Abstract
Expression profiles represent new molecular tools that are useful to characterize the successive steps of tumor progression and the prediction of recurrence or chemotherapy response. In this study, we have used quantitative proteomic analysis to compare different stages of colorectal cancer. A combination of laser microdissection, OFFGEL separation, iTRAQ labeling, and MALDI-TOF/TOF MS was used to explore the proteome of 28 colorectal cancer tissues. Two software packages were used for identification and quantification of differentially expressed proteins: Protein Pilot and iQuantitator. Based on ∼1,190,702 MS/MS spectra, a total of 3138 proteins were identified, which represents the largest database of colorectal cancer realized to date and demonstrates the value of our quantitative proteomic approach. In this way, individual protein expression and variation have been identified for each patient and for each colorectal dysplasia and cancer stage (stages I–IV). A total of 555 proteins presenting a significant fold change were quantified in the different stages, and this differential expression correlated with immunohistochemistry results reported in the Human Protein Atlas database. To identify a candidate biomarker of the early stages of colorectal cancer, we focused our study on secreted proteins. In this way, we identified olfactomedin-4, which was overexpressed in adenomas and in early stages of colorectal tumors. This early stage overexpression was confirmed by immunohistochemistry in 126 paraffin-embedded tissues. Our results also indicate that OLFM4 is regulated by the Ras-NF-κB2 pathway, one of the main oncogenic pathways deregulated in colorectal tumors.
Every year, more than one million individuals around the world are diagnosed with colorectal cancer (CRC),1 and with a death rate ∼33% (1), this disease is an important cause of mortality. CRC diagnosis and prognosis rely on the tumor-node-metastasis and clinical staging systems, which illustrate local lymph node and distal organ invasion. These clinical stages are important prognostic factors because survival rates of 5 years or more are observed for more than 90% of patients diagnosed with Stage I CCR, whereas survival rates drop to only ∼10% for CRC that have metastasized to distant organs (stage IV) (2). As a consequence, early stage detection has the most impact on cancer incidence and mortality in this disease (3, 4). As initially described by Vogelstein et al. (5), colorectal transformation is explained by the sequential accumulation of genetic alterations that generate malignant cells (6). Mutations of the adenomatous polyposis coli gene and the subsequent activation of β-catenin is probably the most common initiating event of CRC, leading to the transformation of normal colonic epithelium into adenomas (7–10). This stage represents an intermediate lesion where cells exhibit autonomous growth and probably genetic instability but are incapable of invasive growth and metastasis. It is estimated that only a small proportion of ∼5% of adenomas will progress to the next CRC stages, implying that the transition from normal cells to adenoma differs from the progression from adenomas to adenocarcinomas. Following the loss of adenomatous polyposis coli, it has been proposed recently that KRAS mutations are essential to allow the nuclear accumulation of β-catenin and the subsequent progression to the adenocarcinoma step (11). Although Ras mutations have probably no prognosis value, this modification is associated with resistance to anti-epidermal growth factor receptor-targeted therapies (12), indicating that this transformation pathway is associated with an intrinsic drug resistance program.
Despite their utility, tumor-node-metastasis and clinical staging remain relatively imprecise and are not well characterized at the molecular level. For this reason, the development of new prognostic tools would be useful to characterize the successive steps of the disease and predict the risk or recurrence of chemotherapy escape. Elegant results have recently identified gene expression profiles that associate with specific oncogenic pathways and can eventually predict chemotherapy sensitivity (13, 14). This approach has also been successfully used in CRC to identify a 50-gene signature that distinguished patients with low or high risks or recurrence at the early stage of the disease (15). These results also led to the identification of a therapeutic approach that would be specific of a particular stage of CRC. Besides these genomic data, proteomics analysis is also a powerful tool for the global evaluation of protein expression and the identification of prognosis or predictive signatures. However, although recent in-depth proteomics analyses have generated large protein data sets, only a few proteins such as carcinoembryonic antigen, CA19.9, and CA125 have been described as potential prognosis or prognostic biomarkers, and none of them are recommended for clinical screening (16–20). These analyses essentially used two-dimensional gel strategies combined with image analysis, thereby limiting the analyses to the more abundant CRC proteins. A few recent studies have combined more targeted approaches with two-dimensional electrophoresis, including studies focusing on membrane proteins (21), basic proteins (22), heparin-affinity isolated proteins (23), or proteasome (24). Some of these studies have identified novel candidate CRC serum biomarkers with comparable or better sensitivity than carcinoembryonic antigen, such as nicotinamide N-methyltransferase (25), proteasome activator complex subunit, PSME3 (26), S100A9, S100A8 (27), and Desmin (28). However, these results are often largely limited to abundant proteins that are commonly overexpressed in cancers (structural proteins, glycolytic enzymes, annexins, cathepsins, and heat shock protein). Because these proteins are probably not specific to CRC, the benefits in clinical staging or in predicting the success of targeted therapies remain to be determined. Tumor analysis is challenging, given the heterogeneity of the colorectal cancer tissue and the limited number of tumor cells generally available. To increase the specificity of the analysis, tumor cells should be ideally isolated from heterogeneous samples by laser capture microdissection (LCM) (29). Although this approach is widely used in the genomic field, proteomic analyses of tumor tissue are limited, and to our knowledge only one study has been performed in CRC, using adenoma tissue (29).
In this study, we have performed comparative proteomic profiling of laser capture microdissected adenoma, stage I–IV adenocarcinoma tissues, and normal colorectal tissues from 28 different human specimens. Using only the limited amount of material collected by LCM, a proteomic profile for each sample has been separately acquired by iTRAQ labeling using our previously developed method (30, 31). 555 proteins were quantified in the different stages. Among these proteins, we focused on OLFM4 (olfactomedin-4), because this protein has recently been shown to be expressed in colorectal stem cells in association with Lgr5 (32, 33). We found that OLFM4 is a secreted protein that is regulated by Ras-NF-κB2, one of the main oncogenic pathway in colorectal cells. These results highlight the power of quantitative proteomic approaches to allow the identification of stage-specific markers in colorectal cancer. To our knowledge, this is the most comprehensive global study that compares proteomic results of phenotypically normal, adenoma, early stage tumor, and metastatic adenocarcinoma in multiple individual samples.
MATERIALS AND METHODS
Sample Collection
The study protocol and patient consent forms were approved by the Angers Hospital and CRLCC Paul Papin Ethic Committee. Twenty-eight colorectal frozen tissue samples, collected between 1999 and 2004, were obtained from the Cancer Center tumor bank. All of the tissue specimens were obtained from surgical resection. Normal colonic tissue was obtained from the distal edge of the resection at least 10 cm from the tumor. After hematoxylin and eosin staining, the paraffin-embedded tissue sections from all of the specimens were evaluated by two experienced pathologists independently according to Union Internationale contre le Cancer staging. Clinical features of tissue candidates are summarized in Table I.
Table I. Clinical features of human tissue samples.
| Clinical features | iTRAQ | IHC |
|---|---|---|
| Gender (male/female) | 16/12 | 70/56 |
| Mean age (years) | 68.7 ± 8.9 | 69.39 ± 10.56 |
| Age range (years) | 48–87 | 41–94 |
| Union Internationale contre le Cancer stage | ||
| Adenoma | 4 (14.3%) | 30 (23.8%) |
| Intramucosal carcinoma | 0 | 12 (9.5%) |
| Adenocarcinoma | ||
| Stage I | 6 (21.5%) | 26 (20.6%) |
| Stage II | 5 (17.8%) | 14 (11.1%) |
| Stage III | 9 (32.1%) | 25 (19.8%) |
| Stage IV | 4 (14.3%) | 19 (15.2%) |
| Location | ||
| Colon | 17 (58.6%) | 92 (73.0) |
| Rectum | 1 (3.4%) | 10 (7.9%) |
| Rectosigmoid hinge | 6 (20.7%) | 9 (7.2%) |
| Sigmoid | 3 (10.4%) | 10 (7.9%) |
| Caecum | 2 (6.9%) | 5 (4.0%) |
Laser Capture Microdissection
Frozen sections (12 μm thick) of either colon cancer or normal colonic mucosa were cut on a cryostat (Bright Instrument Co. Ltd., Huntingdon, UK). Specific sections were stained with toluidine blue for visual reference. Tissue sections were incubated in 70% ethanol for 1 min, 95% ethanol for 2 min, 95% ethanol for 2 min, and finally twice in 100% xylene for 5 min. Xylene was evaporated, and sections were microdissected using a PixCell II laser capture microdissection system (Arcturus Engineering, Mountain View, CA) equipped with the PixCell II image archiving software (Arcturus Engineering). Laser settings were as follows: λ = 810 nm, spot diameter set at 7.5 μm, pulse duration = 70 ms, and power = 70 mW. After microdissection, the plastic film containing the microdissected cells was removed, the film containing the tumor cells was placed in a microcentrifuge tube, and the protein lysis solution was added. Approximately 30,000 cells were captured from either a single or consecutive tissue sections using up to five CapSure LCM caps (Molecular Devices Corporation).
