supplemental Table VI). Of the total of 57 peptide level quantifications in this study, 38 were Type A, 15 were Type B, and 4 were Type C (as defined in at least one turbidostat).
Selection of suitable peptides for any quantification is critical. In some cases, we were limited for sequence choice; for example hexokinase I and II are 77% identical over the whole protein sequences, restricting the choice of isoform specific peptides, and for hexokinase I this was limited still further because peptides were selected to avoid putative phosphorylation sites. Other examples where peptide selection was problematic included glyceraldehyde-3-phosphate dehydrogenase (Tdh1p is over 88% identical to Tdh2p or Tdh3p, and Tdh2p and Tdh3p are over 96% identical, which severely restricts the choice of peptides for quantification of individual isoenzymes); thus, some common peptides were used. Therefore selection of peptides common to more than one isoform depends on separation of the signal by factoring in data for unique peptides, and so a complete data set is desirable. As expected, the peptides common to all three isoforms yielded the strongest ion currents and showed reasonable agreement. Reliable quantification data could not be obtained for the peptide YAGEVSHDDK because of miscleavage, likely occurring as a result of the close proximity of the aspartic acid residues to the cleavage site (21, 22). The peptide DPANLPWASLNIDIAIDSTGVFK partially deamidated at both asparagine residues, an artifact of the sample preparation process that affected both standard and analyte, and this was confirmed by tandem MS sequencing (see supplemental Fig. 1). Quantification of Tdh2p was achieved by subtracting Tdh1p (IDVAVADSTGVFK) and Tdh3p (DPANLPWGSSNVDIAIDSTGVFK) from the data obtained from the peptides VPTVDVSVVDLTVK and VLPELQGK, common to all three isoenzymes. Difficulties in peptide selection caused by high sequence homology also apply in the case of enolase, because enolase 1 and enolase 2 are 95% identical. The isoform-specific peptides were TFAEALR and NVNDVIAPAFVK for Eno1p and IEEELGDK and TAGIQIVADDLTVTNPAR for Eno2p (the latter identified as putatively phosphorylated at both the second and third threonine residues (23)). A fifth peptide, SGETEDTFIADLVVGLR, common to both isoforms, was included as a summation check. The peptide TFAEALR could not be used for quantification because an isobaric peptide TFAEAIR, corresponding to an unrelated protein (ribose-phosphate pyrophosphokinase) might have compromised the analysis. Peptide IEEELGDK was also discounted because sequence verification by tandem MS was unsuccessful. Moreover, the peptide used for summation, SGETEDTFIADLVVGLR, consistently appeared to be under-represented in the data sets, and this might in part be explained by the close proximity of this sequence to the C terminus in the native protein and the possibility of endogenous proteolytic degradation. However, we have not explored this discrepancy further.
Pyruvate kinase has two isoenzymes: pyruvate kinase 1 and pyruvate kinase 2, sharing over 70% identity. Two peptides were used to quantify each of these variants: IENQQGVNNFDEILK and IIYVDDGVLSFQVLEVVDDK for Pyk1p (also known as Cdc19p) and VLQIIDESNLR and FIYVDDGILSFK for Pyk2p. For Pyk1p, IENQQGVNNFDEILK gave a strong signal, but peptide IIYVDDGVLSFQVLEVVDDK was detected by accurate mass/retention time in some, but not all turbidostats. Because we could not obtain tandem MS data to verify sequence authenticity, this peptide was not included in the quantification. For Pyk2p, no analyte signal was detectable, although tandem MS data were obtained for the corresponding heavy peptides, and extracted ion chromatograms of the analyte in the Orbitrap MS1 scans failed to give data. If we assume that we would easily detect 5 fmol of any given peptide on the Orbitrap platform, this would quantify the protein at less than 134,000 molecules/cell (extrapolation of the data based on the proportion represented in M1 Extract 1 scaled up to 100%). The differences in expression levels are consistent with the label-free data (see Fig. 7) (20) that reported 100-fold greater expression for Pyk1p than Pyk2p. Pyruvate kinase 2 is repressed by glucose and may be used by the cell under conditions of low glycolytic flux (24).
