Abstract
Protein phosphorylation modulates a myriad of biological functions, and its regulation is vital for proper cellular activity. Mass spectrometry is the enabling tool for phosphopeptide analysis, where recent instrumentation advances in both speed and sensitivity in linear ion trap and orbitrap technologies may yield more comprehensive phosphoproteomic analyses in less time. Protein phosphorylation analysis by MS relies on structural information derived through controlled peptide fragmentation. Compared with traditional, ion-trap-based collision-induced dissociation (CID), a more recent type of fragmentation termed HCD (higher energy collisional dissociation) provides beam type CID tandem MS with detection of fragment ions at high resolution in the orbitrap mass analyzer. Here we compared HCD to traditional CID for large-scale phosphorylation analyses of murine brain under three separate experimental conditions. These included a same-precursor analysis where CID and HCD scans were performed back-to-back, separate analyses of a phosphotyrosine peptide immunoprecipitation experiment, and separate whole phosphoproteome analyses. HCD generally provided higher search engine scores with more peptides identified, thus out-performing CID for back-to-back experiments for most metrics tested. However, for phosphotyrosine IPs and in a full phosphoproteome study of mouse brain, the greater acquisition speed of CID-only analyses provided larger data sets. We reconciled our results with those in direct contradiction from Nagaraj N, D'Souza RCJ et al. (J. Proteome Res. 9:6786, 2010). We conclude, for large-scale phosphoproteomics, CID fragmentation with rapid detection in the ion trap still produced substantially richer data sets, but the back-to-back experiments demonstrated the promise of HCD and orbitrap detection for the future.
The brain harbors specialized functions such as neural transmission and memory that are contingent upon synchronized phosphorylation events (1, 2), and the identification and characterization of protein phosphorylation events is most commonly accomplished using mass spectrometry (3, 4). Studies cataloging brain phosphorylation events further expand our understanding of specialized signaling events, and previous mouse brain phosphorylation investigations, with extensive fractionation, have sometimes yielded data sets containing thousands of sites (5–8). The large-scale identification of phosphorylation sites is not only biologically important, but presents an analytical challenge that has traditionally stretched the capabilities of mass spectrometry-based proteomics technology. Attaining maximum depth of phosphoproteomic analysis requires continual refinement and optimization of analytical methods as new technologies are introduced.
Hybrid mass spectrometers are becoming increasingly common, including quadrupole-time-of-flight instruments, quadrupoles coupled with ion traps, and ion traps coupled with ion cyclotron resonance cells (9–14). The hybrid linear ion trap-orbitrap combination also enjoys widespread use. For most bottom-up proteomic experiments, intact peptides are initially detected in the orbitrap with high resolution and high mass accuracy. However, MS/MS spectra are typically isolated and detected in the linear ion trap by collision-induced dissociation (CID)1 because of its speed and sensitivity. For phosphopeptide analysis, this combination is especially ubiquitous (6, 15–19). Recently, new fragmentation techniques have emerged that may complement or replace traditional CID and include electron capture dissociation (20), electron transfer dissociation (14), and higher energy collisional dissociation (HCD) (10, 21, 22).
HCD fragmentation is available for the LTQ Orbitrap (21) where ions are fragmented in a collision cell rather than an ion trap and then transferred back through the C-trap for analysis at high resolution in the orbitrap. Compared with traditional ion trap-based collision-induced dissociation, HCD fragmentation with orbitrap detection has no low-mass cutoff, high resolution ion detection, and increased ion fragments resulting in higher quality MS/MS spectra. HCD also employs higher energy dissociations than those used in ion trap CID, enabling a wider range of fragmentation pathways. One drawback, however, is that spectral acquisition times are up to twofold longer because more ions are required for Fourier transform detection in the orbitrap compared with detection of CID spectra in the ion trap via electron multipliers. The LTQ Orbitrap Velos platform offers improvements to the source region, leading to increases in ion current by ∼10-fold (21). When coupled with a more efficient HCD collision cell this has greatly improved the performance of HCD fragmentation, making rapid and routine analysis possible (23).
Recent comparative studies of complex proteomic mixtures aimed at determining the value of HCD with orbitrap detection compared with CID with ion trap detection have found mixed conclusions (23–25). Given that it is still unclear which approach is best suited for proteomics, we set out to design several large-scale comparisons using phosphoproteomics, which is an ideal model system both for its biological importance and because it is analytically challenging. Moreover, phosphopeptides typically provide relatively less informative MS/MS spectra, and localizing individual phosphorylation sites emphasizes the quality of the spectra collected.
In order to evaluate the performance of HCD and CID fragmentation within their respective analysis strategies, we designed three experiments including a back-to-back analysis of same-precursors, which removed the speed advantage of CID, and two real world phosphoproteome analyses (a brain antiphosphotyrosine peptide affinity pull-down and large-scale brain phosphoproteome comparison). We found that despite higher primary scores via HCD, collecting CID-based MS/MS spectra in the ion trap was still superior, allowing the detection of nearly twice as many phosphopeptides. In light of these findings, we then compared our results with those recently claiming that HCD (orbitrap detection) outperformed CID (ion trap detection) in phosphoproteomics to help explain our divergent conclusions (25).
