Abstract
A wide range of biomolecules, including proteins, are excreted and secreted from helminths and contribute to the parasite's successful establishment, survival, and reproduction in an adverse habitat. Excretory and secretory proteins (ESP) are active at the interface between parasite and host and comprise potential targets for intervention. The intestinal nematode Strongyloides spp. exhibits an exceptional developmental plasticity in its life cycle characterized by parasitic and free-living generations. We investigated ESP from infective larvae, parasitic females, and free-living stages of the rat parasite Strongyloides ratti, which is genetically very similar to the human pathogen, Strongyloides stercoralis. Proteomic analysis of ESP revealed 586 proteins, with the largest number of stage-specific ESP found in infective larvae (196), followed by parasitic females (79) and free-living stages (35). One hundred and forty proteins were identified in all studied stages, including anti-oxidative enzymes, heat shock proteins, and carbohydrate-binding proteins. The stage-selective ESP of (1) infective larvae included an astacin metalloproteinase, the L3 Nie antigen, and a fatty acid retinoid-binding protein; (2) parasitic females included a prolyl oligopeptidase (prolyl serine carboxypeptidase), small heat shock proteins, and a secreted acidic protein; (3) free-living stages included a lysozyme family member, a carbohydrate-hydrolyzing enzyme, and saponin-like protein. We verified the differential expression of selected genes encoding ESP by qRT-PCR. ELISA analysis revealed the recognition of ESP by antibodies of S. ratti-infected rats. A prolyl oligopeptidase was identified as abundant parasitic female-specific ESP, and the effect of pyrrolidine-based prolyl oligopeptidase inhibitors showed concentration- and time-dependent inhibitory effects on female motility. The characterization of stage-related ESP from Strongyloides will help to further understand the interaction of this unique intestinal nematode with its host.
The successful establishment of nematodes in the intestinal habitat of a mammalian host and subsequent survival for an extended period of time in the adverse biotope, hinges upon the ability of the parasitic nematode to generate an array of molecules that interfere with the host's defense system endeavored to eliminate the untoward lodger (1). Excretory and secretory (E/S)1 and some somatic products released from living and moribund helminth parasites, respectively, are initial factors, including proteases, enzyme regulators, anti-oxidative proteins, transporters, and various ligand-binding proteins (2). E/S products, active at the interface between the parasite and host are currently intensely investigated as potential targets for therapeutic intervention.
In evolutionary terms, long-lasting interaction between intestinal parasitic nematodes and mammalian hosts has led to increased adaptation and co-evolution (3). The “old friend” hypothesis assumes that the presence of certain helminths and microbes chronically colonizing the intestine stimulates the host′s immunoregulatory system to tolerate these “harmless,” yet foreign, organisms. It is currently hypothesized that increases in chronic inflammatory disorders, such as inflammatory bowel diseases and allergies, in developed countries are partially attributable to diminished exposure to organisms that were part of mammalian evolutionary history (4).
Strongyloides generates infective, parasitic and free-living stages, making this parasite genus ideally suited for investigating E/S products and enabling the identification and characterization of proteins with pivotal relevance for its parasitic lifestyle and putative immune-modulating capability. The human pathogen Strongyloides stercoralis shows several fundamental differences to the other helminths: (1) In contrast to other soil-transmitted helminths, the unique life cycle of S. stercoralis encompasses both, a direct (asexual) and—facultatively—an indirect (sexual) development (5). Thus, in contrast to e.g. Ascaris and hookworm, the Strongyloides larvae can develop ex vivo into adults resulting in sexual reproduction and egg formation; infective larvae (iL3) eventually hatch from these eggs. (2) S. stercoralis exhibits the ability to complete its life cycle within the human host. Accordingly, larvae can develop to the iL3 within the gastrointestinal tract, traverse the intestinal mucosa, migrate through the tissues, and establish again in the small intestine (6). Such cycles of autoinfection can lead to repeated re-infection that can persist for several decades without apparent symptoms. (3) No other human parasitic nematode has been associated with such a broad spectrum of manifestations and clinical syndromes as S. stercoralis. Chronic infections with S. stercoralis are often associated with no or mild cutaneous, gastrointestinal, or pulmonary symptoms. In immune-competent hosts, the disease is generally not life-threatening. However, in immunocompromised patients, e.g. after treatment with immunosuppressive drugs like glucocorticoids, after co-infection with HTLV-1, or tuberculosis, in case of hematologic malignancies, or protein-caloric malnutrition syndrome, an accelerated autoinfection (hyperinfection) normally occurs, leading, in ≥87% of the cases, to life-threatening disseminated infections and death (7, 8). Recent reports have indicated the underestimation of strongyloidiasis and its hyperinfection syndrome, which is now considered an emerging global infectious disease that has migrated from developing regions to industrialized areas (9). More than 100 million people are probably infected, as the current stool diagnostic is insensitive, and as such the number of infected people was grossly underestimated (10, 11).
To investigate E/S products (ESPs) possibly relevant in the parasite-host interaction during Strongyloides infection, we chose the rat-infecting S. ratti as a model parasite, which is genetically closely related to the human parasite S. stercoralis (12). This model system is highly advantageous as the life cycle is short and easily maintained, giving access to infective, parasitic, and free-living stages and their respective ESPs. In this study, the E/S products from the accessible S. ratti stages were collected and comprehensively analyzed using liquid chromatography (LC)/MS-based proteomics strategies. Selected proteins identified in this proteomic study were characterized and studied further. The overall objective of this study was the identification and functional characterization of molecules that are potentially important for the establishment and maintenance of parasitism and the helminth-induced immunosuppression (2, 13).
EXPERIMENTAL PROCEDURES
Maintaining S. ratti Life Cycle
The S. ratti life cycle established in our laboratory was provided by Dr. G. Pluschke (Swiss Tropical Institute, Basel). Wistar rats were used to maintain the life cycle by serial passage, as described previously (14, 15). Approval was obtained from the Animal Protection Board of the City of Hamburg.
Preparation of Infective Larvae (iL3)
For the isolation of iL3, fecal pellets were collected on days 6–16 after subcutaneous infection of male Wistar rats with 1800–2500 iL3s. Charcoal coprocultures (12) were established and incubated at 26 °C. The culture dishes were incubated 5–7 days for the collection of newly generated iL3. For the recovery of iL3, the Baermann method was used (16). After separation of from the charcoal coproculture the larvae were extensively washed (see below).
Preparation of Free-living Stages (flS)
For the flS preparation, fecal pellets were collected at the earliest 6 days after subcutaneous infection of rats with 1800–2500 iL3s. Charcoal coprocultures were prepared and incubated at 26 °C for 24 h. Free-living females were manually isolated from other flS using a light microscope. The free-living females (flF) were isolated from other flS by careful pipeting under the light microscope.