Protein Extraction and Digestion
Protein extraction was carried out using the Liquid Tissue MS protein preparation kit according to manufacturer's protocol (Expression Pathology Inc., Gaithersburg, MD). Briefly, the films from the underside of the caps for all samples were removed, transferred to low binding reaction tubes, incubated with 20 μl of Liquid Tissue extraction, and heated at 95 °C for 90 min. After cooling for 2 min on ice, 5 μl of trypsin reagent was added and incubated at 37 °C for 1 h with vigorous shaking for 30 s at 20 min intervals. The samples were further incubated overnight at 37 °C followed by heating at 95 °C for 5 min. The samples were then harvested via centrifugation at 10,000 × g, dried completely using a SpeedVac, resuspended in 100 μl of 0.5% TFA in 5% acetonitrile, desalted via PepClean C-18 spin columns (Pierce), and dried for iTRAQTM processing.
Peptide Labeling with iTRAQ Reagents
Peptides samples were resuspended with 30 μl of iTRAQ dissolution buffer (AB Sciex). They were reduced with 5 mm tris-(2-carboxyethyl)phosphine at 60 °C for 1 h, and the cysteine-groups were blocked using a 10 mm methyl methanethiosulfonate solution at room temperature for 10 min. Each peptide solution was labeled at room temperature for 2 h with one iTRAQ reagent vial previously reconstituted with 70 μl of ethanol for 4-plex iTRAQ reagent and reconstituted with 50 μl of isopropanol for 8-plex iTRAQ reagent. A mixture containing small aliquots from each labeled sample was analyzed by MS/MS to determine a proper mixing ratio to correct for unevenness in peptide yield from Liquid Tissues procedures. Labeled peptides were then mixed in a 1:1:1:1 (or 1:1:1:1:1:1:1:1) ratio. The peptide mixture was then dried completely using a SpeedVac.
Peptide OFFGEL Fractionation
For pI-based peptide separation, we used the 3100 OFFGEL Fractionator (Agilent Technologies, Böblingen, Germany) with a 24-well set-up. Prior to electrofocusing, the samples were desalted onto a Sep-Pak C18 cartridge (Waters). For a 24-well set-up, peptide samples were diluted to a final volume of respectively 3.6 ml using OFFGEL peptide sample solution. To start, the 24-cm-long IPG gel strip (GE Healthcare) with a 3–10 linear pH range was rehydrated with the peptide IPG strip rehydradation solution according to the protocol of the manufacturer for 15 min. Then 150 μl of sample was loaded in each well. Electrofocusing of the peptides was performed at 20 °C and 50 μA until the 50-kVh level was reached. After focusing, the 24 peptide fractions were withdrawn, and the wells were washed with 200 μl of a solution of water/methanol/formic acid (49:50:1). After 15 min, the washing solutions were pooled with their corresponding peptide fraction. All of the fractions were evaporated by centrifugation under vacuum and maintained at −20 °C. Just prior nano-LC, the fractions were resuspended in 20 μl of H2O with 0.1% (v/v) TFA.
Capillary LC Separation
The samples were separated on an Ultimate 3,000 nano-LC system (Dionex, Sunnyvale, CA) using a C18 column (PepMap100, 3 μm, 100 A, 75 μm inner diameter × 15 cm; Dionex) at 300 nl/min a flow rate. Buffer A was 2% ACN in water with 0.05% TFA, and buffer B was 80% ACN in water with 0.04% TFA. The peptides were desalted for 3 min using only buffer A on the precolumn, followed by a separation for 105 min using the following gradient: 0 to 20% B in 10 min, 20% to 45% B in 85 min, and 45% to 100% B in 10 min. Chromatograms were recorded at the wavelength of 214 nm. Peptide fractions were collected using a Probot microfraction collector (Dionex). We used CHCA (LaserBioLabs, Sophia-Antipolis, France) as MALDI matrix. The matrix (concentration of 2 mg/ml in 70% ACN in water with 0.1% TFA) was continuously added to the column effluent via a micro T mixing piece at 1.2 μl/min flow rate. After 12-min run, a start signal was sent to the Probot to initiate fractionation. Fractions were collected for 10 s and spotted on a MALDI sample plate (1,664 spots/plate; Applied Biosystems, Foster City, CA).
MALDI-MS/MS
MS and MS/MS analyses of off-line spotted peptide samples were performed using the 4800 or 5800 MALDI-TOF/TOF Analyzers (Applied Biosystems/ABsciex) and 4000 Series Explorer software, version 3.5 (with MALDI 4800) and version 4.0 with MALDI 5800). The instrument was operated in a positive ion mode and externally calibrated using a mass calibration standard kit (ABsciex). The laser power was set between 2800 and 3400 for MS and between 3600 and 4200 for MS/MS acquisition. After screening all LC-MALDI sample positions in MS-positive reflector mode using 1500 laser shots, the fragmentation of automatically selected precursors was performed at a collision energy of 1 kV using air as collision gas (pressure of ∼2 × 10–6 Torr) with an accumulation of 2000 shots for each spectrum. MS spectra were acquired between m/z 800 and 4000. For internal calibration, we used the parent ion of Glu-1 fibrinopeptide at m/z 1570.677 diluted in the matrix (30 fmol/spot). Up to 12 of the most intense ion signals per spot position having a S/N of >12 were selected as precursors for MS/MS acquisition. Peptide and protein identification were performed by the ProteinPilotTM software version 3.0 (AB Sciex) using the Paragon algorithm as the search engine (34). Each MS/MS spectrum was searched for Homo sapiens species against the Uniprot/Swissprot database (UniProtKB/Sprot 20090414 release 15.0, with 525,997 sequence entries). The searches were run using with the fixed modification of methylmethanethiosulfate-labeled cysteine parameter enabled. Other parameters such as tryptic cleavage specificity, precursor ion mass accuracy and fragment ion mass accuracy are MALDI 4800 or 5800 built-in functions of ProteinPilot software. The detected protein threshold (unused protscore (confidence)) in the software was set to 2 to achieve 99% confidence, and identified proteins were grouped by the ProGroup algorithm (ABsciex) to minimize redundancy. The bias correction option was executed.
A decoy database (based on a reverse sequence database concatenated with the forward sequence database) search strategy was also used to estimate the false discovery rate (FDR = number of validated decoy hits/(number of validated target hits + number of validated decoy hits) × 100). The FDR was calculated by searching the spectral against the Uniprot H. sapiens decoy database. The FDR for each iTRAQ experiment is indicated in Table II.
Table II. Spectra, peptides, and proteins identified and quantified of eight independent iTRAQ experiments.
| Experiment | iTRAQ | Mass spectrometer | Quantity | Total spectra | Identified peptides (confidence = 95%) | Identified peptides (confidence = 95%, contribution = 2) | Identified proteins (after grouping) |
Quantified proteins (unused >4) | |
|---|---|---|---|---|---|---|---|---|---|
| Unused >2 | Unused >4 | ||||||||
| μg | |||||||||
| iTRAQ-1 | 4-plex | 4800 | 100 | 146,063 | 51,399 | 17,287 | 2175 | 1210 | 1180 |
| iTRAQ-2 | 4-plex | 4800 | 100 | 105,013 | 52,462 | 6638 | 2017 | 1127 | 1102 |
| iTRAQ-7 | 4-plex | 5800 | 100 | 229,545 | 88,106 | 8215 | 2251 | 1253 | 1224 |
| iTRAQ-8 | 8-plex | 5800 | 100 | 338,262 | 67,606 | 6573 | 1937 | 1041 | 1012 |
| iTRAQ-6 | 8-plex | 4800 | 100 | 131,143 | 42,005 | 12,997 | 1406 | 742 | 740 |
| iTRAQ-3 | 4-plex | 4800 | 70 | 86,380 | 23,768 | 8299 | 1214 | 604 | 603 |
| iTRAQ-4 | 4-plex | 4800 | 70 | 97,110 | 46,234 | 6844 | 1075 | 557 | 557 |
| iTRAQ-5 | 4-plex | 4800 | 70 | 57,186 | 26,247 | 3945 | 1242 | 431 | 431 |
Quantification of Relative Protein Expression
We employed a customized software package, iQuantitator (35, 36), to infer the magnitude of change in protein expression. The software infers treatment-dependent changes in expression using Bayesian statistical and Markov Chain Monte Carlo methods. Basically, this approach was used to generate means, medians, and 95% confidence intervals (upper and lower) for each treatment-dependent change in protein expression by using peptide level data for each component peptide and integrating data across the two experiments. For proteins whose iTRAQ ratios were down-regulated in tissues, the extent of down-regulation was considered further if the higher limit of the confidence interval had a value lower than 1. Conversely, for proteins whose iTRAQ ratios were up-regulated in tumors, the extent of up-regulation was considered further if the lower limit of the confidence interval had a value greater than 1. The width of these credible intervals depends on the data available for a given protein. Because the number of peptides observed and the number of spectra used to quantify the change in expression for a given protein are taken into consideration, it is possible to detect small but significant changes in up- or down-regulation when many peptides are available. For each protein and each peptide associated with a given protein, the mean, median, and 95% credible intervals were computed for each of the protein and peptide level treatment effects.