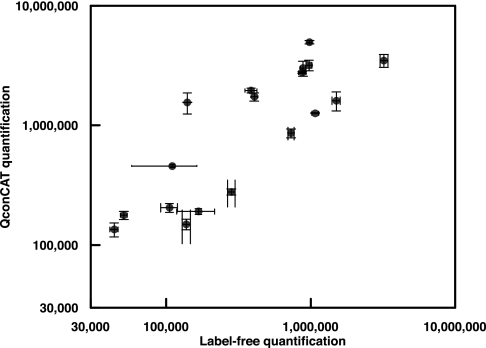
Fig. 7.
Comparison of label-free and QconCAT methodologies. For eighteen of the yeast glycolytic enzymes, data for quantification were obtained by both methodologies. The quantification values are presented as a log-log plot as the average of four chemostat cultures (QconCAT, diploid strain) and the average of five chemostat cultures, haploid strain). In both instances, the error bars are the S.E.
We also encountered modifications of peptides that could not be predicted. A case in point is fructose bisphosphate aldolase, assessed using two peptides (GISNEGQNASIK and EDLYTKPEQVYNVYK). Despite a possible putative phosphorylation site at the first serine residue (23) in the first peptide above, there were clear signals from both heavy and light variants of this protein, yielding an estimate of approximately 3 million molecules/cell. The second peptide yielded substantially lower measures, and database searching of experimental data suggested that the analyte peptide might be internally acetylated on a lysine residue (an internal KP), thus dividing the signal between the modified and unmodified forms. We also noted in some cases that neither standard nor analyte could be detected e.g. YSVWSAIGLSVALYIGYDNFEAFLK from PGI, although the peptides were readily detected in mixtures of recombinant QconCAT heavy and light only. This serves to emphasize the importance of evaluating peptides in the true complex biological background. Discrepancies between the two peptides for the same protein might also be attributed to miscleavage, especially where the terminal lysine and arginine are preceded by an aspartic acid residue. For phosphoglycerate kinase, this is a possibility for the peptide IQLIDNLLDK, which yielded lower values than ALLDEVVK, but we were unable to substantiate this.
The major isoform of pyruvate decarboxylase is Pdc1p, and the least abundant is Pdc5p, and this is consistent with our data (QconCAT). The published tagging data (18) are at variance with this statement, because the ratio of Pdc1p:Pdc5p:Pdc6p is 6:30,000:1. For Pdc1p, the two peptides used were VATTGEWDK and AQYNEIQGWDHLSLLPTFGAK, and both gave consistent data for cells grown in batch culture, with much higher levels obtained than for cultures from turbidostats, although this may be due to differences in media/culture methods. Detection of Pdc5p and Pdc6p has proven challenging in our hands; for the two peptides selected for Pdc5p, LLETPIDLSLKPNDAEAEAEVVR and VATTGEWEK, no analyte signal was detected in any of the analyses undertaken, irrespective of the culture method used. Of the two peptides selected for Pdc6p, IATTGEWDALTTDSEFQK and LPVFDAPESLIK, the former was detected using an SRM approach with 23,400 molecules/cell obtained corresponding to ∼800 amol on column for the most abundant extract. In the label-free analysis, there was evidence for low levels of expression of these two isoforms (approximately 10,000 copies/cell).
We included peptides for seven isoforms of alcohol dehydrogenase. In the previous study (20), all were detected barring Adh1p, with the following numbers of molecules/cell obtained: Adh2p, 1,600; Adh3p, 11,400; Adh4p, 125; Adh5p, 1,300; Adh6p, 21,700; and Adh7p, 28,700. In this study, two peptides were selected for Adh1p quantification, ANELLINVK and GVIFYESHGK, and initial experiments showed detection of heavy ANELLINVK in the turbidostat cultures, with supporting tandem MS data (data not included). The same peptide was also detected in batch (heavy and analyte), but there were issues with overlapping isotopic profiles complicating analysis of the analyte signal in these cultures. Moreover, the second peptide was also observed by accurate mass only in the turbidostat cultures, but there was no supporting tandem MS data in this case. Method transfer to SRM on the triple quadrupole facilitated detection and quantification of both peptides in the turbidostat cultures for ANELLINVK and GVIFYESHGK, with the resulting data showing good agreement; 420,000 and 498,000 molecules/cell were obtained respectively (supplemental Table Vb).