Preparation of Parasitic Females (pF)
For the collection of pF, the rats were infected with about 2500 iL3s. On day six and seven postinfection, the rats were sacrificed, and the small intestines between the stomach and ∼10 cm before the appendix were removed. The intestines were precleaned by emptying the contents through careful squeezing of the intestinal walls. Next, the intestine was opened longitudinally using scissors and cut into strips of about 8–10 cm length. The strips were washed three times by gentle shaking in three different beakers filled with 500 ml of water or PBS to remove residual debris. To separate the female nematodes from the tissue, the strips were placed directly on the sieve without the cotton in a Baermann apparatus and incubated for 3 h. After sedimentation of the females, 50 ml of the solution in the Baermann funnel were transferred into a 50-ml conical tube. After a second sedimentation of the females, they were transferred to a 1.5-ml tube and washed six times in sterile Hanks balanced salt solution (Sigma-Aldrich, Steinheim, Germany) supplemented with 100 U/ml penicillin and 100 mg/ml streptavidin (Sigma). Between the washing steps, the tube was centrifuged at 1000 rpm for 1 min. These repeated centrifugation steps at low speed resulted in a separation of pF from tissue and residual eggs, as well as first-stage larvae as confirmed by microscopy. The suspension of parasitic females was used for in vitro culture or for the preparation of RNA as described (17).
Preparation of Excretory and Secretory Products
Freshly harvested and extensively washed iL3, pF, or flS were carefully suspended in sterile worm culture medium under the laminar flow hood. The washing solution consisted of Hanks Balanced Salt Solution supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml). The culture medium was RPMI 1640 (Sigma-Aldrich) supplemented with penicillin and streptomycin (same concentration as washing solution) and 10 mm HEPES (Sigma-Aldrich). The incubation densities did not exceed 30,000 iL3/ml, 15,000 flS/ml and 100 pF/ml. IL3 and pF were incubated at 37 °C for 24 h and 72 h, respectively. FlS were incubated at 26 °C for 24 h. After the incubation period, vitality and sterility were checked under the microscope. An additional test for sterility was performed by placing 5 μl of each culture medium on blood agar plates and subsequently incubating the plates at 37 °C for 24 h. Only sterile cultures were used for further experiments. All culture supernatants were supplemented with protease inhibitors (Complete protease inhibitor mixture, Roche Diagnostics, Mannheim, Germany) and concentrated about 200-fold using microspin filters with a 10 kDa molecular weight cutoff. For the inhibition of an active astacin metalloprotease, 10 mm phenanthroline solution was added to the supernatants prior to centrifugation. Biological triplicates were generated from all studied stages.
Inhibition of Protein Release
To demonstrate active protein excretion and secretion, cycloheximide or sodium azide were added to inhibit protein synthesis. The inhibitors were added immediately prior to the incubation of iL3: cycloheximide was added to final concentrations of either 50 or 70 mm, and sodium azide was added to final concentrations of either 0.5% or 1.0% (w/v). After two hours of incubation, the culture medium was removed, an equal amount of new medium was added, and the larvae were further incubated for 24 h. In parallel, equal numbers of iL3 were incubated at 4 °C, 37 °C, and 70 °C affecting the metabolic status. After treatment of the nematodes with 70 mm cycloheximide—similarly after treatment either with sodium azide, or at 4 °C and at 70 °C—all proteins, except those proximal to the 16 kDa molecular weight marker, were absent in a silver-stained SDS-PAGE gel. In addition, no metalloprotease activity was detected (see below), confirming the notion of active biosynthesis and secretion of the identified nematode proteins (data not shown).
Protease Activity Assay
To determine whether the excretory and secretory (E/S) products show proteolytic activity, substrate gel electrophoresis was performed (18, 19). Briefly, 1% gelatin was added as a substrate to the acrylamide solution before preparing the gel. The E/S products were separated under nonreducing conditions. Because SDS inhibits the activity of enzymes, the gels were washed three times in Triton X-100 for 20 min, three times in distilled water for 5 min and finally covered with renaturation buffer (Tris-HCl, 50 mm; NaCl, 100 mm; pH 7.5). After overnight staining in Coomassie blue, the bands containing active proteases appear colorless against a blue background.
One-dimensional Electrophoresis and Tryptic Digestion of E/S Proteins (ESP)
All samples were reduced by adding 10 mm dithiotreitol and incubating at 56 °C for 45 min. For alkylation, 55 mm iodoacetamide was added, and the samples were incubated 30 min at room temperature in the dark. Subsequently, NuPAGE® LDS 4 × sample buffer (Invitrogen, Darmstadt, Germany) was added, and the samples were loaded on 12% NuPAGE® Novex® SDS-PAGE gels. After electrophoresis, gels were stained with SimplyBlue (Invitrogen) protein stain. Entire gel lanes were cut into 30–40 pieces using a disposable scalpel. Prior to tryptic digestion, gel pieces were washed 5 min in 0.1 m ammonium bicarbonate, destained in 50 mm ammonium bicarbonate buffer containing 1/3 acetonitrile for 60 min, and finally dehydrated in 100% acetonitrile.
For in-gel tryptic digestion, the gel pieces were hydrated for 45 min in sequencing grade trypsin solution (12.5 ng/μl; Promega) on ice. The remaining trypsin solution was removed, gel pieces were covered with 50 mm ammonium bicarbonate and incubated overnight at 37 °C. After overnight incubation, the supernatants containing the majority of the tryptic peptides were transferred into new 1.5-ml Eppendorf tubes. The gel pieces were washed in ammonium bicarbonate containing 1/3 acetonitrile, and the washing buffer was pooled with the respective supernatant. Remaining peptides were extracted with 100% acetonitrile, which was added to the pooled supernatant and washing solution.
Mass Spectrometric Analysis
Peptides derived from in-gel digested proteins were analyzed by online microscale capillary reversed-phase HPLC hyphenated to a linear ion trap mass spectrometer (LTQ, Thermo Scientific). Samples were loaded onto an in-house packed 100 mm i.d. × 15 cm C18 column (Magic C18, 5 mm, 200 Å, Michrom Bioresource, Auburn, CA) and separated at ∼ 500 nl/min with 34 min linear gradients from 6–32% acetonitrile in 0.4% formic acid. The instrument was operated in data dependent acquisition mode with a dynamic exclusion of 45 s: after each survey spectrum (m/z 350 to 1400), the six most intense ions per cycle were selected for fragmentation by collision-induced dissociation.