The peptide selection criteria for relative quantification were performed as follows. Only peptides unique for a given protein were considered for relative quantification, excluding those common to other isoforms or proteins of the same family. Proteins were identified on the basis of having at least two peptides with an ion score above 95% confidence. The protein sequence coverage (95%) was estimated for specific proteins by the percentage of matching amino acids from the identified peptides having confidence greater than or equal to 95% divided by the total number of amino acids in the sequence.
Functional Annotation and Network Analysis
Gene ontology (GO) terms for identified proteins were extracted, and overrepresented functional categories for differentially abundant proteins were determined by the high throughput GOminer tool (National Cancer Institute, http://discover.nci.nih.gov.gate2.inist.fr/gominer/) (37). All proteins that were subjected to iQuantitator analysis served as the background list, and GO terms with at least five proteins were used for statistical calculations. A p value for each term was calculated via the one-sided Fisher's exact test, and FDR was estimated by permutation analysis using 1000 randomly selected sets of proteins sampled from the background list. Statistically significant (FDR < 25%) GO terms were clustered based on the correlation of associated proteins to minimize potential redundancy in significant GO terms.
Network analyses of protein candidates and the ratios of their expression in tumor and nontumor tissues (obtained from eight independent experiments) were performed using the MetaCoreTM analytical suite version 4.7 (GeneGo, Inc., St. Joseph, MI) and compared using p values of <0.01 as statistical metrics. For enrichment analysis, gene identifiers of the uploaded files were matched with gene identifiers in functional ontologies in MetaCoreTM (38), which included canonical pathway maps (GeneGo maps), GeneGo cellular processes, GO cellular process, and diseases categories.
Western Blot Analysis of Proteins in Normal Tissues and CRC
Whole cell lysates were prepared from normal tissues and tumoral tissues. Frozen tissue samples were homogenized and lysed in a buffer containing 7 m urea, 2 m thiourea, and 4% (w/v) CHAPS at 4 °C for 1 h using a rotary shaker. Lysis was achieved by sonication on ice (three 5-s pulses), and the lysates were clarified by centrifugation at 14,000 × g at 4 °C for 15 min. Protein concentrations were determined using the FluoroProfile protein quantification kit (Sigma-Aldrich), with BSA as the standard, and equal amounts of proteins (80 μg/lane) from the samples tissues were resolved on a 10% SDS-polyacrylamide gel. The proteins were then electrotransferred onto PVDF membranes. After blocking with 3% BSA in TBS (0.1 m, pH 7.4), blots were incubated with the respective primary antibodies (1:200 dilution) at 4 °C overnight. The protein abundance of β-tubulin was used as a control for protein loading and was determined with rabbit polyclonal anti-β-tubulin: (H-235) antibody (sc-9104; Santa Cruz Biotechnology Inc.). The membranes were incubated with the respective secondary antibody, horseradish peroxidase-conjugated rabbit anti-IgG (goat anti-rabbit IgG, 1:5000; Santa Cruz Biotechnology Inc.), and diluted with 1% bovine serum albumin for 1 h at room temperature. After each step, blots were washed three times with 0.05% Tween, TBS. The membrane was probed with the indicated antibodies and developed with the ECL.
Immunohistochemistry
One hundred twenty-six patients with adenoma or colorectal adenocarcinoma were studied by immunohistochemistry. All of the tumors were obtained from the Departments of Pathology at the Paul Papin Cancer Center and at the University Hospital of Angers and from the Center of Pathology of Angers between 2000 and 2005. Some samples were excluded: young patients (under 40 years old) and tumors having received chemotherapy, metastasic tumors. The location was almost colic or from high rectum. There were 30 adenomas with 15 low grade adenomas and 15 high grade adenomas; the dysplasia was classified according to the established criteria of architectural features and cytological atypia. There were 72 colorectal adenocarcinomas. According to the seventh tumor-node-metastasis staging system (39) of the American Joint Committee on Cancer, the depth of tumor invasion in each of the carcinomas was classified into five groups, as follows: Tis, carcinoma in situ or limited to mucosa; T1, invading the submucosa; T2, invading the muscularis propria; T3, invading either the subserosa; and T4, invading through the serosa or invading contiguous organs. The status of lymph node metastasis was therefore stratified as follows: N0, absence of regional lymph node metastasis; N1, one to three regional lymph node metastasis; N2, four or more regional lymph node metastasis. The presence of distant metastasis was noted as follows: M0, absence of distant metastasis; M1, presence of distant metastasis. The clinicopathological parameters are summarized in Table I. One representative slide with a transversal section of the tumor and with safe mucosa for each sample was selected. The immunohistochemistry was carried out on 4-μm-thick paraffin-embedded sections of formalin-fixed tumor samples using an antibody directed against olfactomedin-4 (catalog number ab78496; Abcam, Cambridge, MA; 1:25). The immunolabeling technique was performed by a Benchmark automated tissue staining system (Ventana).
The immunohistochemistry was evaluated semi-quantitatively by the percentage of cytoplasmic staining cells, the intensity, and the presence or lack of secretory granules. To exclude subjectivity, all of the slides were evaluated by two pathologists who had no knowledge of the patients' identities or clinical status. In discrepant cases, the two pathologists reviewed the slides together and reached a consensus.
The percentage of immunopositive stained cells (A) was divided into five grades as: <10% (score 0); 10–30% (score 1); 30–50% (score 2); 50–70% (score 3); and >70% (score 4). Second, the intensity of staining was scored by evaluating the average staining intensity (B) of the positive cells (0, none; 1, weak; 2, intermediate; and 3, strong). The score for each section was measured as A × B, and the result was defined as negative (−, 0), weakly positive (+, 1–3), positive (++, 4–7), and strongly positive (+++, 8–12). The immunohistochemical data were subjected to statistical analysis. All of the quantitative data were recorded as the means ± S.D. Comparison between multiple groups were performed by one-way analysis of variance and Wilcoxon rank tests (p < 0.05).
Specificity of the OLFM4 Antibody
The specificity of the OLFM4 antibody was determined by Western blot analyses using protein extracts from four different cell lines (supplemental Fig. 1, A and B). RNA interference data were obtained from colorectal cell lines transfected with OLFM4 specific or control siRNA oligonucleotides (supplemental Fig. 1, C and D). The validation of the OLFM4 antibody was also determined by Western blot analysis using cell extract from COS7 cells overexpressing OLFM4 (supplemental Fig. 1E). Finally, in situ hybridization for OLFM4 in the human colon realized in Clevers's group (9) and our immunohistochemistry (IHC) images were compared (supplemental Fig. 1F).
Cell Lines and Transfection
Human cell lines (American Type Culture Collection) were maintained in antibiotic-free RPMI 1640 medium (Lonza). The cultures were supplemented with 10% fetal bovine serum. The cell lines were maintained at 37 °C in 5% carbon dioxide and were tested to rule out mycoplasma contamination. For transfection experiments, the cells were seeded into 60-mm culture dishes and grown until 80% confluence. The empty plasmid pcDN4/T0 and the pcDNA4/OLFM4 plasmids were stably cotransfected with the pcDNA6/TR using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. The cells were selected with 100 μg/ml blasticidin (Sigma-Aldrich) and 500 μg/ml zeocin (Invitrogen) for 2 weeks and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum containing 100 μg/ml zeocin and 2.5 μg/ml blasticidin.
Chromatin Immunoprecipitation Assays
ChIP experiments were performed as previously described (40–43). Briefly, HT29 cells were fixed with 1% formaldehyde. After 10 min, the cells were washed with ice-cold Tris-buffered saline and lysed with 500 μl of ChIP buffer (50 mm Tris-HCl, pH 8.1, 1% SDS, 10 mm EDTA, 1 mm PMSF, 10 μg/ml aprotinine, 10 μg/ml leupeptine, 10 μg/ml pepstatine, 1 mm Na3VO4, 50 mm NaF). Chromatin was sheared by sonication to an average size of 500 bp. The chromatin solution was diluted with 1 volume of dilution buffer (2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 1% Triton X-100, 0.1% Nonidet P-40, 150 mm NaCl, 1 mm PMSF, 10 μg/ml aprotinine, 10 μg/ml leupeptine, 10 μg/ml pepstatine, 1 mm Na3VO4, 50 mm NaF) and incubated 1 h at 4 °C on a rotating platform with protein A-agarose and protein G-Sepharose that was pretreated with sheared DNA salmon sperm. Chromatin was then incubated overnight at 4 °C on a rotating platform with 1 μg of the indicated antibodies or anti-GAL4 antibodies. Following precipitation with protein A-agarose and protein G-Sepharose (pretreated with sheared DNA salmon sperm), chromatin was eluted with elution buffer (1% SDS, 100 mm NaHCO3) for 5 h at 65 °C. DNA was extracted with phenol chloroform, precipitated with ethanol, allowed to air dry, and then dissolved in 100 μl of sterile H2O. Four μl of the DNA samples were then subjected to PCR amplification.