MATERIALS AND METHODS
Tissue Preparation
Murine brain tissues were prepared as described (6). Each 10-mg aliquot of lysate was incubated overnight with 150 μg of sequencing grade trypsin (Promega, Madison, WI). Trypsin digestion was terminated by adding trifluoroacetic acid to a final concentration of 0.2%. The digest was desalted over a 500 mg C18solid-phase extraction (SPE) cartridge (Sep-Pak; Waters, Milford, MA) and lyophilized.
Phosphopeptide Enrichment
Phosphopeptides were enriched from tryptic digests by strong cation exchange (SCX) chromatography and immobilized metal affinity chromatography (IMAC) as previously described (26). Ten SCX fractions were collected, desalted and further enriched by IMAC. Enriched phosphopeptides were desalted using C18StageTips (27) and re-suspended in 10 μl of 5% formic acid (FA)/5% acetonitrile (ACN), of which 4 μl were analyzed by liquid chromatography-tandem MS (LC-MS/MS) using each fragmentation technique separately.
Phosphopeptide enrichment using TiO2 was performed as described by Thingholm and coworkers (28) using Titansphere TiO2-beads (GL Sciences, Japan). Phosphopeptides were desalted using C18StageTips. TiO2-enriched samples were resuspended with 12 μl of 5% FA/5% ACN, and 4 μl were used for each LC-MS/MS analysis for back-to-back comparison of CID and HCD fragmentation as shown in Fig. 1 and supplemental Fig. S3.
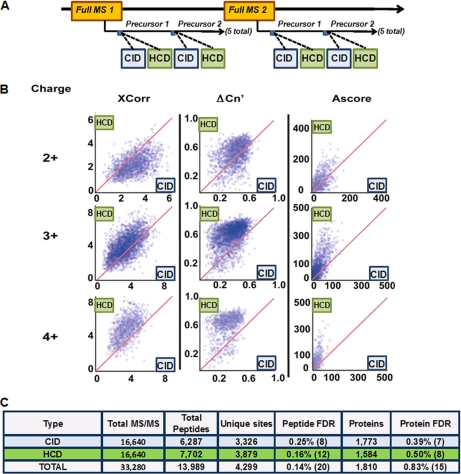
Fig. 1.
Phosphopeptide analysis by a back-to-back, alternating CID- and HCD-type fragmentation for same-precursor ions. A, Workflow for the data-dependent scans using the hybrid LTQ Orbitrap Velos mass spectrometer. The five most abundant ions from each full MS cycle were subjected to sequential CID (ion-trap detection) and HCD (orbitrap detection) fragmentation. A titanium dioxide-enriched sample from 1 mg proteolyzed mouse brain was analyzed by LC-MS/MS techniques using this method. B, Scatter plot distributions of XCorr, ΔCn′, and Ascore values for phosphopeptides identified by both HCD (y axis) and CID (x axis) for 2+, 3+, and 4+ charged ions, showing generally higher trends for HCD. C, Table summarizing peptide and protein identifications from this experiment. Matches to reversed (decoy) sequences are shown in parentheses.
Antibody Enrichment of Phosphotyrosine-containing Peptides
The procedure for phosphotyrosine enrichment was adapted from the description by Rush and coworkers (29). Tryptic peptides (10 mg) were dissolved in 2 ml of immunoprecipitation buffer: 50 mm MOPS-NaOH, pH 7.2, 10 mm Na2HPO4, and 50 mm NaCl. The solution was sonicated at room temperature for 30 min and centrifuged at 4 °C for 10 min at 15,000 × g to remove any insoluble debris. To the supernatant, 50 μg of pY100 anti-phosphotyrosine antibody (Cell Signaling Technology, Danvers, MA) coupled to 50 μl of protein-A Sepharose (Amersham Biosciences/GE Healthcare, Piscataway, NJ) was added, and the mixture was incubated overnight at 4 °C. Immune complexes were washed three times with cold immunoprecipitation buffer and once with water. Peptides were eluted with two volumes of 40 μl of 0.15% trifluoroacetic acid for 10 min at room temperature. They were desalted using C18StageTips and re-suspended in 10 μl of 5% FA/5% ACN. A volume of 4 μl was analyzed for each LC-MS/MS analysis by HCD- and CID-type fragmentation.
Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry (LC-ESI-MS/MS)
We performed LC-ESI-MS/MS on a hybrid dual-pressure linear ion trap/orbitrap mass spectrometer (LTQ Orbitrap Velos, Thermo Scientific, San Jose, CA) equipped with a Famos autosampler (LC Packings, Sunnyvale, CA) and an Agilent 1200 binary HPLC pump (Agilent Technologies, Palo Alto, CA). Peptide mixtures were fractionated on a 100 μm I.D. microcapillary column packed first with ∼0.5 cm of Magic C4 resin (5 μm, 100 Å, Michrom Bioresources, Auburn, CA) followed by 20 cm of Maccel C18AQ resin (3 μm, 200 Å, Nest Group, Southborough, MA). Separation was achieved through applying a gradient from 8% ACN to 30% ACN in 0.125% FA over an 85- or 145-min gradient at a flow rate of ∼300 nL/min.