Protein Identification and Sequence Analysis
The .raw files were converted into .mgf files using in-house written scripts (20, 21). For each fragment ion spectrum, only the 200 most intense fragment ions were exported into the mgf file. The mass spectrometry data were searched against a custom database comprising translated expressed sequence tags (EST) contig sequences and protein sequences from S. ratti and S. stercoralis (22) available at www.nematode.net (23), and other protein sequences from Brugia malayi and Caenorhabditis elegans (supplemental Table S1), totalling 39,318 sequences. Searches were performed using ProteinPilotTM (version 2.0.1; AB/Sciex) using the following search parameters: Sample Type, Identification; Cys Alkylation, Iodoacetamide; Digestion, Trypsin; Instrument, LTQ; Special Factors, Gel-based ID; ID Focus, Amino acid substitutions; Search Effort, Thorough. Proteins were identified based on a minimum unused protein score (UPS) of 4.00 equal to two or more unique peptides of 99% confidence. Protein inference is based on the ProGroup algorithm integrated into ProteinPilot. Using a mixed model approach to estimate the confidence of the protein identifications (21), we calculated an overall protein false discovery rate of 2.17% for the cutoff score applied to the protein identifications.
EST contig sequences were subjected to Basic Local Alignment Search Tool (BLAST) searches with NCBI protein BLAST algorithm (blastp) using the default setting against the nonredundant database without any species restrictions. The top hits, irrespective of E-value and/or species are listed in the tables. The resulting sequences were screened for signal peptide for secretion using SignalP 3.0 (24).
Supernatant Versus Extract Protein Abundance Correlations
We used a rank-order correlation analysis in order to robustly determine the degree of similarity between relative protein abundances in the supernatant and the extract. Briefly, we (1) extracted total protein abundance measurements (spectral counting and/or total score) for all proteins observed in the supernatant as well as in the extract; (2) determined the rank of each measurement, accounting for ties where necessary; and (3) calculated the Spearman rank correlation coefficient between supernatant and extract ranks. All calculations were performed in Microsoft Excel.
Analysis of Relative Gene Expression Levels by qRT-PCR
Real-time PCR was used to measure relative levels of expression of selected genes from iL3, pF, and free-living females (flF). Ten genes were selected from the 25 highest scoring ESPs, including two ESP from iL3, five ESP from pF and three ESP from flS (Table I) and in addition two proteins found in all studied stages (see supplemental Table S2) were selected for quantitative RT-PCR analysis. For constitutive protein control, the S. ratti glyceraldehyde-3-phosphate dehydrogenase (Sr-gapdh) and Sr-actin were included. Total RNA was extracted, quantified, purified, and reverse transcribed separately from two different biological samples of the developmental stages (iL3, pF and flF) after 7 days of infection as described (17). An aliquot of ∼5 μg purified parasite RNA with confirmed quality via the detection of discrete 18 S and 28 S ribosomal RNA bands on ethidium bromide-stained gel was used to confirm equally in each developmental stage. The forward and reverse primers for the Sr-genes were designed with primer3plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) such that each amplicon was around 150 bp. Primers were analyzed using the Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html) to avoid secondary structures such as hairpins, loops, and complementarity. The RT-PCR products were evaluated by agarose gel electrophoresis.
Table I. Genes and dedicated primers applied in the qRT-PCR.
| Gene | Cluster | Accession N° | Stage: Protein list (N°) | Forward primer | Reverse primer | Amplicon (bp) |
|---|---|---|---|---|---|---|
| Sr-astacin | SR11111 | AAK55800 | iL3:Table II A (5) | TTGATACAGGAGTA | CCAACATATGA | |
| AATGAAACTACAG | TCGACAACCA | 145 | ||||
| Sr-ZK1073.1 | SR02886 | XP_001899587 | iL3: Table II A (2) | TTGTAGATTTGC | TCCCAAGCACTTT | |
| CATTGCTCATCC | GAGTCATAATTC | 150 | ||||
| Sr-PSC-1 | SR01641 | NP_971802 | pF: Table II B (7) | TGATGGTAAATTA | CATAATGTTGGATA | |
| GATCGTGATGAGA | ATCTCTTCTGGTGA | 150 | ||||
| Sr-calumenin | SR00564 | NP_001024806 | pF: Table II B (8) | TGATGGTAAATTA | CATAATGTTGGATA | |
| GATCGTGATGAGA | ATCTCTTCTGGTGA | 149 | ||||
| Sr-chitin binding protein | SR04455 | XP_001664881 | pF: Table II B (10) | ATGATACTAAGAA | GTATTGACCATCA | |
| ACCTTTTACTCAAG | GGACATGAACTG | 152 | ||||
| Sr-trypsin inhibitor-like protein | SR02054 | XP_001866937 | pF: Table II B (11) | CTTCCAACTGTC | CAGAAATACACTC | |
| CAACAACTCAAA | ACATTTTGGTGGT | 150 | ||||
| Sr-phosphoribosyl-transferase | SR02118 | XP_001895434 | pF: Table II B (22) | GGAACTGATTCA | TTGATGCTCCATT | |
| ACTGGACATTTAC | GTCATTAACTGT | 151 | ||||
| Sr-MFP2B | SR00863 | AAP94889 | flS: Table II C (2) | ATGCCAAATCTTAA | AGCTCTTCCATGA | |
| ACCAGCTAAAGAAG | ATTGGTTTTCCAT | 150 | ||||
| Sr-CBG22129; Y51F10.7 | SR02091 | XP_001667627 | flS: Table II C (4) | TCTCAAGGATTAG | TCCTTTATCATCAG | |
| TACTTCCAAAAAC | TAATTTGAGCTTT | 150 | ||||
| Sr-lysozyme family | SR00671 | NP_502193 | flS: Table II C (5) | TTACTGGATTCG | ACCAGCTTTCACA | |
| ATGCCATTGGAA | GCATTTTTTATATT | 155 | ||||
| Sr-galectin-2 | SR00627 | AAF63405 | All stages: Suppl. Table II (66) | CAAGCTGGAGAA | ATCACAACGATGA | |
| TGGGGTAATGAGG | GCAAAAGTGCAG | 150 | ||||
| Sr-galectin-1 | SS00840 | AAD39095 | All stages: Suppl. Table II (64) | GGAATGCCTGAAA | CTCTCTCTTCATT | |
| AAAAAGGTAAACG | ACCCCATTCACC | 150 | ||||
| Sr-GAPDH | SR00526 | NP_508534 | - | GTACCACTAAC | GCACCTCTTC | |
| TGTTTAGCTCC | CATCTCTCC | 154 |
ABI PRISM® 7000 SDS/Relative quantification system (Applied Biosystems, Foster City, CA, USA) was used to quantify the relative expression of the selected genes. Sr-GAPDH, which showed a constant and equal expression within the three stages, was included as a housekeeping gene for normalization purposes. PCR was performed using the qPCR Core kit for SYBR® Green I (Eurogentec S.A.) following the manufacturer's protocol. Serial cDNA dilution curves were produced to calculate the amplification efficiency for all genes (25).