Some isoforms were only detected in the batch cultures, e.g. Adh3p and Adh4p were both detected in the cells grown in batch (see supplemental Table 5). We detected very low levels of analyte corresponding to GIDLINESLVAAYK from Adh4p as a Type A in batch culture, but for the turbidostat cultures, only a signal for the standard was obtained (Type B). It was not possible to obtain quantitative data for Adh5p, Adh6p, or Adh7p. In all cases, m/z levels corresponding to the standard peptides were detectable. The absence of a corresponding analyte signal (Type B) precluded quantification, although because there is some duplication of function between the respective isoforms, it is likely that not all are expressed.
DISCUSSION
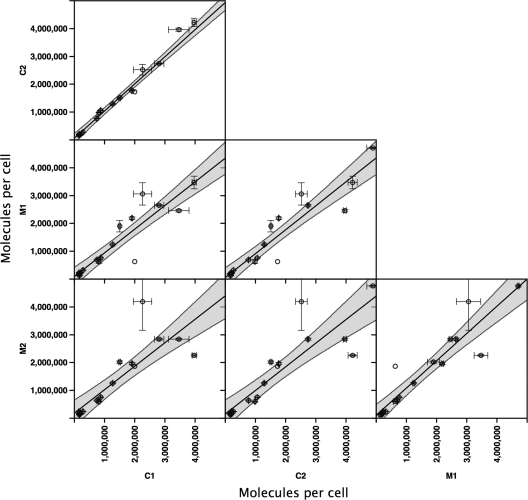
One of the goals of quantitative proteomics must be consistency. The availability of four sets of quantification data (technically triplicated additionally) from different biological replicates permitted assessment of the reproducibility of the expression data (Fig. 6). Not only were the pairs of turbidostats very comparable (M1 versus M2, R2 = 0.9325, gradient = 1.0169 (n = 22 peptides); C1 versus C2, R2 = 0.9887, gradient = 0.9729 (n = 21), but the M and C data sets were highly correlated, with a slope approaching unity (C1 versus M1; R2 = 0.9474, gradient = 0.8663 (n = 21) and C2 versus M2; R2 = 0.8735, gradient = 0.8993 (n = 21). We are confident that carefully grown cells can generate reproducible protein expression profiles that are consistent across laboratories. It remains to be seen whether batch-grown cells can offer the same robust expression analyses, because there is so much potential for variability in growth rate, sampling time, cell density, medium utilization, etc.
Fig. 6.
Reproducibility of expression levels. Full absolute quantification analyses were completed for cell lysates from duplicate parallel turbidostat cultures prepared at two independent sites: Cambridge (termed C1 and C2) and Manchester (termed M1 and M2). For each protein, absolute quantification was expressed as molecules/cell where n = 3 technical replicates (digest + MS analyses); the error bars are the S.E. of these analyses. The shaded area highlights the 95% confidence limits of the quantification comparisons.
It is interesting to contemplate the abundances of the glycolytic pathway proteins in the context of the overall protein complement of the yeast cell. There is some uncertainty about the precise protein content of a cell of S. cerevisiae, but a haploid cell is reported to contain 6 pg of protein (25), although here, we estimate between 3 and 4 pg/cell (it is important to note that for the label-free analyses with haploid cells, we do not centrifuge the broken cell preparation; all of the protein in the cell enters the analytical workflow). From the current study, we estimate 6 pg/cell for a diploid cell (other references in the literature suggest 8 pg/cell for a diploid cell) (25). Assuming an average molecular mass of approximately 50 kDa (26), the diploid yeast cell containing 6 pg of protein has an approximate constituency of 120 amol of protein, or approximately 70 million protein molecules. The glycolytic enzymes quantified in the soluble extracts account for 27.3 ± 1.3 million molecules (mean ± S.E., n = 4 biological replicates) or about one-third of the total proteome, a value consistent with previous analyses (27). The frequent appearance of some of the glycolytic enzymes as the most abundant spots on two-dimensional gel electrophoresis of soluble S. cerevisiae extracts further attests to the preponderance of some members of this pathway (28, 29). This figure is further borne out by label-free quantification (8), where 31% of the total molecules quantified are derived from the glycolytic pathway. Approximately a further 16 million protein molecules are engaged in the ribosome, and thus, to a first approximation, these two cellular components account for about half of the total yeast proteome by number.