The LTQ Orbitrap Velos MS was used in the data-dependent mode. Methods using exclusively CID-fragmentation when acquiring MS/MS spectra consisted of an orbitrap full MS scan followed by up to 20 LTQ MS/MS experiments (TOP20) on the most abundant ions detected in the full MS scan. Essential MS settings were as follows: full MS (AGC 3 × 106; resolution 6 × 104; m/z range 300–1500; maximum ion time 1000 ms); MS/MS (AGC 2 × 103; maximum ion time 150 ms; minimum signal threshold 500; isolation width 2 Da; dynamic exclusion time setting 30 s (±10 ppm relative to the precursor ion m/z); singly charged ions and ions for which no charge state could be determined were excluded from selection. Normalized collision energy was set to 35%, and activation time to 20 ms.
For methods where exclusively HCD MS/MS spectra were acquired an orbitrap full MS scan was followed by up to 10 HCD-orbitrap MS/MS spectra on the most abundant ions detected in the full MS scan. The resolution for full MS scans was set to 3 × 104. The AGC for MS/MS experiments was set to 3 × 104 at a maximum ion accumulation time of 250 ms. Normalized collision energy was set to 45%, and HCD fragmentation ions were detected in the orbitrap at a resolution setting of 7.5 × 103. All other settings were as described for the method using exclusively CID fragmentation.
For back-to-back comparisons of CID and HCD spectra (Fig. 1 and supplemental Table S4) the five most abundant ions detected in an orbitrap full MS spectrum were selected for MS/MS (TOP5). Each peptide ion was first selected for CID-fragmentation and then for an HCD-experiment before the next ion was subjected to CID-MS/MS. CID fragment ions were either detected in the LTQ ion trap or the orbitrap. MS settings were as described above.
Experiments on an LTQ Orbitrap XL (85 min gradients, supplemental Table S1) were performed using exclusively CID fragmentation in the ion trap when acquiring MS/MS spectra. Settings were described as above except for MS/MS AGC, which was set to 5 × 103 and the maximum MS/MS ion accumulation time, set to 120 ms.
Data Analysis
After spectral data acquisition, RAW files were converted into mzXML format and prepared for database searching as previously described (6). MS/MS spectra were searched using the Sequest (version 28 revision 13) or MASCOT (version 2.3) algorithms (30, 31). Spectra were searched against a database containing sequences of all proteins in the mouse IPI database (version 3.60, 56,738 protein entries, downloaded July 2009) and common contaminant sequences (e.g. human keratins and trypsin) in both forward and reversed orientations. The following parameters were used to identify phosphopeptides applying either search algorithm: 10 ppm precursor mass tolerance, 0.8 Da product ion mass tolerance, fully tryptic digestion, up to two missed cleavages, variable modifications: oxidation of methionines (+15.9949) and phosphorylation of serines, threonines, and tyrosines (+79.9663); fixed modifications: carbamidomethylation of cysteine (+57.0214). For all HCD spectra and the high resolution CID spectra in supplemental Table S3, fragment ion tolerances were set to 0.02 Da.
The target-decoy approach was applied to control peptide and protein level false discovery rates (FDRs) (32). Linear discriminant analysis (LDA) was employed to distinguish correct from incorrect peptide identifications using the following variables: XCorr (0.8 score minimum), ΔCn′, precursor mass error, charge state, and solution charge state for SCX/IMAC. The variable ΔCn′ gives the relative difference in XCorr values for the highest scoring peptide match found by Sequest for an MS/MS spectrum and for the next highest scoring match with a distinct amino acid sequence without taking post-translational modifications into account. LDA was also used to control the FDR of data sets from Mascot searches by substituting Mascot's Ion Score and Delta-Ion Score for XCorr and ΔCn′, respectively. Phosphopeptides shorter than six amino acids in length were removed and peptide spectral matches were filtered to a 1% FDR at the peptide-level based on the number of decoy sequences in the remaining data set. After all phosphopeptides were grouped with their corresponding proteins, proteins were scored based on their multiplied peptide LDA probabilities. The sorted list was filtered based on reversed protein hits to maximally contain 1% false positive protein identifications. Any protein redundancy is clearly noted in each supplementary peptide table.
We used the Ascore algorithm to quantify the confidence with which each phosphorylation modification could be assigned to a particular residue in each peptide (33). Phosphopeptides with Ascore values above 13 (p ≤ 0.05) were considered confidently localized to a particular residue. To calculate the number of unique sites for each experiment, all localized sites were counted once. Nonlocalized sites were only counted if they could not be assigned to any localized site (32). Multiple nonlocalized sites occupying overlapping sequence ranges were assessed as a single phosphorylation event. Therefore, the calculated unique sites render a strict minimal estimate for all identified phosphorylation sites.