For measurement of gene expression levels, each sample was tested in triplicate, using positive controls, template-free controls, negative reverse transcription control and the resulting threshold cycle value Cts recorded. The specificity and identity of individual amplicons were verified by melting curve analysis. Relative transcriptional differences were calculated from normalized values following the protocol described by Livak and Schmittgen (26). The data were expressed as the relative quantity of transcripts using the gene-specific transcription levels at the free-living females stage as baseline (value = 1). All of the samples were normalized to the expression levels of the constantly and equally expressed gene Sr-GAPDH.
Identification of the Full-length Gene Sequence of a Prolyl Oligopeptidase (Prolyl Serine Carboxypeptidase, Sr-PSC)
To obtain the 3′-cDNA end, 3′-Rapid Amplification of cDNA Ends (RACE) was performed using the GeneRace Kit (Invitrogen). The 3′-RACE method generates full-length cDNA by utilizing 3′-oligo-dT-containing primer complementary to the poly(A) tail of mRNA at the first strand cDNA synthesis. RACE fragments were then amplified by PCR using Taq polymerase, the gene-specific forward primer PSC f (GGAAAT TTAATGGAAATGAAACATGGT) and the oligo dT-T7II (GAGAGAGGATCCAAGTACTAATACGACTCACTATAGG) as reverse primer. 5′ RACE was performed according to the manufacturer's protocol using the gene-specific primer PSC r (ATTGGAATCATTGTACCATCTTT). The full-length sequence was obtained by comparing the cloned (pGEM-T Easy vector, Promega) and sequenced (AGOWA-Germany) fragments. Primers including the 3′- and the 5′-ends were designed, and the full length of the Sr-PSC-1 cDNA was captured by PCR. The result was confirmed by sequencing using the M13 forward-, the M13 reverse- and gene-specific primers on the fragment that has previously cloned into the pGEM-T Easy vector.
Immune Recognition by ELISA and Immunoblotting with Rat Sera
Sera from 10 rats taken before and 32 days after infection with S. ratti iL3 were analyzed by ELISA (27) for IgG antibodies reactive against S. ratti proteins in the E/S products from iL3, pF and flS. Polystyrene microtiter plates (Maxi-Sorb, Nunc) were coated with Strongyloides protein samples at a concentration of about 200 ng/well in carbonate buffer (pH 9.6), sealed with Saran wrap and incubated overnight at 4 °C. After removal of unbound protein and washing three times with PBS/0.05% (v/v) Tween 20, the plate was blocked with 5% (w/v) bovine serum albumin in PBS for one hour at 37 °C. Different dilutions (1:100 to 1:500) of rat sera were prepared in PBS/0.5% bovine serum albumin, added to the wells and incubated at 37 °C for one hour. Nonspecifically bound proteins were removed, and the wells were washed three times with Tris-buffered saline in 0.05% Tween 20. For detection of bound rat IgG, peroxidase-conjugated anti-rat IgG antibody was applied to a final concentration of 1:5000. Tetramethylbenzidine was used as peroxidase substrate. Data are expressed as end point titers derived from titration curves (1:100 to 1:500) (27). In addition, sera from 10 humans living in areas in West Africa endemic for Strongyloides stercoralis infection (28) and from two healthy Europeans were tested for IgG recognition of Strongyloides proteins applying peroxidase-conjugated anti-human IgG antibody (CalBiochem, San Diego, CA). Collecting blood samples for research purpose was approved by the Ethics Commission of the Medical Board, Hamburg and by ethic committees and medical authorities in the respective countries.
Inhibition Tests for Prolyl Oligopeptidase Activity
For inhibition experiments, 0.5 m prolyl oligopeptidase inhibitor stock solutions of following compounds were applied: (1A) isophthalic acid 2(S)-(cyclopentanecarbonyl) pyrrolidine-l-prolyl-2(S)-cyanopyrrolidine amide; (1B) isophthalic acid 2(S)-(cyclopentanecarbonyl) pyrrolidine-l-prolyl-2(S)-(hydroxyacetyl)-pyrrolidine amide; (2A) 4-phenylbutanoyl-l-prolyl-pyrrolidine; and (2B) 4-phenylbutanoyl-l-prolyl-2(S)-cyanopyrrolidine (supplemental Fig. S1) (29). The compounds were dissolved in PBS and sterile filtered. The stock solutions were added directly before the incubation of pF at seven different final concentrations between 1–10 mm. The cultures were incubated at 36 °C, and the motility of the exposed females was monitored at different time points under the microscope.
Statistical Analysis
Statistical differences between the test groups of sera investigated for IgG recognition were analyzed by the Mann-Whitney U test. Statistical significance was considered for p < 0.05.
RESULTS
Proteomic Analysis of S. ratti ESP
Comparison of ESP Pattern from Different Life Cycle Stages of S. ratti—Active biosynthesis and excretion/secretion were confirmed under our experimental conditions by incubating the worm cultures with cycloheximide and/or sodium azide. Subsequently, we prepared life cycle stage-specific ESP samples from the following S. ratti life cycle stages: iL3, pF, and flS. Each sample type was analyzed in three biological replicates, and proteins were considered to be present when discovered in one of the replicates. SDS gel electrophoretic analysis revealed protein mixtures of different complexities based on the differing banding patterns in the various ESP. The majority of the ESPs from iL3 appeared to be bimodally distributed between 10–30 kDa and 40–100 kDa, whereas ESPs in the pF sample were predominantly observed at 40–100 kDa. The ESPs from the flS sample were mainly of low-molecular weight between 10 and 15 kDa with an extensive smearing above 40 kDa (Fig. 1). The pattern of protein bands differed strongly between ESPs and somatic extracts of the stages (supplemental Fig. S2).
Fig. 1.
One-dimensional SDS-PAGE of E/S proteins from infective larvae, parasitic females and free-living stages showing after Coomassie stain characteristic protein band patterns of the respective stages. The lanes are assigned to the respective stages in the life cycle (CDC, www.dpd.cdc.gov/dpdx/HTML/Strongyloidiasis.htm).
A total of 586 S. ratti proteins were identified in the ESP, 450 proteins in ESP from iL3, 335 in pF, and 217 in flS. Approximately a quarter of the identified proteins were found in all the samples from the studied life-cycle stages, i.e. in iL3, pF, and in flS (140 proteins, 23.8%, Fig. 2). Proteins abundantly observed in all three investigated developmental stages were heat-shock proteins, galectins, enzymes, fatty acid binding protein, as well as distinct structural proteins (supplemental Table S2).
Fig. 2.