When the QconCAT data are compared with the label-free quantitative data (Fig. 7), there is a general trend toward underestimation of protein abundance by label-free methods; most of the quantification data by QconCAT lie above the values obtained by label-free quantification. This suggests that there may be a systematic suppression of abundance in label-free approaches that is particularly prominent for high abundance proteins,. Label-free quantification is also obtained by reference to (usually one) standard proteins, and there may be scope for adoption of more accurate standards for this type of analysis.
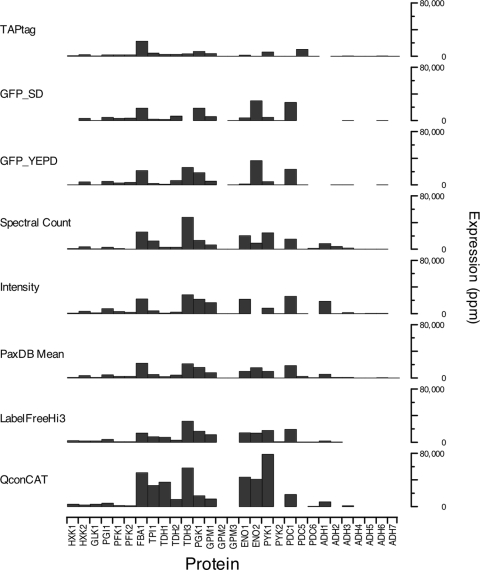
The quantification data described herein can also be compared with other studies, based on green fluorescent protein (30) or TAP (Tandem Affinity Purification) tagging (20) or label-free quantification based on spectral counting or ion intensity (Yeast PeptideAtlas build April 2009) (31). In addition, we completed a 5-fold biologically replicated label-free analysis using the Hi3 approach in an MSE LC-MS/MS workflow (8). Quantification data for the proteins studied here were derived from the integrated data sets in the Pax-DB database (20, 30, 32) (Yeast PeptideAtlas build April 2009 (31). The Pax-DB is developed and maintained by the Swiss Institute for Bioinformatics), as well as from our own quantification (Fig. 8). Comparison of such disparate data sets is fraught with complications, and there is a danger of overinterpretation of the differences. The Pax-DB data sets are normalized to parts per million, and we converted our data to the same parameter, assuming 70 million protein molecules in a diploid yeast cell. As can be seen, there are some notable discrepancies between the different quantitative approaches and without overinterpretation, the following observations are germane. First, QconCAT yields higher estimates in general than all other methods, which would suggest the value of an examination of the ability of these methods to quantify high abundance proteins without introducing range compression; the TAP-tagged protein quantification seems particularly prone to this compression. Second, the overall pattern of expression was reasonably consistent across the markedly different methodologies, suggesting that all such data sets could be internally recalibrated. As more and more quantitative data becomes available, this can be explored more formally. When all of the quantification data sets were ranked according to relative abundance, the overall picture was of consistent ranking (Friedman test, chi-squared = 168.2, d.f. = 28, p < 0.0001), although there were one or two notable exceptions where proteins were ranked at very different abundances. Aldolase, glyceraldehyde-3-phosphate dehydrogenases, and enolases were judged to be expressed at high levels by all methodologies. At this juncture we would, however, venture to suggest that none of the approaches have been demonstrated to be sufficiently robust and independently verified to permit their use for absolute quantification, or indeed, use of such data in modeling studies.
Fig. 8.
Comparison of methods for global proteome quantification. Expression data for the yeast glycolytic enzymes were abstracted from Pax-DB, a collation of expression data from published data sets using different methodologies. In addition, the quantification data from the label-free approach used in this paper and the QconCAT quantification data are included for comparison. Expression levels are normalized to ppm (see text). The data set labeled PaxDB Mean is the mean of the methods collated in Pax-DB.