Analysis of HCD and CID Raw Files From Mann and Coworkers (25)
Xcalibur .RAW files acquired were downloaded from ProteomeCommons.org (34) at the hash below.
/Gyf6Csx8Xlx8aUTof4/OcFDVdL3TDb6J4UPLceZSTXL2kdZr9OUb5j6NdIduK6+ehqHJ3Td9GZSQTKaDtUM4/gMsNYAAAAAAAATLQ ==
We examined all critical spectral acquisition parameters that are embedded in the instrument method within each .RAW file. All .RAW files were then searched by the same parameters as described in the data analysis section of our methods. In addition, we re-examined phosphorylation site lists that were provided as supplemental Materials accompanying the manuscript: pr100637q_si_002.txt, pr100637q_si_003.txt, and pr100637q_si_004.txt.
Data Dissemination
All data are made available including all RAW data files
Fig. 1: /sF+UqXTLzCQn2oo3KKJbkut24ogz+PG5uRn2hHaXZZ8ZMCii31qGwR5Q4n4LgEvWczgt61Mjgg4aLdRLbzOlobUr2QAMAAAAAAALmw ==
Fig. 2: RqQSjWSKodoE55Ftml8pxhwujwu/V1PZMfzgHReZvC44e6/Q1mHHtfOX1HzfJuk84ZrJHMVcVg6pJ8cqFc7lGpKsDpgAAAAAAAAMRQ ==
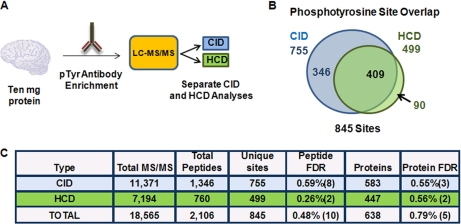
Fig. 2.
CID- and HCD- type fragmentation for phosphotyrosine antibody-enriched peptides. A, Workflow for phosphotyrosine analysis. B, Venn diagram of phosphotyrosine containing peptides and their overlap between CID and HCD. C, Table summarizing this experiment. Matches to reversed (decoy) sequences are in parentheses.
Fig. 3: 1V7/zDAFlz1dDcmkfgiwTj8EWvmK5ycfXvbbNpRZYrtjhm5td8qtj60SEQb5DFlmXPsA4K8MpfDeeDCGCIIBH4Sbd8IAAAAAAAARzA ==
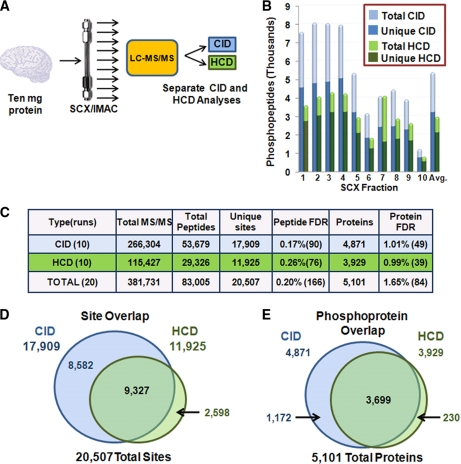
Fig. 3.
Full phosphoproteome analysis of mouse brain by CID- and HCD-type fragmentation. A, Workflow for phosphoproteomic analysis. Brain phosphopeptides were enriched with the SCX-IMAC approach (26), and ten fractions were analyzed by separate 85-min CID and HCD runs. B, Phosphopeptides identified in SCX fractions. C, Summary of these studies. Matches to reversed (decoy) sequences are in parentheses. D, E, Venn diagrams of site and phosphoprotein overlaps between CID and HCD experiments.
Supplemental Fig. S3: N0kSl0hRjSOIsx8tG7Hp5sLijVkQ/f3PjeSs+jqecfSxsCJT09Z+FF5ufUiU/kNgJzo9nMcYa1tKe+aOYWO3cXa8V3gAAAAAAAALfA ==
Supplemental Table S1: POPJa9xrVWQy671CyxSseYuqvKR0gxNAjrqd6q2mWfvQlGYS2zsrNky5xSlvc9ZHYasK//FWqYleO0yIXB0nDJimiUAAAAAAAAAPAQ ==
In addition, all MS/MS spectra are also made available via hyperlink in supplementary Tables S1 to S12. An annotated list of their content is located in the Supplementary Outline of Tables.
RESULTS AND DISCUSSION
HCD-type fragmentation, as employed on the LTQ Orbitrap Velos, is a powerful technique for generating higher resolution MS/MS spectra. However, the acquisition speed of HCD MS/MS Orbitrap spectra is about half of what is found for traditional lower resolution CID ion trap spectra. In addition, HCD with ORBITRAP detection has lower sensitivity than electron multiplier-based CID techniques. Despite these limitations, one very useful feature of HCD is that the low mass region is well represented in the MS/MS spectra when compared with CID, allowing HCD to be combined with isobaric tagging reagents (e.g. TMT, iTRAQ) which produce small m/z reporter ions (10, 35, 36). Moreover, HCD fragmentation supports multiple cleavage events which may result in richer fragmentation of phosphopeptides where the neutral loss of phosphoric acid is unproductive and common (37). Given the progress toward making HCD a mainstream analysis technique and recent conflicting reports of comparisons between HCD and CID (10, 38–40), we designed three experiments to rigorously evaluate each technique's performance with large-scale phosphoproteomic data.