Venn diagram showing the distribution of the identified S. ratti E/S proteins of the studied developmental stages: iL3, pF and flS. The numbers in brackets show the quantities of the proteins in each stage(s) total.
The fraction of stage-specific proteins varied significantly: 33.4% (196 proteins) of the identified ESPs were found exclusively in the iL3 stage, compared with 13.5% (79 proteins) found in only in pF ESP and only 6.0% (35 proteins) in flS. The majority of ESPs, which were observed in two studied stages, occurred in iL3 and pF (16.0%) in comparison to 3.4% and 3.8% found in flS as well as in iL3 or in flS and pF, respectively.
To further confirm active biosynthesis and excretion/secretion, and to exclude the possibility of accidental worm lysis resulting in the presence of cytoplasmic proteins in the supernatant, we also analyzed the proteomes of worm extracts prepared from the corresponding life cycle stages. A rank-order correlation of the identified ESP and extract protein resulted in Spearman rank correlation coefficients between 0.13 and 0.2, indicating little to no rank-order correlation, thus providing evidence in support of excretion/secretion and against worm lysis.
Identification of Stage-specific Proteins-ESP Specific for S. ratti iL3
In total, 196 proteins were identified as iL3-specific (supplemental Table S 3). Of these, 170 proteins were identified as present in either a S. ratti or S. stercoralis EST cluster. It is expected that many more proteins would have been identified if the Strongyloides databases had been more complete. This notion was underscored by the fact that 26 (∼13%) additional proteins were identified, which were assigned to proteins from Onchocerca volvulus (1), Trichuris trichiura (1), B. malayi (1), Haemonchus contortus (1), or C. elegans (22) because the sequences of the respective Strongyloides orthologs were missing in the database. The 25 highest-scoring (based on ProteinPilot's “unused ” score) iL3-specific proteins are listed in Table IIA. GO annotation of these iL3 specific ESPs assigns these proteins to a wide range of functional categories, including protein digestion and folding (n = 7), fatty-acid binding (n = 2), carbohydrate metabolism (n = 8), and cytosol energy metabolism (n = 14).
Table II. A, B, C. The Tables list the 25 highest scoring proteins of E/S products from (A) infective larvae (iL3), (B) parasitic females (pF), and (C) free-living stages (flS) of S. ratti. The lists include the cluster numbers of S. ratti or S. stercoralis ESTs (Cluster), the highest scoring putative proteins identified in a BLAST search (BLAST alignment), the species in which these proteins occurs (Species), the NCBI protein accession numbers of these proteins (Accession Number, the expectation value (E), the presence of a predicted signal peptide in the proteins (SP), the EST length (EST Lgt.), the percentage coverage (% Cov.), the number of peptides found within the EST cluster sequence (# Pep.) and the unused protein scores (UPS).
Table II A. The 25 highest scoring proteins in the E/S products of S. ratti infective larvae. E, expectation value; SP, signal peptide; Lgt., EST length; % Cov., percentage coverage; # Pep., number of peptides; UPS, unused protein scores
| N°. | Cluster | BLAST alignment | Species | Accession number | E | SP | EST Lgt. | % Cov. | # Pep. | UPS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SS01511 | RAS-related protein RAB-1A | Brugia malayi | XP_001901944 | 7e−104 | No | 205 | 57.5 | 8 | 16.05 |
| 2 | SR02886 | Hypothetical 35.6 kDa protein | B. malayi | XP_001899587 | 1e−71 | No | 180 | 55.0 | 7 | 14.10 |
| 3 | SR01803 | Thiosulfate sulfuryltrans ferase | B. malayi | XP_001901653 | 1e−15 | No | 174 | 53.4 | 7 | 14.03 |
| 4 | SS00138 | Adenylate kinase | B. malayi | XP_001894222 | 4e−62 | No | 149 | 49.6 | 6 | 14.00 |
| 5 | SR11111 | Metalloprotease precursor | Strongyloides stercoralis | AAK55800 | 2e−61 | Yes | 265 | 43.8 | 6 | 13.72 |
| 6 | SR01001 | Myosin-filarial antigen | B. malayi | AAB35044 | 0.0 | Truncated | 490 | 41.2 | 13 | 13.67 |
| 7 | SR02558 | Lethal family member (let-805) | Caenorhabditis elegans | NP_001022641 | 1e−69 | Yes | 190 | 37.8 | 5 | 13.52 |
| 8 | SR00386 | L3NieAg.01 | S. stercoralis | AAD46493 | 0.15 | Truncated | 112 | 50.8 | 5 | 13.50 |
| 9 | SR00366 | Hypothetical protein DDBD-RAFT_0217849 | Dictyostelium discoi-deum | XP_642992 | 5e−08 | No | 190 | 40.5 | 6 | 13.15 |
| 10 | SR00901 | TPR domain containg protein | B. malayi | XP_001902724 | 5e−48 | No | 226 | 34.9 | 6 | 12.73 |
| 11 | SS02590 | Sensory Axon guidance family member | C. elegans | NP_001033397 | 3e−42 | Yes | 169 | 59.7 | 6 | 12.00 |
| 12 | SR03037 | Transthyretin related family member | C. elegans | NP_499054 | 1e−30 | Yes | 147 | 50.3 | 5 | 11.80 |
| 13 | SR03119 | Short chain reductase/dehydrogenase | B. malayi | XP_001900343 | 1e−46 | No | 179 | 30.1 | 5 | 11.57 |
| 14 | SS01266 | Myosin-4 | C. elegans | P02566 | 3e−94 | No | 258 | 26.0 | 6 | 11.45 |
| 15 | SR00998 | Myosin light chain family member | C. elegans | NP_510828 | 2e−73 | No | 170 | 31.8 | 4 | 10.60 |
| 16 | SR04474 | Peptidase family M1 containing protein | B. malayi | XP_001897028 | 2e−64 | No | 196 | 32.1 | 4 | 10.36 |
| 17 | SR03753 | K02D10.1b | C. elegans | NP_498936 | 5e−30 | No | 154 | 14.3 | 3 | 10.05 |
| 18 | SR02741 | Fatty acid retinoid binding protein | Wuchereria bancrofti | AAL33794 | 0.37 | No | 139 | 15.8 | 4 | 10.02 |
| 19 | SS01256 | Hypothetical protein Bm1_13900 | B. malayi | XP_001894244 | 1e−26 | Yes | 228 | 39.0 | 5 | 10.00 |