There are inherent difficulties reconciling data from different analyses. A recent SRM study (33), which mirrored the yeast growth conditions of the Western blot study (20), gave similar results following a single round of protein extraction; however we were unable to verify how cell numbers were determined. In a recent stable isotope labeling by amino acids in cell culture study (32), relative quantification of haploid and diploid strains suggested similar amounts of glycolytic enzymes (expressed as molecules/cell) present in both haploid and diploid cells. However, this study used equivalent amounts of extracted protein from both cell types, but the cell numbers were not reported for either cell type. Different yeast strains, growth conditions, extraction methods, and analytical workflows make convergence and comparison of different data sets far from trivial.
This study has served to illustrate the challenges that are attendant upon full quantitative characterization of entire pathways using stable isotope-labeled internal standards. We used a strategy of nomination of standard peptides based on the expectation of efficient cleavage from the analyte protein and high quality MS signals. In several instances, these expectations were confounded. On 21 occasions (of which eight were for enzymes catalyzing the nine steps up to pyruvate and 13 were for enzymes catalyzing the two steps post-pyruvate in the pathway), a strong signal for the standard was not matched by an expected signal from the analyte. In the case of the latter, the expression levels may be low post-pyruvate, or another possibility is the presence of unknown post-translational modifications in the peptides selected.
A commitment step in a QconCAT workflow is the selection of the peptides to be built into the concatamer. The development of proteotypic peptide databases such as Global Proteome Machine (34), PeptideAtlas (31, 35), PRIDE (PRoteomics IDEntifications) (36), SBEAMS (Systems Biology Experiment Analysis Management System), and SRMAtlas (37) is a significant step forward, but these peptides are selected based on observations in MS/MS studies. At present, these peptides are not defined as formally representative of the parent protein, because there is no established resource to show the completeness of proteolysis, the lack of post-translational modification, or, indeed, the uniqueness of the peptide and freedom from isobaric and isomeric peptides derived from other proteins. Large scale quantification studies must develop a workflow that takes into account these considerations.
From the pool of standard peptides that we nominated, the attrition rate was significant, and only 25 peptides yielded reliable quantification data, of which eight were only detectable by SRM analysis (one each for Pfk1p and Pfk2p, and the remainder being enzymes that operated post-pyruvate). Barring the complex cases of isozyme-common peptides (e.g. as applies in the case of enolase and glyceraldehyde-3-phosphate dehydrogenase), for 12 proteins, we were reduced to a single peptide for quantification (six of which were obtained by SRM and of these, four were post-pyruvate). This was either because data were obtained for only one of the peptides or because the data for two peptides from the same protein did not agree (as applies in four cases: Fba1p, Tpi1p, Pgk1p, and Pyk1p), which, while defining the practice in many other quantification studies, does not give the reliability that a duplicate assessment would offer. For the three proteins for which similar quantitative data were obtained for both peptides (Hxkp1, Hxkp2, and Gpm1p), the agreement between the two peptides was very good (with a discrepancy of <4%).
Although we surmised that the glycolytic pathway would be mediated by high concentrations of most of the enzymes, an AMRT strategy was inadequate to permit quantification of all of the proteins. Additional data were acquired using SRM on a triple quadrupole instrument, which provided enhanced selectivity and increased sensitivity. This was particularly useful in cases of overlapping isotopes in the heavy or light, complicating the analysis. In terms of the AMRT data from the LTQ-Orbitrap platform, we note with interest however, that there is much greater concordance between different peptides from the same protein when there is tandem MS data available to verify the sequence authenticity. We rationalize that relying on the accurate mass alone in complex proteome samples may not be sufficient, because a potential miscleavage or unforeseen post-translational modification of the analyte cannot be ruled out and might lead to erroneous calculations based on incorrect peptide selection, i.e. ambiguity where, by chance, different peptides have the same mass, and the co-elution of irrelevant peptides that show the same mass difference as the label.