Comparing Same-Precursor-Ion Spectra
To compare HCD (orbitrap detection) and CID (ion trap detection) fragmentation techniques for spectral differences, we designed a back-to-back method that would equalize experimental variability because of the stochastic nature of data-dependent LC-MS/MS analyses and account for differential rates of HCD and CID acquisition by ensuring that identical precursors were sequentially examined under matched experimental conditions. This experimental design allowed us to compare the relative utility of HCD and CID spectra for proteomic identifications independent of their inherent disparity in acquisition speed. Fig. 1A shows a schematic representation of the method employed on a sample of phosphopeptides enriched from murine brain cell lysates via titanium dioxide stationary phase enrichment. An LC-MS/MS analysis (155-min) acquiring both HCD and CID spectra sequentially on the same precursor ions resulted in the collection of 16,640 MS/MS spectra. All spectra were searched using Sequest (30) and filtered to a 1% protein FDR. More spectra (19%) and more sites (18%) were matched by HCD than CID techniques (Fig. 1C), and similar results were found when searched by MASCOT (supplemental Table S2).
Using all peptides simultaneously identified, we plotted values for XCorr, ΔCn′, and Ascore for phosphopeptides matched by both HCD (orbitrap detection) and CID (ion trap detection) fragmentation for 2+, 3+, and 4+ charged peptides. Although XCorr is the primary score for Sequest, the ΔCn′ value for a peptide is defined as the difference between XCorr values for the top Sequest match and the next highest XCorr value for a peptide with a different amino acid sequence (33). For matched spectra from doubly charged precursors, CID overwhelmingly returned higher XCorr values, yet HCD identified more 2+ peptides overall (Fig. 1B, supplemental Fig. S5). However, for 3+ and 4+ peptides, XCorr values favored the HCD platform while ΔCn′ values always favored the HCD strategy and returned higher XCorr values (Fig. 1B, supplemental Fig. S5). Given that we saw different XCorr trends by charge state, we profiled all phosphopeptides identified in Fig. 1 by a multivariate analysis with respect to 49 different chemical properties in order to elucidate the etiology of these differences (supplemental Fig. S6). Notably, there were little or no observable differences between peptides identified by either fragmentation approach with respect to the parameters used. This suggests that peptides identified uniquely by one technique likely reflect stochastic differences and do not indicate preferences for either fragmentation strategy with respect to a peptide's physical or chemical properties. Thus either platform enables comparable coverage of the phosphoproteome.
We also examined an identical mouse brain sample collecting both the CID and HCD spectra in the orbitrap at high resolution (supplemental Table S4). Each fragmentation method identified similar numbers of phosphopeptides and unique sites, suggesting that the improved mass accuracy of MS/MS ions is at least partially responsible for any differences seen in Fig. 1.
Although search algorithms such as Sequest and MASCOT are able to identify peptide sequences with high confidence, they often struggle to distinguish among isomeric forms of peptides that differ only in the localization of post-translational modifications. In most cases these algorithms provide no direct measures of confidence for localization of individual sites of post-translational modification. The Ascore algorithm gives a probabilistic score that reflects the relative confidence with which each site can be assigned to a specific position on its substrate peptide (33). Ascore values were higher for HCD-based spectra compared with MS/MS obtained by CID (median = 39.6 versus 28.9, respectively). Example spectra are shown in supplemental Fig. S1, and all MS/MS spectra are available via hyperlinks in supplemental Tables S5–S8. Overall, this same-precursor-ion experiment allowed us to assess differences in data quality in the absence of acquisition speed. It is important to note that these results are based on enforcing the same CID and HCD spectral collection rates because of the back-to-back nature of the experiment.
Evaluating Phosphotyrosine Affinity-Enriched Samples
We next examined the differences between HCD (orbitrap detection) and CID (ion trap detection) considering phosphotyrosine- (pY) containing peptides which are typically underrepresented in most phosphoproteomic data sets, usually comprising only ∼1–2% of the total sites detected (6). We and others have previously used antibody immunopurifications (IPs) to selectively enrich for pY-containing peptides (16, 29, 41, 42), and we therefore implemented this approach to address how well each strategy performed on these pY-enriched samples (Fig. 2A). A pY IP from 10 mg of proteolyzed mouse brain lysate was split equally and subsequently analyzed using separate CID and HCD methods. In total, we identified 845 sites (0.46% peptide FDR) by combining these two strategies. CID analysis, however, detected 40% more pY-containing peptides and 34% more sites than HCD (Fig. 2C). Moreover, the site overlap revealed that only 11% of all pY sites were unique to HCD (Fig. 2B). We conclude that larger data sets are produced from pY peptide samples by low resolution, faster acquisition speed a CID-based strategy than by high a resolution, slower acquisition speed HCD-based strategy.