| 20 | SR01321 | Hypothetical protein Bm1_36850 | B. malayi | XP_001898817 | 1e−12 | No | 176 | 43.2 | 5 | 10.00 |
| 21 | SR00700 | Na, K-ATPase alpha subunit | C. elegans | AAB02615 | 3e−101 | No | 233 | 29.2 | 4 | 9.71 |
| 22 | SR02060 | Cell division cycle related family member | C. elegans | NP_495705 | 3e−92 | No | 193 | 32.6 | 4 | 9.50 |
| 23 | SR03954 | CAP protein | B. malayi | XP_001891888 | 2e−53 | No | 242 | 28.1 | 4 | 9.22 |
| 24 | SS01276 | F55F3.3 | C. elegans | NP_510300 | 2e−104 | No | 317 | 12.0 | 3 | 9.17 |
| 25 | SR00383 | Propionyl Coenzyme A Carboxylase | C. elegans | NP_509293 | 2e−17 | No | 76 | 50.0 | 3 | 9.10 |
| 1 | SR01608 | EF hand family protein | Brugia malayi | XP_001901161 | 2e−37 | Yes | 158 | 69.0 | 8 | 23.28 |
| 2 | SR03191 | Prolyl endopeptidase | Rattus norvegicus | EDL99674 | 3e−19 | No | 189 | 51.3 | 6 | 16.69 |
| 3 | SR03587 | Metalloprotease | Nasonia vitripennis | XP_001606489 | 9e−06 | Yes | 166 | 42.8 | 6 | 14.23 |
| 4 | SR03901 | Aspartyl protease (asp-2) | C. elegans | NP_505384 | 4e−43 | No | 191 | 56.5 | 5 | 13.20 |
| 5 | SR04847 | Acetylcholinesterase 2 | Ditylenchus destructor | ABQ58116 | 1e−44 | No | 192 | 36.5 | 5 | 12.4 |
| 6 | SR03310 | mp1 | Onchocerca volvulus | AAV71152 | 2e−13 | Yes | 189 | 39.2 | 5 | 11.64 |
| 7 | SR01641 | Prolyl endopeptidase | Treponema denticola | NP_971802 | 2e−12 | No | 126 | 31.0 | 4 | 11.26 |
| 8 | SR00564 | Calumenin | C. elegans | NP_001024806 | 2e−134 | Yes | 286 | 19.2 | 4 | 10.87 |
| 9 | SR00984 | Small heat-shock protein | Trichinella spiralis | ABJ55914 | 2e−21 | No | 160 | 35.6 | 4 | 10.67 |
| 10 | SR04455 | Hypothetical protein CBG05204 | Caenorhabditis briggsae | XP_001664881 | 2.0 | Yes | 88 | 56.8 | 5 | 10.64 |
| 11 | SR02054 | Scavenger receptor cysteine-rich protein | Culex pipiens quinque-fasciatus | XP_001866937 | 3e−24 | Yes | 328 | 14.6 | 4 | 10.44 |
| 12 | SR03349 | Heat-shock protein HSP17 | C. elegans | NP_001023958 | 2e−20 | No | 157 | 39.5 | 5 | 10.04 |
| 13 | SR01073 | Ribosomal protein (rpl-5) | C. elegans | NP_495811 | 2e−119 | No | 290 | 19.3 | 4 | 9.47 |
| 14 | SR00396 | Endoplasmin precursor | B. malayi | XP_001899398 | 7e−98 | Yes | 231 | 16.5 | 3 | 8.56 |
| 15 | SR00986 | 60S ribosomal protein L10 | B. malayi | XP_001898297 | 2e−85 | No | 189 | 17.5 | 3 | 8.26 |
| 16 | SR01297 | Immunosuppressive ovarian message protein | Ascaris suum | CAK18209 | 2e−17 | Yes | 324 | 19.8 | 4 | 8.15 |
| 17 | SR04713 | Surface antigen BspA-like | Trichomonas vaginalis | XP_001315000 | 5.3 | No | 55 | 89.1 | 3 | 7.27 |
| 18 | SR02663 | Metalloprotease precursor | S. stercoralis | AAK55800 | 6e−12 | Truncated | 182 | 19.2 | 3 | 6.91 |
| 19 | SR01002 | Ribosomal protein (rps-18) | C. elegans | NP_502794 | 5e−75 | No | 154 | 15.6 | 2 | 6.64 |
| 20 | SR00979 | Ribosomal protein L9 | Strongyloides papillosus | ABK55147 | 2e−73 | Truncated | 166 | 21.7 | 2 | 6.47 |
| 21 | SR02153 | Hypothetical protein CBG20335 | C. briggsae | CAP37373 | 2e−34 | No | 178 | 13.5 | 2 | 6.34 |
| 22 | SR02118 | Phosphoribosyl transfe-rase | B. malayi | XP_001895434 | 2e−27 | No | 152 | 31.6 | 3 | 6.24 |
| 23 | SS03220 | Intermediate filament protein (ifa-3) | C. elegans | NP_510649 | 6e−38 | No | 141 | 24.1 | 3 | 6.23 |
| 24 | SS03344 | SPARC precursor | B. malayi | XP_001897784 | 2e−74 | Yes | 175 | 20.6 | 3 | 6.23 |
| 25 | SR01499 | Troponin family protein | B. malayi | XP_001898461 | 5e−50 | No | 257 | 14.0 | 3 | 6.19 |
| 1 | SR02994 | Hypothetical protein Y49E10.18 | Caenorhabditis elegans | NP_499623 | 2e−27 | Yes | 143 | 51.0 | 7 | 19.13 |
| 2 | SR00863 | MFP2b | Ascaris suum | AAP94889 | 5e−71 | No | 173 | 52.0 | 7 | 14.85 |
| 3 | SR00375 | Hypothetical protein CBG05949 | Caenorhabditis briggsae | XP_001670383 | 5e−09 | Yes | 148 | 31.1 | 5 | 13.91 |
| 4 | SR02091 | Hypothetical protein CBG22129 | C. briggsae | XP_001667627 | 1e−35 | Yes | 174 | 36.8 | 6 | 13.27 |
| 5 | SR00671 | Lysozyme family member (lys-5) | C. elegans | NP_502193 | 4e−37 | Yes | 160 | 23.1 | 4 | 12.70 |
| 6 | SR02511 | Acyl sphingosine amino hydrolase | C. briggsae | CAP33700 | 2e−48 | Yes | 184 | 27.2 | 4 | 8.35 |
| 7 | SR01169 | Aminotransferase | Clostridium botulinum | ZP_02614737 | 0.53 | No | 176 | 31.8 | 4 | 8.14 |
| 8 | SR00576 | MSP domain protein | B. malayi | XP_001899679 | 3e−32 | No | 97 | 47.4 | 4 | 8.02 |
| 9 | SR00479 | Hexosaminidase B | Pantroglodytes | XP_517705 | 2e−46 | Yes | 167 | 16.8 | 2 | 7.13 |
| 10 | SR00767 | F25A2.1 | C. elegans | NP_503390 | 6e−25 | No | 178 | 17.4 | 2 | 6.67 |
| 11 | SS01173 | Enoyl-CoA reductase | A. suum | AAC48316 | 1e−104 | No | 299 | 10.0 | 2 | 6.41 |
| 12 | SR00821 | Saposin-like protein | C. elegans | NP_491803 | 5.4 | Yes | 86 | 54.7 | 3 | 6.39 |
| 13 | SR00750 | Similar to mannose receptor | Gallus gallus | XP_418617 | 2e−07 | Yes | 174 | 21.3 | 3 | 6.35 |
| 14 | SR00354 | Acid sphingomyelinase | C. elegans | NP_001040996 | 2e−89 | Yes | 269 | 19.3 | 3 | 6.09 |
| 15 | SR01936 | Hypothetical protein CBG21853 | C. briggsae | XP_001672742 | 1e−21 | Yes | 190 | 13.7 | 2 | 5.98 |
| 16 | SR00380 | Hypothetical protein EUBVE N 01944 | Eubacterium ventriosum | ZP_02026680 | 7.9 | Yes | 154 | 14.9 | 2 | 5.78 |
| 17 | SR05257 | Putative serine protease F56F10.1 | C. elegans | P90893 | 2e−24 | Yes | 185 | 25.4 | 2 | 5.52 |
| 18 | SR02550 | Putative serine protease F56F10.1 | C. elegans | P90893 | 2e−35 | Yes | 239 | 11.7 | 2 | 5.22 |