The objective of global quantification of a complete proteome is unlikely to be easily achieved. There are several reasons for this. The most sensitive methodology (far exceeding the sensitivity of label-free quantification) is based on single reaction monitoring in a triple quadrupole instrument, and at the moment, this can attain sensitivities of 10 amol on column. However, these sensitivities are often achieved with pure peptides, and sensitivity, which is largely a function of signal: noise ratio, is compromised when the SRM is conducted in a complex biological background; future developments are as likely to emphasize reduction of background as enhancement of signal. Also, the choice of peptides for such quantification studies is not unambiguous. The definition of “proteotypic” peptides is often driven by the frequency of observation of peptides for that particular protein, and it has rarely been demonstrated that these peptides are formally representative of the absolute quantity of the protein. For this, the peptides would have to be isobarically unique in the proteome, devoid of variable (and preferably, fixed) post-translational modifications, and should generate unique peptides (in both unlabeled and stable isotope labeled forms). Also, peptide level quantification (whether label-free or label-mediated) is only valid if it can be demonstrated that the peptides are excised from the parent protein completely; it is again rare for the abundances of partial cleavage products to be formally examined and interpreted. We propose that the term “quantotypic peptides” or Q-peptides should be reserved for those digestion products that are formally and quantitatively representative of a protein, whether in label-free or label-mediated workflows. As evidenced here, “best guess” methods can be compromised, and several optimally chosen peptides did not deliver useful quantitative data.
Deep quantification is exacerbated when one moves away from relatively simple organisms. For this study on S. cerevisiae, the protein load applied to the column (150 ng) was representative of about 30,000 cells (at 5 pg/cell). Higher loads (1500 ng is feasible, at the expense of chromatographic quality) or prefractionation (33) can enhance this performance, of course. At a limit of quantification of 100 amol, this sets a limit of detection at 2,000 copies/cell at 150 ng, or 200 copies/cell at 1500 ng. In mammalian cells, the protein content/cell is much higher, at typical values of 250 pg/cell. Thus, a column load of 1000 ng would be equivalent to the loading of only 6000 cells. At current instrumentation sensitivities, the ability to reach deeply in the proteome seems to be unattainable for any cell type other than simple organisms. An instrument with the rather exceptional performance of a good signal: noise ratio at 1 amol (as a limit of quantification rather than limit of detection, and assessed in the context of a complete biological background), capable of delivering high quality chromatography and mass spectrometry on a column load of 2500 ng of cell digest, would attain a copy number of 60 for a typical mammalian cell; this seems unreachable at present. Sample prefractionation and concentration (such as isoelectric focusing of peptides) can give at least an order of magnitude of sensitivity gain, at the expense of multiple LC-MS/MS runs (38–43). Prefractionation protocols couple with instruments capable of low attomole quantification (at signal: noise ratios of >10), delivering this level of routine performance for every Q-peptide, are not yet in common usage, but awareness of such issues can assist in defining the performance that should be sought.
Footnotes
* This work was supported by Biotechnology and Biological Sciences Research Council/Engineering and Physical Sciences Research Council Grant BB/C008219/1 from the Manchester Centre for Integrative Systems Biology (KMC, WD, CLW, and NM); Biotechnology and Biological Sciences Research Council Grants BB/C007433/1 and BB/G00912 (to R. J. B.), BB/C007735/1 (to S. J. G.), and BB/C505140/1 (to S. G. O.); and a contract from the European Commission under the FP7 Collaborative Programme, UNICELLSYS (to S. G. O.).
 The on-line version of this article (available at http://mcponline.org) contains supplemental Tables I–VI and Figs. 1–4.
The on-line version of this article (available at http://mcponline.org) contains supplemental Tables I–VI and Figs. 1–4.
1 The abbreviations used are:
- AMRT
- accurate mass/retention time
- SRM
- selected reaction monitoring
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight.