Evaluating the Mouse Brain Phosphoproteome
We next performed a full phosphoproteome analysis of mouse brain using separate CID (ion trap detection) and HCD (orbitrap detection) analyses to address whether higher search engine scores from HCD spectra can overcome the speed advantages of traditional CID. In total, ten IMAC-enriched SCX fractions were split in half and analyzed by HCD and CID (Fig. 3A). Within their respective strategies, CID analyses collected 2.3-fold more MS/MS spectra, matched 1.8-fold more peptides, and identified 1.5-fold more sites than HCD (Fig. 3C). Although HCD fragmentation performed better in later fractions, where larger peptides and higher charge states dominated, it still did not render more peptide identifications than CID for any fraction (Fig. 3B). Moreover, the HCD results are more representative of the numbers we accumulated from a matched brain SCX/IMAC experiment analyzed by CID using the previous generation LTQ Orbitrap XL mass spectrometer (supplemental Table S1) and a previously published study using the mice from the same litter as this one (6).
Further examination of site and phosphoprotein overlap between CID and HCD (Figs. 3D, 3E) revealed the vast majority of sites (87%) and phosphoproteins (95%) identified were not exclusive to HCD fragmentation. Altogether, when the two data sets were combined, we detected 83,005 phosphopeptides yielding 20,506 unique sites on 5101 proteins, representing the largest brain phosphorylation data set to date. In addition, we attempted to create a decision tree that would choose the optimal fragmentation technique based on charge state and m/z value as described in the literature (40, 43). We found, irrespective of the charge state and m/z value, CID would be preferred over HCD when success probabilities were adjusted for acquisition speed (supplemental Fig. S7). We conclude that traditional CID (ion trap detection) allowed much deeper coverage of the phosphoproteome than high mass accuracy HCD spectra, demonstrating a clear advantage to CID.
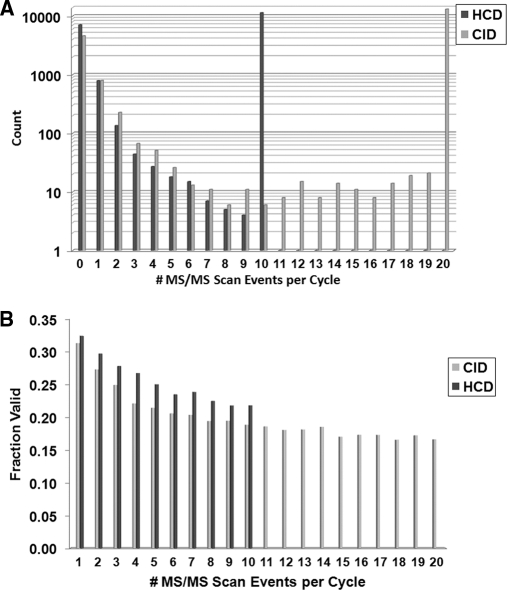
Interestingly, our findings directly contradict the results recently reported by Mann and coworkers (25) who performed a similar large-scale phosphoproteomic analysis from HeLa cells (also using 10 fractions) to evaluate HCD- and CID-type fragmentation. In contrast to our data, their study reported that HCD with orbitrap detection identified more phosphorylation sites than traditional CID (16,559 versus 11,893). We initially questioned whether this large difference could be attributed to the search algorithm used (Sequest and MASCOT, respectively). However, when comparing MASCOT and Sequest for our back-to-back experiment (Fig. 1), we found only small differences in algorithm performance (supplemental Table S2). Therefore, to determine the key differences between our respective studies (especially in light of the highly similar methods used for the collection of both data sets), we downloaded the .RAW files provided by the authors on Tranche (32). Importantly, these files contain not only the raw MS data from the experiments, but also include the actual methods and instrument settings used for data acquisition. Among these parameters was a significant difference between the reported and actual settings for the exclusion width mass tolerance for the CID analyses. As shown in supplemental Fig. S3, the exclusion mass width window after selecting a peak for MS/MS was set to ±10 ppm for all HCD files but to −5 and +10 m/z for all CID files (±10 ppm was reported for both in the manuscript). Thus, a CID-specific, 15 m/z (instead of 20 ppm) window was blocked from re-analysis for every MS/MS that was acquired for up to 1 min (a 750-fold difference in window size at m/z 1000). Since hundreds of MS/MS spectra can be collected per minute, this difference likely resulted in the majority of ions present in the spectrum being unavailable for selection by the software. Hints of this problem were apparent in the data as the authors noted that their cycle times were extremely inefficient, stating that “… in particular, the instrument only performed 10 or more CID scans in 1% of the cases and in most cases only fragmented none or one precursor ion” (Page 6792). This led to a dramatic decrease in the number of MS/MS events triggered for CID fragmentation and is likely the main reason why HCD outperformed CID in their hands. Fig. 4 contains the number of MS/MS spectra acquired per cycle in our experiments. The vast majority (∼90%) of cycles with MS/MS scans contained the full limit of MS/MS scans.