| 19 | SS01082 | Hypothetical 86.9 kDa protein | B. malayi | XP_001896095 | 5e−51 | No | 309 | 6.8 | 2 | 5.22 |
| 20 | SR02018 | Yeast Glc seven-like Phosphatase | C. elegans | NP_491237 | 2e−94 | No | 184 | 13.0 | 2 | 4.74 |
| 21 | SR00716 | F09C8.1 | C. elegans | NP_510636 | 3e−45 | Yes | 170 | 22.4 | 2 | 4.66 |
| 22 | SR01063 | Aspartyl protease precursor | C. briggsae | CAP30637 | 2e−98 | Yes | 359 | 10.9 | 2 | 4.49 |
| 23 | SS00929 | High mobility group protein | C. elegans | NP_496970 | 5e−21 | No | 94 | 20.2 | 2 | 4.02 |
| 24 | SR00223 | Hypothetical protein C50B6.7 | C. elegans | NP_506303 | 6e−51 | Yes | 192 | 18.2 | 2 | 4.02 |
| 25 | SR00899 | Hypothetical protein CBG09313 | C. briggsae | XP_001674244 | 4e−11 | No | 231 | 13.9 | 2 | 4.01 |
Several nominally cytosolic proteins were identified among the 25 highest-scoring iL3-specific ESPs, including thiosulfate sulfuryltransferase (SR01803), short chain reductase/dehydrogenase (SR03119), and propionyl coenzyme A carboxylase (SR00383). The above mentioned control experiments, however, convinced us that these nominally cytosolic proteins are indeed bona fide ESPs. Other identified proteins include the nematode-specific transthyretin-like protein family member with a suggested role in the nervous system (30).
Homologs to previously reported ESP from other nematodes included a fatty-acid retinoid binding protein (SR02714) (31) and astacin, a metalloproteinase (SR11111), related to the proteases reported for the infective larval stages of related skin-invasive nematodes, such as S. stercoralis, Ancylostoma caninum, or Onchocerca volvulus (18, 32, 33). The full-length sequences of the S. ratti and the Onchocerca astacins have first been described by Borchert et al. (19). The notion that astacins were secreted was supported by the presence of a canonical signal peptide for secretion sequence. The release of metalloprotease from S. ratti iL3 was shown to be completely inhibited by cycloheximide (data not shown), confirming that astacin was indeed actively translated and secreted.
A third example of such ESP homologs was the EST cluster SR00386. BLAST search of this EST cluster identified, as best matching homolog, the third larva (L3)-specific L3NieAg from S. stercoralis, which was reported as a highly specific immunodiagnostic antigen of S. stercoralis (34) and appears to be weakly related to a group of proteins comprising the secretory vespid venom allergen family. However, the BLAST match was only scored with an E-value of 0.15 indicating either very little homology for this particular protein even between the two related Strongyloides species, or that the real ortholog has not yet been sequenced. These facts might indicate that Nie family proteins are possibly genus-specific among the Strongyloides spp.
ESP Specific for S. ratti Parasitic Females
In total, 79 proteins were identified solely in the parasitic female ESP. These proteins were assigned to protein digestion and folding (n = 11), heat-shock proteins (n = 3), carbohydrate metabolism (n = 4), nucleic acid metabolism (n = 17), structural proteins (n = 3), and proteins of other putative functions (n = 20). Interestingly, 11 proteins were not assigned to any function or specific protein. Nine of the pF-specific ESPs did not relate to any S. ratti or S. stercoralis EST clusters, i.e. they were not covered by the current Strongyloides EST cluster database. Instead, only homologous sequences from other nematodes e.g. C. elegans, Necator americanus, and Heterodera glycines were identified (see supplemental Table S4).
The 25 highest-scoring pF-specific proteins were listed in Table IIB. One of the most abundant proteins shows homology to a B. malayi-derived EF-hand protein family member that had putative calcium-binding activity. Another calcium-binding pF-specific ESP was the secreted protein acidic and rich in cysteine (SPARC), which had been shown to be an extracellular calcium-binding protein (35). The EST cluster corresponding to this SPARC homolog covered the N terminus of the protein and included a predicted a signal peptide for secretion, corroborating the fact that SPARC is a bone fide ESP.
Another interesting group of pF-specific proteins were heat shock proteins (HSPs). These included two small HSP proteins (SR00984, SR03349) homologous to proteins from C. elegans and the parasitic nematode T. spiralis, which was also detected in Trichinella pseudospiralis-derived excretome/secretome specimens (36). In addition, a protein with homology to C. elegans endoplasmin was identified. The endoplasmins belong to the group of HSPs and were important for the processing and transport of secreted proteins. The human endoplasmin precursor was also termed HSP-90 beta.
One protein function was particularly enriched in the pF-derived E/S proteome compared with the iL3 excretome/secretome, namely protein digestion and folding. The number of proteins assigned to this category increased from 3.6% (7/196) in iL3 to 13.9% (11/79) in pF. Within these 11 proteins, six putative proteases were identified as pF-specific E/S proteins: three metalloproteinases with homologues in S. stercoralis, A. caninum (37) and Nasonia vitripennis, respectively (it should be noted that the metalloproteinases identified as pF-specific differ from the astacin found abundantly in the iL3 E/S proteome fraction (see above)); one aspartyl proteinase-like protein (APR-2); and two EST clusters (SR03191 and SR01641) with pronounced homology to prolyl oligopeptidases (prolyl serine carboxypeptidase; E.C. 3.4.21.26, PSC).