REFERENCES
- 1. Neilson K. A., Ali N. A., Muralidharan S., Mirzaei M., Mariani M., Assadourian G., Lee A., van Sluyter S. C., Haynes P. A. (2011) Less label, more free: Approaches in label-free quantitative mass spectrometry. Proteomics 11, 535–553 [DOI] [PubMed] [Google Scholar]
- 2. Sandin M., Krogh M., Hansson K., Levander F. (2011) Generic workflow for quality assessment of quantitative label-free LC-MS analysis. Proteomics 11, 1114–1124 [DOI] [PubMed] [Google Scholar]
- 3. Brun V., Dupuis A., Adrait A., Marcellin M., Thomas D., Court M., Vandenesch F., Garin J. (2007) Isotope-labeled protein standards: Toward absolute quantitative proteomics. Mol. Cell. Proteomics 6, 2139–2149 [DOI] [PubMed] [Google Scholar]
- 4. Dupuis A., Hennekinne J. A., Garin J., Brun V. (2008) Protein Standard Absolute Quantification (PSAQ) for improved investigation of staphylococcal food poisoning outbreaks. Proteomics 8, 4633–4636 [DOI] [PubMed] [Google Scholar]
- 5. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beynon R. J., Doherty M. K., Pratt J. M., Gaskell S. J. (2005) Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods 2, 587–589 [DOI] [PubMed] [Google Scholar]
- 7. Pratt J. M., Simpson D. M., Doherty M. K., Rivers J., Gaskell S. J., Beynon R. J. (2006) Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat. Protoc. 1, 1029–1043 [DOI] [PubMed] [Google Scholar]
- 8. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 9. Davey H. M., Davey C. L., Woodward A. M., Edmonds A. N., Lee A. W., Kell D. B. (1996) Oscillatory, stochastic and chaotic growth rate fluctuations in permittistatically controlled yeast cultures. Biosystems 39, 43–61 [DOI] [PubMed] [Google Scholar]
- 10. Allen J., Davey H. M., Broadhurst D., Heald J. K., Rowland J. J., Oliver S. G., Kell D. B. (2003) High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 21, 692–696 [DOI] [PubMed] [Google Scholar]
- 11. Baganz F., Hayes A., Farquhar R., Butler P. R., Gardner D. C., Oliver S. G. (1998) Quantitative analysis of yeast gene function using competition experiments in continuous culture. Yeast 14, 1417–1427 [DOI] [PubMed] [Google Scholar]
- 12. Wiechelman K. J., Braun R. D., Fitzpatrick J. D. (1988) Investigation of the bicinchoninic acid protein assay: Identification of the groups responsible for color formation. Anal. Biochem. 175, 231–237 [DOI] [PubMed] [Google Scholar]
- 13. Brown R. E., Jarvis K. L., Hyland K. J. (1989) Protein measurement using bicinchoninic acid: Elimination of interfering substances. Anal. Biochem. 180, 136–139 [DOI] [PubMed] [Google Scholar]
- 14. Kiyonami R., Schoen A., Prakash A., Peterman S., Zabrouskov V., Picotti P., Aebersold R., Huhmer A., Domon B. (July 27, 2010) Increased selectivity, analytical precision, and throughput in targeted proteomics. Mol. Cell. Proteomics 10.1074/mcp.M110.002931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yates J. R., Eng J. K., Clauser K. R., Burlingame A. L. (1996) Search of sequence databases with uninterpreted high-energy collision-induced dissociation spectra of peptides. J. Am. Soc. Mass Spectrom. 7, 1089–1098 [DOI] [PubMed] [Google Scholar]
- 16. Eng J. K., Fischer B., Grossmann J., Maccoss M. J. (2008) A fast SEQUEST cross correlation algorithm. J. Proteome Res. 7, 4598–4602 [DOI] [PubMed] [Google Scholar]
- 17. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 18. Brownridge P., Holman S. W., Gaskell S. J., Grant C. M., Harman V. M., Hubbard S. J., Lanthaler K., Lawless C., O'Cualain R., Sims P., Watkins R., Beynon R. J. (2011) Global absolute quantification of a proteome: Challenges in the deployment of a QconCAT strategy. Proteomics 11, 2957–2970 [DOI] [PubMed] [Google Scholar]
- 19. Rivers J., Simpson D. M., Robertson D. H., Gaskell S. J., Beynon R. J. (2007) Absolute multiplexed quantitative analysis of protein expression during muscle development using QconCAT. Mol. Cell. Proteomics 6, 1416–1427 [DOI] [PubMed] [Google Scholar]
- 20. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 21. Siepen J. A., Keevil E. J., Knight D., Hubbard S. J. (2007) Prediction of missed cleavage sites in tryptic peptides aids protein identification in proteomics. J. Proteome Res. 6, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brownridge P., Beynon R. J. (2011) The importance of the digest: Proteolysis and absolute quantification in proteomics. Methods 54, 351–360 [DOI] [PubMed] [Google Scholar]