RESULTS
iTRAQ Analysis of Colorectal Tumors
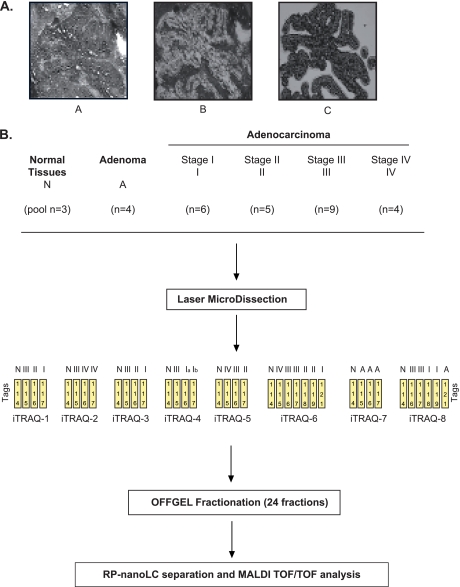
Given the cellular heterogeneity of colorectal cancer, LCM was first applied on tissue to obtain a highly purified population of tumor cells. Representative images of pre- and postmicrodissected tissue images as well as purified cells are presented Fig. 1A. Note that UV laser capture induced cell damage and reduced protein yield during microdissection so that all experiments were performed using infrared laser capture. Approximately 100,000 cells were collected from multiple consecutive tissue sections, and quantitative expression profiles were obtained using iTRAQ labeling coupled with OFFGEL fractionation and off-line nanoLC/MS/MS as we previously described (30). To obtain proteomic maps of the successive steps of colorectal cancer, four adenomas and 24 adenocarcinomas representing the four clinical stages of the disease were subjected to eight different iTRAQ experiments (six 4-plex and two 8-plex; see Fig. 1B). To compare the different results, a pool of three different normal tissues was included in each experiment, labeled with a 114 tag. Finally, among these eight experiments, five were performed using 100 μg of tissue, and three were conducted with less than 70 μg, for each sample.
Fig. 1.
iTRAQ-based quantitative proteomic profiling of colorectal cancer progression. A flow chart of the steps involved in quantitative proteomic profiling of colon-derived tissues is shown. A, this study includes tissue procurement, histopathological examination, and microdissection of the cells using infrared laser capture from frozen tissue sections (10 μm). Panel A, image of premicrodissection; Panel B, post-microdissection; Panel C, isolated cells. B, lysis of captured cells was performed directly on LCM caps. The proteins were extracted, reduced, alkylated, trypsin-digested, and iTRAQ-labeled. Labeled peptides were separated by OFFGEL fractionation. Finally, the proteins were identified and quantified by MALDI-TOF/TOF experiments.
Proteomic Analyses of Colorectal Cancer
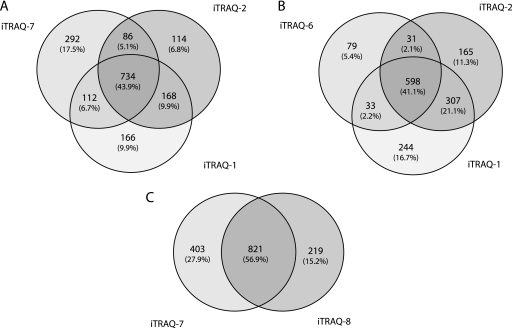
We used the ProteinPilot algorithm (34) to characterize the number of identified proteins (see the identification criteria under “Materials and Methods”). In a first three 4-plex analyses (iTRAQ-1, -2, and -7; supplemental Tables 1–3), 1672 unique proteins were identified (more than 1100 unique proteins by experiment; Table II). Among these, 734 (43.9%) were detected in each of the three iTRAQ experiments, whereas 366 (21.9%) were common to at least two analysis (Fig. 2A). This indicates that approximately two-thirds of the identified proteins can be detected in at least two of the three iTRAQ experiments. To extend these results, we then compared iTRAQ results, using either a 4800 (Fig. 2B) or 5800 (Fig. 2C) MALDI TOF/TOF. Using the 4800 approach, we were able to identify 1457 unique proteins with two 4-plex and one 8-plex experiments. 598 (41.0%) common proteins were detected in all three experiments, and 371 (25.4%) were shared by at least two experiments, indicating again that two-third of the identified proteins can be detected in at least two of the three iTRAQ experiments (Fig. 2B). Using the 5800 MALDI TOF/TOF and one 4-plex and one 8-plex experiments, we were able to identify 1443 unique proteins; among which 821 (56.9%) were common to the two experiments (Fig. 2C).
Fig. 2.
Venn diagrams for the distribution of proteins between the different iTRAQ experiments. A, comparison between 4-plex experiments (two MALDI 4800 and one MALDI 5800 experiments). B, comparison between MALDI 4800 experiments (two 4-plex and one 8-plex experiments). C, comparison between MALDI 5800 experiments (one 4-plex and one 8-plex experiments).
To define a colorectal cancer proteome, we then determined the total number of unique proteins that can be identified from the 28 tumor samples. As a first attempt, according to the criteria, two peptides/protein with score >95% for each iTRAQ experiment, 2141 unique proteins were identified (Table II and supplemental Table 4). We then repeated this identification but combined the eight iTRAQ experiments into a single group. In this case, 3138 unique proteins were identified with at least two peptides (confidence score > 99%) (supplemental Table 5). We then classified this colorectal cancer proteome using GOminer software. 17 GO cellular components terms were defined among the 3138 proteins (supplemental Fig. 2A), and the majority (37%) was attributed to membrane proteins from cytoplasm, nucleus, mitochondrion, endoplasmic reticulum, and Golgi apparatus. This demonstrated the effectiveness of the reported approach for the identification of hydrophobic species. Proteins were also grouped based on their biological functions (supplemental Fig. 2B): 68% were involved in metabolism, 39% were involved in the regulation of biological process, and 23% were transport proteins. In addition, 21 and 19% were involved in development and cell communication, respectively. GO molecular functions annotation terms indicated that 64% were associated with protein binding, 47% had a catalytic activity, 23% were involved in nucleotide, and 27% were involved in nucleic acid binding (supplemental Fig. 2C).
Technical and Experimental Reproducibility of iTRAQ Experiments
To estimate the analytical reproducibility of our results, two experimental replicates (identical sample in two different iTRAQ sets) of a technical duplicate (two identical samples in an iTRAQ set) were run (supplemental Fig. 3A). In total, 1282 proteins were identified (with at least two peptides) across both iTRAQ experiment replicates using Protein Pilot (1222 in replicate A1 and 1175 in replicate A2) (supplemental Table 6). The Venn diagram provided in supplemental Fig. 3B shows that 1115 (87%) of these proteins were common to both sets. Linear regression analyses were performed on ratios obtained from the duplicate analyses. Pearson correlation coefficients between both technical samples were 0.94 and 095, and those between the technical duplicate were 0.91 and 0.92 (ρ < 0.0001) (supplemental Fig. 3C). Thus, the duplicate ratios were significantly positively correlated, indicating a good technical sample preparation and a good analytical reproducibility of the OFFGEL-LC/MS/MS approach.
Proteomic Analysis of the Different Stages of Colorectal Cancer
We then used the iQuantitator software to quantify protein expression between the different stages of colorectal cancer. This approach generates variation means and 95% credible intervals for each expression change. For proteins whose iTRAQ ratios were down-regulated, down-regulation was considered to be significant if the upper limit of the credible interval was below 1. Conversely, for proteins whose iTRAQ ratios were up-regulated, up-regulation was considered to be significant if the lower limit of the credible interval had a value greater than 1. By taking into consideration both the peptide and spectra numbers, this approach allowed us to detect small but significant expression changes, provided that several peptides are detected. Using this analysis, we were able to obtain a list of quantified proteins from the eight iTRAQ experiments (supplemental Table 7). This allowed us to determine the individual protein expression of each patient (supplemental Table 8) and consequently to define the variation of protein expression between the different stages of colorectal cancer (adenoma, stages I–IV). 555 proteins were identified that vary significantly between these different steps of the disease (supplemental Table 9). At the adenoma stage, 164 proteins were expressed differently as compared with normal tissue, and significant variations were also observed for the other stages (stage 1, 183 proteins; stage 2, 304 proteins; stage 3, 194 proteins; and stage 4, 69 proteins; in each case the variation is expressed as compared with normal tissue). Note that in each condition, both up- or down-regulated proteins can be detected, indicating that this approach is suitable to identify proteins that are inactivated during the transformation process and not only oncogenes that are overexpressed (supplemental Table 9). Using the Metacore data mining tool (http://www.genego.com/metacore.php), we then analyzed the signaling pathways represented at the different stages. The cell adhesion-cell matrix pathway was identified as the most significant network associated with adenoma and stage I (p = 4,20348E-10 and 1,81119E-12, respectively), the cytoskeleton-actin filament pathway was significantly associated with stage II (p = 1,5857E-19), the integrin-mediated cell adhesion and migration pathway was correlated with stage III (p = 1,41513E-07), and the cytoskeleton-intermediate filament pathway was significantly associated with stage IV (p = 1,642E-09). Using this analysis, we also found as expected that the adenoma signature was associated with intestinal diseases (p = 3,4659E-21); stage 1 was associated with gastrointestinal neoplasms (p = 2,0057E-16); stage II was associated with digestive system neoplasms (p = 7,7799E-24); stage III was associated with pathologic processes (p = 2,0623E-28); and stage IV was associated with intestinal diseases (p = 7,1607E-19) (supplemental Tables 10–14).