Fig. 4.
Cycle depth (A) and validation rate (B) for the CID and HCD methods. The data from Fig. 3 were analyzed for cycle depth by computing the number of MS/MS spectra triggered from each full MS scan across all ten SCX runs. A TOP10 and TOP20 method were used for HCD and CID analyses, respectively. When MS/MS were triggered, the vast majority of cycles contained either 10 or 20 MS/MS events, correspondingly. HCD spectra had higher validation rates at all MS/MS positions within a cycle. Fraction valid denotes the fraction containing phosphopeptides at the 1% FDR.
For completeness, we searched the MS/MS spectra from the .RAW files provided by Mann and coworkers (25) (including MASCOT searches as well as the Ascore algorithm to localize sites). Here we again encountered an apparent discrepancy from the published results. Their report claimed 16,559 sites detected in a data set containing 10 HCD analyses. However, our re-analysis assigned only 8910 sites (55% of their value) in those same runs (supplemental Tables S3, S9). We then attempted to reconcile our results with their supplementary tables. Unfortunately, there was not enough information shared to fully evaluate the difference in site counting. For example, two pieces of information were missing: (1) the total number of phosphopeptides identified by HCD analysis, and (2) a list of the sequences of these identified phosphopeptides. One of their supplementary tables contains 16,559 entries to represent the sites determined in one replicate of their analysis. However, many entries cite the same singly phosphorylated peptide from the same scan and the same file and then assign several sites to it (supplemental Figs. S4A, 4B). Based on these data, we can confirm that over-counting occurred, but we have no way to determine its actual extent without the complete list of phosphopeptides identified. To address this issue, we have provided all data for this reanalysis in supplemental Table S9 in the hope that this can be resolved at a later date.
In conclusion, we evaluated CID (ion trap detection) versus HCD (orbitrap detection) for phosphopeptide analysis in a full phosphoproteome setting. Using a same-precursor analysis (Fig. 1), HCD demonstrated superior ΔCn′ and Ascore values and identified more phosphopeptides. However, CID was greatly favored for pY IP samples and for the comprehensive analysis of the mouse brain phosphoproteome. At this point, the speed benefits of CID (ion trap detection) outweigh the higher search engine scores from high resolution HCD for in depth phosphoproteomics, despite a previous report in the literature (25). Nevertheless, HCD's true experimental benefit may lie in the realm of multiplexed mass spectrometry analyses using isobaric tags such as TMT or iTRAQ where its ability to measure low m/z products is clearly superior to CID-based methods. Finally, newer and faster orbitrap-based HCD analyses have been reported which may negate the speed advantage for CID with ion trap detection (44).
Acknowledgments
We would like to thank Deepak Kolippakkam for help with the bioinformatics aspects of this manuscript. We also thank John Rush of Cell Signaling Technology for pY antibody method help.
Footnotes
* This work was supported in part by a grant from the National Institutes of Health to SPG (HG3456).
 This article contains supplemental Tables S1 to S12 and Figs. S1 to S7.
This article contains supplemental Tables S1 to S12 and Figs. S1 to S7.
1 The abbreviations used are:
- CID
- collision-induced dissociation
- HCD
- higher energy collisional dissociation
- SCX
- strong cation exchange
- IMAC
- immobilized metal affinity chromatography
- FA
- formic acid
- ACN
- acetonitrile
- FDR
- false discovery rate.