- 23. Albuquerque C. P., Smolka M. B., Payne S. H., Bafna V., Eng J., Zhou H. (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics 7, 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boles E., Schulte F., Miosga T., Freidel K., Schlüter E., Zimmermann F. K., Hollenberg C. P., Heinisch J. J. (1997) Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J. Bacteriol. 179, 2987–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sherman F. (2002) Getting started with yeast. Methods Enzymol. 350, 3–41 [DOI] [PubMed] [Google Scholar]
- 26. Lodish H., Berk A., Zipursky S. L., Matsudaira P., Baltimore D., Darnell J. (2000) Hierarchical Structure of Proteins. In Molecular Cell Biology, 4th Ed., W. H. Freeman, New York [Google Scholar]
- 27. Fraenkel D. G. (2003) The top genes: On the distance from transcript to function in yeast glycolysis. Curr. Opin. Microbiol. 6, 198–201 [DOI] [PubMed] [Google Scholar]
- 28. Pratt J. M., Petty J., Riba-Garcia I., Robertson D. H., Gaskell S. J., Oliver S. G., Beynon R. J. (2002) Dynamics of protein turnover, a missing dimension in proteomics. Mol. Cell. Proteomics 1, 579–591 [DOI] [PubMed] [Google Scholar]
- 29. Futcher B., Latter G. I., Monardo P., McLaughlin C. S., Garrels J. I. (1999) A sampling of the yeast proteome. Mol. Cell Biol. 19, 7357–7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Newman J. R., Ghaemmaghami S., Ihmels J., Breslow D. K., Noble M., DeRisi J. L., Weissman J. S. (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441, 840–846 [DOI] [PubMed] [Google Scholar]
- 31. King N. L., Deutsch E. W., Ranish J. A., Nesvizhskii A. I., Eddes J. S., Mallick P., Eng J., Desiere F., Flory M., Martin D. B., Kim B., Lee H., Raught B., Aebersold R. (2006) Analysis of the Saccharomyces cerevisiae proteome with PeptideAtlas. Genome Biol. 7, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 33. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Craig R., Cortens J. P., Beavis R. C. (2004) Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 35. Desiere F., Deutsch E. W., Nesvizhskii A. I., Mallick P., King N. L., Eng J. K., Aderem A., Boyle R., Brunner E., Donohoe S., Fausto N., Hafen E., Hood L., Katze M. G., Kennedy K. A., Kregenow F., Lee H., Lin B., Martin D., Ranish J. A., Rawlings D. J., Samelson L. E., Shiio Y., Watts J. D., Wollscheid B., Wright M. E., Yan W., Yang L., Yi E. C., Zhang H., Aebersold R. (2005) Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 6, R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vizcaíno J. A., Côté R., Reisinger F., Foster J. M., Mueller M., Rameseder J., Hermjakob H., Martens L. (2009) A guide to the Proteomics Identifications Database proteomics data repository. Proteomics 9, 4276–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Picotti P., Lam H., Campbell D., Deutsch E. W., Mirzaei H., Ranish J., Domon B., Aebersold R. (2008) A database of mass spectrometric assays for the yeast proteome. Nat. Methods 5, 913–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geiser L., Vaezzadeh A. R., Deshusses J. M., Hochstrasser D. F. (2011) Shotgun proteomics: A qualitative approach applying isoelectric focusing on immobilized pH gradient and LC-MS/MS. Methods Mol. Biol. 681, 449–458 [DOI] [PubMed] [Google Scholar]
- 39. Wang H., Chang-Wong T., Tang H. Y., Speicher D. W. (2010) Comparison of extensive protein fractionation and repetitive LC-MS/MS analyses on depth of analysis for complex proteomes. J. Proteome Res. 9, 1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heller M., Ye M., Michel P. E., Morier P., Stalder D., Jünger M. A., Aebersold R., Reymond F., Rossier J. S. (2005) Added value for tandem mass spectrometry shotgun proteomics data validation through isoelectric focusing of peptides. J. Proteome Res. 4, 2273–2282 [DOI] [PubMed] [Google Scholar]
- 41. Elschenbroich S., Ignatchenko V., Sharma P., Schmitt-Ulms G., Gramolini A. O., Kislinger T. (2009) Peptide separations by on-line MudPIT compared to isoelectric focusing in an off-gel format: Application to a membrane-enriched fraction from C2C12 mouse skeletal muscle cells. J. Proteome Res. 8, 4860–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 43. Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P. (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: The yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]