Validation of the Proteomic Analysis Using the Human Atlas Protein
The verification of proteomic results involves IHC analysis on tumor tissue where only a few proteins are generally examined. Rather than performing IHC analysis on a limited number of proteins, we took advantage of IHC data available in the Human Protein Atlas (http://www.proteinatlas.org, 8832 antibodies and 7,334,244 images). As an unbiased approach, we selected all of proteins for which expression differed significantly as compared with normal tissue and assessed HPA IHC data. Because all of these proteins were not represented in the HPA, most of the time as a consequence of missing data, we further selected within this list 83 proteins that met the following criteria: 1) IHC expression reported in more than one normal colorectal tissue; 2) IHC expression reported in more than eight colorectal cancer samples; and 3) the HPA antibody verification score was moderate or high (see http://www.proteinatlas.org). HPA images were then manually inspected to confirm that the protein of interest was overexpressed, unchanged, or underexpressed in tumor cells as compared with normal tissue. Of the 83 selected proteins, 44 (53%) presented consistent expression ratios between iTRAQ and IHC results (supplemental Table 15). Twenty-seven proteins (33%) were determined to be unchanged by IHC, whereas they were down- or up-regulated in tumor samples, and 12 proteins (14%) in IHC did not fit with the expression in our study. Examination of the literature allowed the confirmation of our iTRAQ results for 21 of 39 proteins (unchanged or no fit by IHC); no information was found for the 16 remaining proteins. Only the tenascin result does not seem to match the information in the literature and the IHC results. Note that the different colon cancer stages are not specified in the HPA database. This prevents the detection of variation that would be stage-specific and might explain these discrepancies.
OLFM4 Is Expressed at the Early Stages of Colorectal Cancer
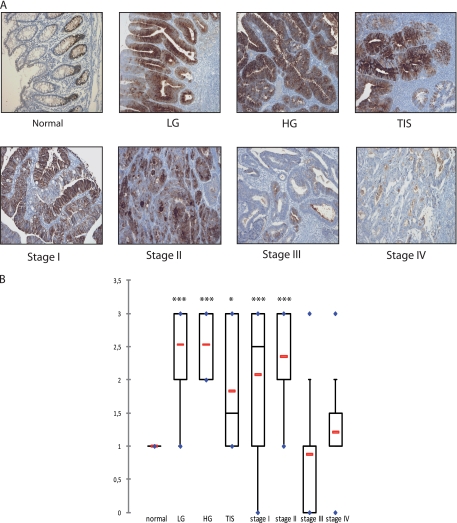
We then asked whether we could identify some proteins that are specific to the early stages of colorectal cancer. To this end, proteins were classified with the following criteria: 1) significant up-regulation in adenoma and stages I/II according to iQuantitator analysis and 2) not significantly expressed or underexpressed in stages III/IV. With this approach, only four proteins were identified (supplemental Table 9), the aldehyde dehydrogenase (ALDH1A1), the heat shock protein 1 (HSPE1), the sorbitol dehydrogenase (SORD), and OLFM4. Interestingly, OLFM4 encodes a protein that has been recently described as a specific marker of colorectal stem cells in association with Lgr5 (32, 33). In our experimental conditions, OLFM4 was detected with the highest statistical confidence, and given its importance in colorectal cancer, we focused on this protein for the following part of the study. To confirm its dysregulation in adenoma and in the early stage of CCR, its expression was first analyzed by immunohistochemistry using paraffin-embedded tissues isolated from 126 patients. Representative pictures of OLFM4 staining in adenomas, early or metastatic CRC cases are shown Fig. 3A. Whereas normal intestinal crypts showed moderate nuclear staining, results showed that the cytoplasmic and nuclear staining increased significantly in dysplasia tissue and in noninvasive tumors. OLFM4 was found to be significantly up-regulated in low grade adenoma, high grade adenoma, in situ adenoma, and stages I and II as compared with normal crypts (one-way analysis of variance test, p < 0.05). By contrast, OLFM4 expression was not significantly different between invasive tissues (stage III/IV) and normal tissues (Fig. 3B).
Fig. 3.
The validation of OLFM4 in human CRC tissue. A, the immunohistochemistry staining of the different colorectal tumor stages (100×). LG, low grade adenoma; HG, high grade adenoma; TIS, carcinoma in situ. Stages I and II, early stage carcinomas; stages III and IV, metastatic carcinomas. B, different distribution of OLFM4 IHC scores (staining intensity × staining percentage) between normal crypts, adenomas, carcinomas in situ, and different colorectal tumor stages (p < 0.05, significant for normal versus low grade adenoma, high grade adenoma, carcinoma in situ, stage I, and stage II, not significant for normal versus stages III and IV). The red lines represent the mean, and the blue points represent the minimum and maximum values. Dunnett's multiple comparison tests were performed; q = 3.20 (*) for normal versus carcinoma in situ; q = 6.35 (***) for normal versus low and high grade adenoma; q = 5.21 and 5.49 for normal versus stages I and II, respectively.
OLFM4 Expression Is Activated by the Ras-NF-κB2 Pathway
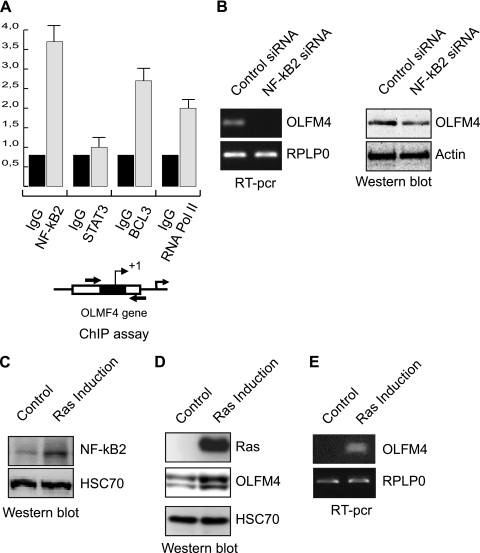
These observations suggested to us that OLFM4 is expressed at the early stages of colorectal cancer, probably in response to oncogenes that are involved in the initial step of cell transformation. Although it is well known that the adenomatous polyposis coli/β-catenin pathway plays an important role in the initial transformation of intestinal crypts, we and others have also shown that the STAT3 and NF-κB transcription factor plays an essential role in this disease (43–45). Because STAT3 and NF-κB2 subunits are known to interact in tumor cells, we investigated the role of these two transcription factors in the regulation of OLFM4 expression. Transcription factor recognition site analysis of the OLFM4 promoter revealed the presence of several potential binding sites for these proteins. To determine whether STAT3 and NF-κB2 can be found associated with the OLFM4 gene, ChIP experiments were performed in growing HT29 cells using pair of primers corresponding to the proximal promoter. Although we were not able to detect any association of STAT3 with this region, ChIP results showed that NF-κB2 and its cofactor BCL3 are associated with the OLFM4 proximal promoter and that this was correlated with the binding of the RNA polymerase II (Fig. 4A). Note that this effect was noticed on endogenous proteins and not following overexpression. In addition, using RNA interference, we also noticed that the down-regulation of NF-κB2 inhibits OLFM4 expression at the protein and mRNA levels (Fig. 4B).
Fig. 4.
NF-κB2 regulates OLFM4 expression following Ras induction. A, soluble chromatin from growing HCT116 cells was prepared and immunoprecipitated with antibodies targeted against NF-κB2, Bcl3, and the RNA polymerase II. DNA was amplified using pair of primers that cover the NF-κB proximal binding site of the OLFM4 promoter. ChIP assays were then quantified by real time RT-PCR as compared with the signal obtained on a control sequence with a control IgG. The schematic representation of the potential NF-κB binding sites of the OLFM4 promoter is presented. The reading frame of the binding site is indicated by + for 5′-3′ and − for 3′-5′. B, HCT116 colorectal cell lines were either transfected with NF-κB2 specific or control siRNA oligonucleotides as indicated. The mRNA extracts (left panels) and whole cell extracts (right panels) were processed, and OLFM4 expression was analyzed 48 h after siRNA transfection. C, HT29 colorectal cell lines expressing an incible vector of the RasV12 oncogene were treated or not with doxycyclin for 48 h, and whole cell extracts were prepared and analyzed using antibodies directed against NF-κB2 and HSC70 as a loading control. D and E, cells were treated as above, and the expression of OLFM4 or Ras was evaluated by Western blot analysis (D) or RT-PCR (E).
It has been shown recently that the Ras oncogene plays an important role in the initial stages of colorectal cancer and that this signaling pathway can deregulate the NF-κB transcription factor to allow abnormal cell cycle progression and survival (44, 46, 47). To determine whether Ras regulates OLFM4, we used stable HT29 cells expressing the RasV12 oncogene under the control of a doxycycline-inducible promoter (48). As expected, Ras was up-regulated in response to doxycycline, and a significant activation of the NF-κB2 transcription factor was detected (Fig. 4C). Interestingly, the induction of Ras was correlated with an up-regulation of OLFM4 at the protein level (Fig. 4D). In addition, semi-quantitative PCR experiments showed that this effect was regulated at the transcriptional level (Fig. 4D).