REFERENCES
- 1. Greengard P. (2001) The neurobiology of slow synaptic transmission. Science 294, 1024–1030 [DOI] [PubMed] [Google Scholar]
- 2. Lisman J., Schulman H., Cline H. (2002) The molecular basis of CaMKII function in synaptic and behavioural memory. Nat. Rev. Neurosci. 3, 175–190 [DOI] [PubMed] [Google Scholar]
- 3. Annan R. S., Carr S. A. (1997) The essential role of mass spectrometry in characterizing protein structure: mapping posttranslational modifications. J. Protein Chem. 16, 391–402 [DOI] [PubMed] [Google Scholar]
- 4. Grimsrud P. A., Swaney D. L., Wenger C. D., Beauchene N. A., Coon J. J. (2010) Phosphoproteomics for the masses. ACS Chem. Biol. 5, 105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wisniewski J. R., Nagaraj N., Zougman A., Gnad F., Mann M. (2010) Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J. Proteome Res. [DOI] [PubMed] [Google Scholar]
- 6. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Trinidad J. C., Thalhammer A., Specht C. G., Lynn A. J., Baker P. R., Schoepfer R., Burlingame A. L. (2008) Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics 7, 684–696 [DOI] [PubMed] [Google Scholar]
- 8. Tweedie-Cullen R. Y., Reck J. M., Mansuy I. M. (2009) Comprehensive mapping of post-translational modifications on synaptic, nuclear, and histone proteins in the adult mouse brain. J. Proteome Res. 8, 4966–4982 [DOI] [PubMed] [Google Scholar]
- 9. Makarov A., Denisov E., Kholomeev A., Balschun W., Lange O., Strupat K., Horning S. (2006) Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 78, 2113–2120 [DOI] [PubMed] [Google Scholar]
- 10. McAlister G. C., Phanstiel D., Wenger C. D., Lee M. V., Coon J. J. (2010) Analysis of tandem mass spectra by FTMS for improved large-scale proteomics with superior protein quantification. Anal. Chem. 82, 316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yates J. R., Cociorva D., Liao L., Zabrouskov V. (2006) Performance of a linear ion trap-Orbitrap hybrid for peptide analysis. Anal. Chem. 78, 493–500 [DOI] [PubMed] [Google Scholar]
- 12. Kristensen D. B., Imamura K., Miyamoto Y., Yoshizato K. (2000) Mass spectrometric approaches for the characterization of proteins on a hybrid quadrupole time-of-flight (Q-TOF) mass spectrometer. Electrophoresis 21, 430–439 [DOI] [PubMed] [Google Scholar]
- 13. Belov M. E., Nikolaev E. N., Anderson G. A., Udseth H. R., Conrads T. P., Veenstra T. D., Masselon C. D., Gorshkov M. V., Smith R. D. (2001) Design and performance of an ESI interface for selective external ion accumulation coupled to a Fourier transform ion cyclotron mass spectrometer. Anal. Chem. 73, 253–261 [DOI] [PubMed] [Google Scholar]
- 14. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Villén J., Beausoleil S. A., Gerber S. A., Gygi S. P. (2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 18. Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Palumbo A. M., Smith S. A., Kalcic C. L., Dantus M., Stemmer P. M., Reid G. E. (2011) Tandem mass spectrometry strategies for phosphoproteome analysis. Mass Spectrom. Rev. [DOI] [PubMed] [Google Scholar]
- 20. Zubarev R. A., Horn D. M., Fridriksson E. K., Kelleher N. L., Kruger N. A., Lewis M. A., Carpenter B. K., McLafferty F. W. (2000) Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem. 72, 563–573 [DOI] [PubMed] [Google Scholar]
- 21. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 22. McAlister G. C., Phanstiel D. H., Westphall M. S., Coon J. J. (2011) Higher-energy collision-activated dissociation without a dedicated collision cell. Mol. Cell. Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Second T. P., Blethrow J. D., Schwartz J. C., Merrihew G. E., MacCoss M. J., Swaney D. L., Russell J. D., Coon J. J., Zabrouskov V. (2009) Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal. Chem. 81, 7757–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nagaraj N., D'Souza R. C., Cox J., Olsen J. V., Mann M. (2010) Feasibility of large-scale phosphoproteomics with higher energy collisional dissociation fragmentation. J. Proteome Res. 9, 6786–6794 [DOI] [PubMed] [Google Scholar]
- 26. Villén J., Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 28. Thingholm T. E., Jørgensen T. J. D., Jensen O. N., Larsen M. R. (2006) Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protocols 1, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 29. Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 30. Eng J. K., McCormack A. L., Yates J. R. (1994) An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 31. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 32. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 33. Beausoleil S. A., Villén J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 34. Hill J. A., Smith B. E., Papoulias P. G., Andrews P. C. (2010) ProteomeCommons.org collaborative annotation and project management resource integrated with the Tranche repository. J. Proteome Res. 9, 2809–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boja E. S., Phillips D., French S. A., Harris R. A., Balaban R. S. (2009) Quantitative mitochondrial phosphoproteomics using iTRAQ on an LTQ-Orbitrap with high energy collision dissociation. J. Proteome Res. 8, 4665–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pichler P., Köcher T., Holzmann J., Möhring T., Ammerer G., Mechtler K. (2011) Improved precision of iTRAQ and TMT quantification by an axial extraction field in an Orbitrap HCD cell. Anal. Chem. 83, 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nagaraj N., D'Souza R. C., Cox J., Olsen J. V., Mann M. (2010) Feasibility of large-scale phosphoproteomics with higher energy collisional dissociation fragmentation. J. Proteome Res. 9, 6786–6794 [DOI] [PubMed] [Google Scholar]
- 39. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frese C. K., Altelaar A. F., Hennrich M. L., Nolting D., Zeller M., Griep-Raming J., Heck A. J., Mohammed S. (2011) Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J. Proteome Res. 10, 2377–2388 [DOI] [PubMed] [Google Scholar]
- 41. Ballif B. A., Carey G. R., Sunyaev S. R., Gygi S. P. (2008) Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain. J. Proteome Res. 7, 311–318 [DOI] [PubMed] [Google Scholar]
- 42. Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 43. Swaney D. L., McAlister G. C., Coon J. J. (2008) Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat. Methods 5, 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Michalski A., Damoc E., Hauschild J. P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., Horning S. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]