OLFM4 Is Overexpressed in Ras-mutated Tumors
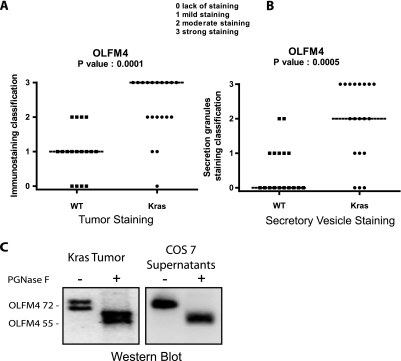
These observations suggested to us that the expression of OLFM4 might be enhanced in tumor samples expressing the Ras-NF-κB2 pathway. Because NF-κB2 is generated as a cleavage product of its p100 precursor, the detection of the active form of this transcription factor is difficult in tumor samples. However, the presence of the Rasv12 oncogene can be determined by DNA sequencing. For this reason, we then analyzed OLFM4 expression in tumors expressing or not a mutated form of this oncogene. Interestingly, results presented Fig. 5A indicate that OLFM4 expression was significantly enhanced in Ras-mutated tumors (p < 0.0001) as compared with wild type tumor samples.
Fig. 5.
OLFM4 is glycosylased and secreted in KRAS tumors. A and B, OLFM4 IHC scores (staining intensity × staining percentage) between WT and KRAS tissue in tumor tissue (A) and in secretory vesicles (B). C, deglycosylation analysis of OLFM4. OLFM4 was significantly secreted from the COS-7 cells; the secreted form at 72 kDa was treated with (+) or (−) peptide:N-glycosidase F (PGNase F). The size of deglycosylated OLFM4 is consistent with the molecular mass of calculated OLFM4 gene product. Treatment of the 72-kDa band in KRAS tumor leads to the same band at 55 kDa.
OLFM4 Is Present in the Cytoplasm and as a Secreted Protein
During the course of the IHC experiments, we noticed that OLFM4 was expressed in the cytoplasm, and there was significant expression in the secretory vesicles (Fig. 5B). This result suggested to us that this protein might be secreted, and OLFM4 was effectively detected in the cell supernatants (data not shown and see Fig. 5C). In addition, we also noticed that two bands can be detected by Western blot, one at the expected molecular mass of 55 kDa and another band ∼72 kDa. This observation suggested to us that OLFM4 was modified by glycosylation. To verify this hypothesis, extracts obtained from tumor samples were incubated with the peptide:N-glycosidase F deglycosylating enzyme. Following incubation, a shift in the molecular mass from 72 to 55 kDa was observed, indicating that the protein is effectively modified by N-glycosylation (Fig. 5C). To confirm this observation, COS cells were transfected with a vector allowing OLFM4 expression, and its potential secretion was analyzed by Western blot. Interestingly, results showed that only the 72-kDa band was detected in the supernatant, suggesting that OLFM4 is secreted as a glycosylated protein. Following peptide:N-glycosidase F treatment, the same migration shift was observed, and OLFM4 migrated as a 55-kDa protein (Fig. 5C, right panel). Importantly, in Ras-mutated tumors, OLFM4 was essentially detected at 72 kDa, suggesting that this protein is effectively secreted in vivo.
DISCUSSION
Following the initial description of the genetic modifications occurring during colorectal cancer transformation, several studies have clearly shown that CRC results from multiple mutations that induce the deregulation of cell cycle and cell death pathways. This led to the important conclusion that CRC is a heterogeneous disease, which certainly explains why patients suffering from the same apparent disease have distinct outcome and different responses to the same anti-cancer treatment. Molecular clustering has therefore become an essential goal of cancer treatment, not only to establish tumor prognosis but also to identify the specific addictive oncogenic pathways that should be targeted (49–51). Recent results have shown the interest of using genomic signatures to identify these deregulated pathways and characterize prognosis markers. These gene signatures can also be used to characterize predictive markers that reflect the response to a particular treatment, but in this case, the predicting value of this approach remains to be fully validated (13, 52). In addition to these genomic experiments, quantitative proteomics also appears as a powerful tool to define cancer signatures that would identify disease subtypes, predict tumor escape, or characterize new molecular targets.
In this study, we provide what is to our knowledge the most extensive proteome database established so far for colorectal cancer and illustrate the value of using the combination of OFFGEL-iTRAQ labeling and MALDI-TOF/TOF approaches to explore the deep proteome of frozen tissues. It should be noted that the use of LCM favored the identification of many low concentrated proteins by removing abundant stromal proteins. This approach enables the identification and quantification of ∼1100 proteins by patient, allowing the identification of a proteomic map for each tumor, which could be used in the future for individual clinical monitoring.
From a technical point of view, this study allowed us to compare the two MALDI-TOF/TOF 4800 and 5800 from AB sciex. With the iTRAQ technology, the 5800 MALDI seems to be slightly more sensitive than the 4800 MALDI (increase in identification of ∼6% of proteins). However, in quite a surprising way, with the iTRAQ 8-plex, we can identify 35% more proteins with the MALDI 5800 than with the MALDI 4800. Although it is it well known that the number of identified proteins and peptides is larger when using iTRAQ 4-plex than with iTRAQ 8-plex (53), it seems that the use of the MALDI 5800 lessens this difference. The difference in proteins identified changes from 35% in favor of the 4-plex when using the MALDI 4800 as compared with 18% with the 5800, thereby reviving interest in this reactive agent that can compare eight samples instead of four.
Starting from a cohort of 28 frozen tumors, we compared the protein profiles of adenomas or adenocarcinomas at the early stages (I and II) or metastatic stages (III and IV). The results indicate that the expression of a total of 555 proteins was significantly over- or underexpressed in colorectal cancer as compared with normal tissue, representing 16% of the total identified proteome. This approach allowed us to characterize the proteins that expression varied significantly between the different stages (adenoma, stages I–IV) and to establish what is to our knowledge the first proteomic analysis of these different steps. As compared with normal tissue, it is interesting to note that the most important variation was observed between stages II and III, which corresponds to the transition between a nonmetastatic and metastatic tumor. In this case, 304 proteins were found to vary significantly, whereas 180 were differently regulated in adenomas and stages I and III, and only 68 were modified in stage IV. Although this remains to be demonstrated, it is tempting to speculate that some proteins expressed in the stage II are necessary for invasive migration. In line with this hypothesis, we observed using gene ontology analysis that the expression of extracellular matrix proteins varies significantly. Among these proteins, we have focused on secreted proteins present at the early stage of the disease because they can be easily detected by ELISA, and they can be useful to distinguish these aggressive cancers from early stage cases. It is important to consider that patients with stage I/II cancer are believed to be cured after surgery but that ∼20% of them will relapse. The distinction between these two cases is actually difficult, and for this reason, the identification of specific biomarkers of stage II colorectal tumors is an important goal that would allow the prediction of recurrence events. A 50-gene signature has been recently described that can separate early tumors depending on their relapse probability (15). Therefore, it will be interesting to determine whether the protein list identified in this study can be used to improve this early stage stratification to predict tumor relapse.
Among these proteins, we focused on OLFM4 because its expression was significantly up-regulated in adenomas, further increased in stage I, and maximum in stage II before dropping considerably in stages III and IV. Importantly, OLFM4 has recently been shown to be expressed in colorectal stem cells in association with Lgr5 (32, 33), further confirming its expression at the early stages of the disease. This protein plays probably an important role in cancer because it has been recently shown in myeloid cells that its promoter is probably methylated. Its re-expression induced cell cycle arrest and cell death in myeloid cells (54). Interestingly, the same effect has also been observed in prostate cancer cells where OLFM4 levels are down-regulated during cancer progression, most significantly in tumors with high Gleason scores (55). Importantly, restoring the OLFM4 level through overexpression led to reduced proliferation and invasiveness. Because this protein is known to interact with lectins as well as cadherin (56), this effect might be explained by a better adhesion to the extracellular matrix or to the surrounding cells. However, this study also proposed that this effect is related to an inhibition of cathepsin D expression and an enhanced autophagic activity of prostate cancer cells. Because autophagy plays an important role in tumor suppression (57), it will be interesting to determine whether this effect of OLFM4 can be extended to other experimental models. Moreover, it should be noted that OLFM4 is a member of the olfactomedin domain-containing protein. This family includes Noelin (OLFM1), which prolongs neural crest production. The involvement of nervous system proteins, such as neurotrophins, in carcinogenesis has been reported for prostate (58) and breast (59) cancers, and it will be interesting to test whether OLFM4 behaves in the same way.
In our experimental conditions, it is striking to note that OLFM4 level was very significantly down-regulated in stages III and IV compared with stages I/II tumors and that a reduced expression of this protein has been recently correlated with poor prognosis. In light of these observations, it is tempting to speculate that the inactivation of OLFM4 is a necessary event to prevent cell death and allow tumor progression and metastasis in colorectal cancer. In line with this hypothesis, it has been recently reported that OLFM4 expression is reduced in poorly differentiated colon cancers, as well as at the late tumor-node-metastasis stage (60). If feasible, the detection of OLFM4 variations in the serum of the patients might therefore be an interesting tool to follow the evolution of stage II tumors. Interestingly, we also noticed that OLFM4 was regulated by the Ras-NF-κB2 pathway and that the expression of this protein was significantly enhanced in Ras-mutated tumors. We have recently shown that senescence is induced in response to the Ras oncogene in colorectal cell lines (48). Oncogene-induced senescence (OIS) is a powerful antitumor mechanism that induces permanent cell cycle arrest in response to abnormal proliferative signals (61). Originally described in cell culture, OIS has been recently shown to occur also in vivo as an early protection against carcinogenesis. In light of these observations, one can speculate that OLFM4 overexpression is an early event occurring in response to OIS in Ras-expressing cells. Further experiments are therefore necessary to characterize the effect of OLFM4 on cell cycle and cell death pathways in colorectal cell lines and to determine whether this effect is deregulated by the Ras oncogene to allow OIS escape and tumor progression.
Through the characterization of the molecular aberrations present in cancer cells, it is now widely accepted that the identification of new biomarkers will improve the outcome prognosis or the prediction of therapy response. In addition, it is also expected that molecular clustering will help to separate apparently similar tumors to provide rationale treatments. Besides genomic approaches, our results indicate that proteomic analysis can be used on tumor samples to provide not only a better understanding of cell transformation in colorectal cancer but also to identify new biomarkers of the different tumor stages such as OLFM4.
Acknowledgments
We thank John Schwacke for help with iQuantitator software.
Footnotes
* This work was supported by grants from the Ligue Contre le Cancer (Comité du Maine et Loire), the Institut du Cancer, the Region Pays de Loire, and Amgen France. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental Tables 1–15 and Figs. 1–3.
This article contains supplemental Tables 1–15 and Figs. 1–3.
1 The abbreviations used are:
- CRC
- colorectal cancer
- IHC
- immunohistochemistry
- HPA
- Human Protein Atlas
- LCM
- laser capture microdissection
- FDR
- false discovery rate
- GO
- gene ontology
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- ChIP
- chromatin immunoprecipitation
- STAT
- signal transducers and activators of transcription
- OIS
- oncogene-induced senescence.
REFERENCES
- 1. Cunningham D., Atkin W., Lenz H. J., Lynch H. T., Minsky B., Nordlinger B., Starling N. (2010) Colorectal cancer. Lancet 375, 1030–1047 [DOI] [PubMed] [Google Scholar]
- 2. Jemal A., Siegel R., Xu J., Ward E. (2010) Cancer statistics, 2010. CA Cancer J. Clin. 60, 277–300 [DOI] [PubMed] [Google Scholar]
- 3. Huang C. S., Lal S. K., Farraye F. A. (2005) Colorectal cancer screening in average risk individuals. Cancer Causes Control 16, 171–188 [DOI] [PubMed] [Google Scholar]
- 4. van Rossum L. G., van Rijn A. F., Laheij R. J., van Oijen M. G., Fockens P., van Krieken H. H., Verbeek A. L., Jansen J. B., Dekker E. (2008) Random comparison of guaiac and immunochemical fecal occult blood tests for colorectal cancer in a screening population. Gastroenterology 135, 82–90 [DOI] [PubMed] [Google Scholar]
- 5. Vogelstein B., Fearon E. R., Hamilton S. R., Kern S. E., Preisinger A. C., Leppert M., Nakamura Y., White R., Smits A. M., Bos J. L. (1988) Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319, 525–532 [DOI] [PubMed] [Google Scholar]
- 6. Pino M. S., Chung D. C. (2010) The chromosomal instability pathway in colon cancer. Gastroenterology 138, 2059–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clevers H. (2006) Wnt/beta-catenin signaling in development and disease. Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 8. Reya T., Clevers H. (2005) Wnt signalling in stem cells and cancer. Nature 434, 843–850 [DOI] [PubMed] [Google Scholar]
- 9. Barker N., Ridgway R. A., van Es J. H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A. R., Sansom O. J., Clevers H. (2009) Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 [DOI] [PubMed] [Google Scholar]
- 10. van der Flier L. G., Clevers H. (2009) Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 71, 241–260 [DOI] [PubMed] [Google Scholar]
- 11. Phelps R. A., Chidester S., Dehghanizadeh S., Phelps J., Sandoval I. T., Rai K., Broadbent T., Sarkar S., Burt R. W., Jones D. A. (2009) A two-step model for colon adenoma initiation and progression caused by APC loss. Cell 137, 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lièvre A., Bachet J. B., Boige V., Cayre A., Le Corre D., Buc E., Ychou M., Bouché O., Landi B., Louvet C., André T., Bibeau F., Diebold M. D., Rougier P., Ducreux M., Tomasic G., Emile J. F., Penault-Llorca F., Laurent-Puig P. (2008) KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J. Clin. Oncol. 26, 374–379 [DOI] [PubMed] [Google Scholar]
- 13. Bild A. H., Yao G., Chang J. T., Wang Q., Potti A., Chasse D., Joshi M. B., Harpole D., Lancaster J. M., Berchuck A., Olson J. A., Jr., Marks J. R., Dressman H. K., West M., Nevins J. R. (2006) Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439, 353–357 [DOI] [PubMed] [Google Scholar]
- 14. Chang J. T., Carvalho C., Mori S., Bild A. H., Gatza M. L., Wang Q., Lucas J. E., Potti A., Febbo P. G., West M., Nevins J. R. (2009) A genomic strategy to elucidate modules of oncogenic pathway signaling networks. Mol. Cell 34, 104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garman K. S., Acharya C. R., Edelman E., Grade M., Gaedcke J., Sud S., Barry W., Diehl A. M., Provenzale D., Ginsburg G. S., Ghadimi B. M., Ried T., Nevins J. R., Mukherjee S., Hsu D., Potti A. (2008) A genomic approach to colon cancer risk stratification yields biologic insights into therapeutic opportunities. Proc. Natl. Acad. Sci. U.S.A. 105, 19432–19437 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Crawford N. P., Colliver D. W., Galandiuk S. (2003) Tumor markers and colorectal cancer: Utility in management. J. Surg. Oncol. 84, 239–248 [DOI] [PubMed] [Google Scholar]
- 17. Sidransky D. (2002) Emerging molecular markers of cancer. Nat. Rev. Cancer 2, 210–219 [DOI] [PubMed] [Google Scholar]
- 18. Duffy M. J., van Dalen A., Haglund C., Hansson L., Holinski-Feder E., Klapdor R., Lamerz R., Peltomaki P., Sturgeon C., Topolcan O. (2007) Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur. J. Cancer 43, 1348–1360 [DOI] [PubMed] [Google Scholar]
- 19. Locker G. Y., Hamilton S., Harris J., Jessup J. M., Kemeny N., Macdonald J. S., Somerfield M. R., Hayes D. F., Bast R. C., Jr. (2006) ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J. Clin. Oncol. 24, 5313–5327 [DOI] [PubMed] [Google Scholar]
- 20. Jimenez C. R., Knol J. C., Meijer G. A., Fijneman R. J. (2010) Proteomics of colorectal cancer: Overview of discovery studies and identification of commonly identified cancer-associated proteins and candidate CRC serum markers. J. Proteomics 73, 1873–1895 [DOI] [PubMed] [Google Scholar]
- 21. Alfonso P., Cañamero M., Fernández-Carbonié F., Núñez A., Casal J. I. (2008) Proteome analysis of membrane fractions in colorectal carcinomas by using 2D-DIGE saturation labeling. J. Proteome Res. 7, 4247–4255 [DOI] [PubMed] [Google Scholar]
- 22. Bi X., Lin Q., Foo T. W., Joshi S., You T., Shen H. M., Ong C. N., Cheah P. Y., Eu K. W., Hew C. L. (2006) Proteomic analysis of colorectal cancer reveals alterations in metabolic pathways: Mechanism of tumorigenesis. Mol. Cell. Proteomics 5, 1119–1130 [DOI] [PubMed] [Google Scholar]
- 23. Rho J. H., Qin S., Wang J. Y., Roehrl M. H. (2008) Proteomic expression analysis of surgical human colorectal cancer tissues: Up-regulation of PSB7, PRDX1, and SRP9 and hypoxic adaptation in cancer. J. Proteome Res. 7, 2959–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ducoux-Petit M., Uttenweiler-Joseph S., Brichory F., Bousquet-Dubouch M. P., Burlet-Schiltz O., Haeuw J. F., Monsarrat B. (2008) Scaled-down purification protocol to access proteomic analysis of 20S proteasome from human tissue samples: Comparison of normal and tumor colorectal cells. J. Proteome Res. 7, 2852–2859 [DOI] [PubMed] [Google Scholar]
- 25. Roessler M., Rollinger W., Palme S., Hagmann M. L., Berndt P., Engel A. M., Schneidinger B., Pfeffer M., Andres H., Karl J., Bodenmüller H., Rüschoff J., Henkel T., Rohr G., Rossol S., Rösch W., Langen H., Zolg W., Tacke M. (2005) Identification of nicotinamide N-methyltransferase as a novel serum tumor marker for colorectal cancer. Clin. Cancer Res. 11, 6550–6557 [DOI] [PubMed] [Google Scholar]
- 26. Roessler M., Rollinger W., Mantovani-Endl L., Hagmann M. L., Palme S., Berndt P., Engel A. M., Pfeffer M., Karl J., Bodenmüller H., Rüschoff J., Henkel T., Rohr G., Rossol S., Rösch W., Langen H., Zolg W., Tacke M. (2006) Identification of PSME3 as a novel serum tumor marker for colorectal cancer by combining two-dimensional polyacrylamide gel electrophoresis with a strictly mass spectrometry-based approach for data analysis. Mol. Cell. Proteomics 5, 2092–2101 [DOI] [PubMed] [Google Scholar]
- 27. Kim H. J., Kang H. J., Lee H., Lee S. T., Yu M. H., Kim H., Lee C. (2009) Identification of S100A8 and S100A9 as serological markers for colorectal cancer. J. Proteome Res. 8, 1368–1379 [DOI] [PubMed] [Google Scholar]
- 28. Ma Y., Peng J., Liu W., Zhang P., Huang L., Gao B., Shen T., Zhou Y., Chen H., Chu Z., Zhang M., Qin H. (2009) Proteomics identification of desmin as a potential oncofetal diagnostic and prognostic biomarker in colorectal cancer. Mol. Cell. Proteomics 8, 1878–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu B. J., Li J., Beauchamp R. D., Shyr Y., Li M., Washington M. K., Yeatman T. J., Whitehead R. H., Coffey R. J., Caprioli R. M. (2009) Identification of early intestinal neoplasia protein biomarkers using laser capture microdissection and MALDI MS. Mol. Cell. Proteomics 8, 936–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ernoult E., Gamelin E., Guette C. (2008) Improved proteome coverage by using iTRAQ labelling and peptide OFFGEL fractionation. Proteome Sci. 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ernoult E., Bourreau A., Gamelin E., Guette C. (2010) A proteomic approach for plasma biomarker discovery with iTRAQ labelling and OFFGEL fractionation. J. Biomed. Biotechnol. 2010, 927917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barker N., van Es J. H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P. J., Clevers H. (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 [DOI] [PubMed] [Google Scholar]
- 33. van der Flier L. G., Haegebarth A., Stange D. E., van de Wetering M., Clevers H. (2009) OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 137, 15–17 [DOI] [PubMed] [Google Scholar]
- 34. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 35. Schwacke J. H., Hill E. G., Krug E. L., Comte-Walters S., Schey K. L. (2009) iQuantitator: A tool for protein expression inference using iTRAQ. BMC Bioinformatics 10, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grant J. E., Bradshaw A. D., Schwacke J. H., Baicu C. F., Zile M. R., Schey K. L. (2009) Quantification of protein expression changes in the aging left ventricle of Rattus norvegicus. J. Proteome Res. 8, 4252–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zeeberg B. R., Qin H., Narasimhan S., Sunshine M., Cao H., Kane D. W., Reimers M., Stephens R. M., Bryant D., Burt S. K., Elnekave E., Hari D. M., Wynn T. A., Cunningham-Rundles C., Stewart D. M., Nelson D., Weinstein J. N. (2005) High-Throughput GoMiner, an “industrial-strength ” integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID). BMC Bioinformatics 6, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ekins S., Nikolsky Y., Bugrim A., Kirillov E., Nikolskaya T. (2007) Pathway mapping tools for analysis of high content data. Methods Mol. Biol. 356, 319–350 [DOI] [PubMed] [Google Scholar]
- 39. Sobin L. H., Gospodarowicz M. K., C. W. (eds.) (2009) TNM Classification of Malignant Tumours, 7th Ed., Wiley-Blackwell, Chichester, UK [Google Scholar]
- 40. Bienvenu F., Barré B., Giraud S., Avril S., Coqueret O. (2005) Transcriptional regulation by a DNA-associated form of cyclin D1. Mol. Biol. Cell 16, 1850–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Courapied S., Sellier H., de Carn é Trécesson S., Vigneron A., Bernard A. C., Gamelin E., Barré B., Coqueret O. (2010) The cdk5 kinase regulates the STAT3 transcription factor to prevent DNA damage upon topoisomerase I inhibition. J. Biol. Chem. 285, 26765–26778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Courapied S., Cherier J., Vigneron A., Troadec M. B., Giraud S., Valo I., Prigent C., Gamelin E., Coqueret O., Barré B. (2010) Regulation of the Aurora-A gene following topoisomerase I inhibition: Implication of the Myc transcription factor. Mol. Cancer 9, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vigneron A., Gamelin E., Coqueret O. (2008) The EGFR-STAT3 oncogenic pathway up-regulates the Eme1 endonuclease to reduce DNA damage after topoisomerase I inhibition. Cancer Res. 68, 815–825 [DOI] [PubMed] [Google Scholar]
- 44. Greten F. R., Eckmann L., Greten T. F., Park J. M., Li Z. W., Egan L. J., Kagnoff M. F., Karin M. (2004) IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 118, 285–296 [DOI] [PubMed] [Google Scholar]
- 45. Bollrath J., Phesse T. J., von Burstin V. A., Putoczki T., Bennecke M., Bateman T., Nebelsiek T., Lundgren-May T., Canli O., Schwitalla S., Matthews V., Schmid R. M., Kirchner T., Arkan M. C., Ernst M., Greten F. R. (2009) gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell 15, 91–102 [DOI] [PubMed] [Google Scholar]
- 46. Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R. S., Karantza-Wadsworth V., White E. (2009) Autophagy suppresses tumorigenesis through elimination of p62. Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barré B., Perkins N. D. (2010) The Skp2 promoter integrates signaling through the NF-kappaB, p53, and Akt/GSK3beta pathways to regulate autophagy and apoptosis. Mol. Cell 38, 524–538 [DOI] [PubMed] [Google Scholar]
- 48. de Carn é Trécesson S., Guillemin Y., Bélanger A., Bernard A. C., Preisser L., Ravon E., Gamelin E., Juin P., Barré B., Coqueret O. (2011) Escape from p21-mediated oncogene-induced senescence leads to cell dedifferenciation and dependency on anti-apoptotic Bcl-xL and Mcl1 proteins. J. Biol. Chem. 286, 12825–12838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bernards R. (2010) It's diagnostics, stupid. Cell 141, 13–17 [DOI] [PubMed] [Google Scholar]
- 50. Chin L., Gray J. W. (2008) Translating insights from the cancer genome into clinical practice. Nature 452, 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van't Veer L. J., Bernards R. (2008) Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature 452, 564–570 [DOI] [PubMed] [Google Scholar]
- 52. Sawyers C. L. (2008) The cancer biomarker problem. Nature 452, 548–552 [DOI] [PubMed] [Google Scholar]
- 53. Pichler P., Köcher T., Holzmann J., Mazanek M., Taus T., Ammerer G., Mechtler K. (2010) Peptide labeling with isobaric tags yields higher identification rates using iTRAQ 4-plex compared to TMT 6-plex and iTRAQ 8-plex on LTQ Orbitrap. Anal. Chem 82, 6549–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu W., Lee H. W., Liu Y., Wang R., Rodgers G. P. (2010) Olfactomedin 4 is a novel target gene of retinoic acids and 5-aza-2′-deoxycytidine involved in human myeloid leukemia cell growth, differentiation, and apoptosis. Blood 116, 4938–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen L., Li H., Liu W., Zhu J., Zhao X., Wright E., Cao L., Ding I., Rodgers G. P. (2011) Olfactomedin 4 suppresses prostate cancer cell growth and metastasis via negative interaction with cathepsin D and SDF-1. Carcinogenesis 32, 986–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grover P. K., Hardingham J. E., Cummins A. G. (2010) Stem cell marker olfactomedin 4: Critical appraisal of its characteristics and role in tumorigenesis. Cancer Metastasis Rev. 29, 761–775 [DOI] [PubMed] [Google Scholar]
- 57. White E., Lowe S. W. (2009) Eating to exit: Autophagy-enabled senescence revealed. Genes Dev. 23, 784–787 [DOI] [PubMed] [Google Scholar]
- 58. Thiele C. J., Li Z., McKee A. E. (2009) On Trk: The TrkB signal transduction pathway is an increasingly important target in cancer biology. Clin. Cancer Res. 15, 5962–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vanhecke E., Adriaenssens E., Verbeke S., Meignan S., Germain E., Berteaux N., Nurcombe V., Le Bourhis X., Hondermarck H. (2011) Brain-derived neurotrophic factor and neurotrophin-4/5 are expressed in breast cancer and can be targeted to inhibit tumor cell survival. Clin. Cancer Res. 17, 1741–1752 [DOI] [PubMed] [Google Scholar]
- 60. Liu W., Liu Y., Zhu J., Wright E., Ding I., Rodgers G. P. (2008) Reduced hGC-1 protein expression is associated with malignant progression of colon carcinoma. Clin. Cancer Res. 14, 1041–1049 [DOI] [PubMed] [Google Scholar]
- 61. Roninson I. B. (2003) Tumor cell senescence in cancer treatment. Cancer Res. 63, 2705–2715 [PubMed] [Google Scholar]