ESP Specific for S. ratti Free-living Stages
In the E/S samples derived from the flS, only 35 stage-specific proteins were identified (supplemental Table S5). Interestingly, two of the 25 highest-scoring ESTs (Table IIC) did not share any significant homology with proteins in the public database (E < 10−5). These proteins might not only be stage- but also organism-specific. Another eight proteins showed sequence similarity to hypothetical proteins from C. elegans, C. briggsae, and/or B. malayi, i.e. proteins for which not much additional information is available. Taking together the proteins without similarity and the large number of hypothetical proteins highlights the fact that currently, there is very limited knowledge about the functions of the excretome/secretome of free-living stages in S. ratti.
The list of flS-specific ESPs featured several hydrolases, including two serine proteases, one aspartic protease, a lysozyme family member and a carbohydrate-hydrolyzing enzyme. One protein (SS00929) showed similarity to a high-mobility group box (HMGB) protein from C. elegans. However, it had been shown in C. elegans and B. malayi that such HMGB proteins were predominantly expressed in developing larvae (38); thus, the identification of this protein might indicate the presence of small amounts of contaminating L1 and L2 larvae in the cultures.
Verification of Stage Relation of S. ratti Proteins by Differential Gene Expression Through qRT-PCR Analysis
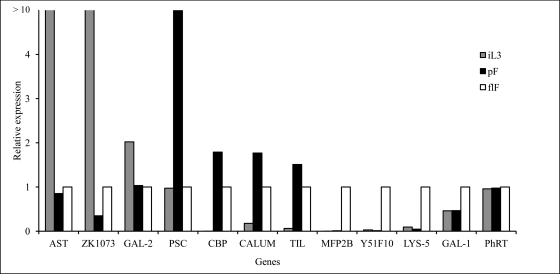
Quantitative real-time PCR (qRT-PCR) was used to measure relative transcription levels of 10 selected genes (Table I), whose products, i.e. proteins, were found to be of higher abundance in iL3, pF and flSs. These proteins could be analyzed in this experiment. The testing of stage-specific transcripts included (1) Sr-astacin (SR11111) and ZK1073.1 (SR02886) for the iL3 stage, (2) Sr-PSC-1 (SR01641), Sr-calumenin (SR00564), Sr-chitin-binding protein (SR04455), Sr-trypsin inhibitor-like protein (SR02054), and Sr-phosphoribosyl-transferase (SR02118) for the pF stage, and (3) MFP2B (SR00863), Sr-CBG22129 or Y51F10 (SR02091), and a Sr-lysozyme family protein (SR00671) representing the flS stage. The qRT-PCR results for each transcript were expressed relative to the respective transcript level of the flF stage. As a control, we used proteins that are constantly expressed during the life-cycle: Sr-gapdh and actin. The efficiency and linearity of qRT-PCR reactions were examined using 10-fold serial dilutions, indicating efficient amplification. Furthermore, each qRT-PCR reaction was performed on two biological replicates. The qRT-PCR reactions corroborated the findings of the proteomics experiments, i.e. increased transcript levels for those proteins that showed exclusive presence in the respective E/S proteome (Fig. 3). One exception was the Sr-galectin-2 (Sr-GAL-2; SR00627), which was observed in the ESP from all stages, but was found 2.1-fold up-regulated in iL3 whereas another galectin (Sr-Gal-1; SS00840) was found up-regulated in flF. In contrast, the phosphoribosyltransferase (Sr-PhRT) was identified as pF-specific. At the transcript level, however, no differential abundance could be measured for Sr-PhRT, indicating that the secretion of this protein was post-transcriptionally regulated either at the translational or post-translational level. The transcript levels for Sr-PhRT—although found as protein only in pF—showed such little variation that it can serve as control.
Fig. 3.
Stage-specific gene expression confirming stage-related occurrence of secreted proteins. The testing of stage-specific transcripts included the genes of Sr-astacin (SR11111; AST), ZK1073.1 (SR02886; ZK1073), Sr-galectin-2 (SS00627; GAL-2), Sr-PSC-1 (SR01641; PSC), Sr-chitin-binding protein (SR04455; CBP), Sr-calumenin (SR00564; CALUM), Sr-trypsin inhibitor-like protein (SR02054; TIL), MFP2B (SR00863), Sr-Y51F10.7 or CBG22129 (SR02091; Y51F10), a Sr-lysozyme family protein (SR00671; LYS-5), galectin-1 (SS00840; GAL-1), and Sr-phosphoribosyl-transferase (SR02118; PhRT). cDNA from infective larvae, parasitic and free-living females were used as a template in real-time quantitative PCR, applying a SYBR Green assay with gene-specific primers. Data were expressed as relative quantity of gene-specific transcription levels of free-living female stage as baseline (value = 1). The results of all samples were normalized to the expression levels of the constitutively expressed gene GAPDH. The experiments were performed twice.
Immune Recognition of S. ratti Proteins
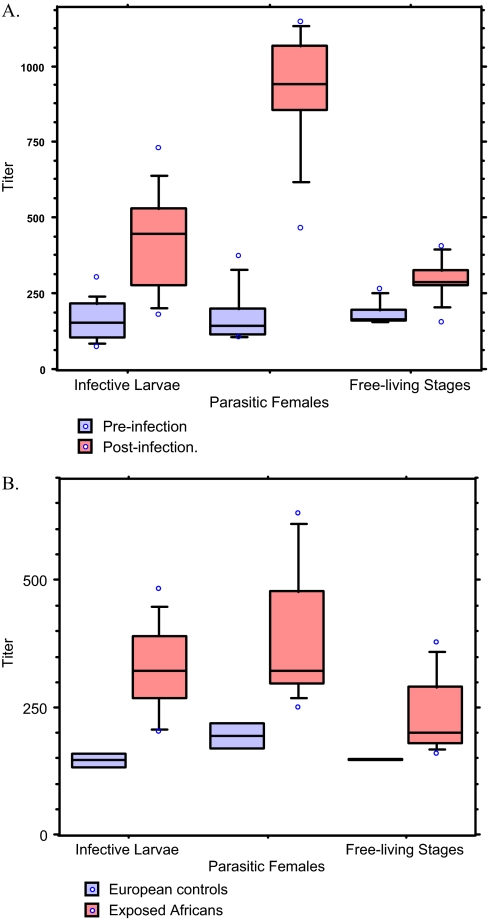
ELISA was performed to determine if the E/S proteins from iL3, pF and flS were targets for immune recognition. Sera from 10 rats were taken before and 32 days after subcutaneous infection with 1500 iL3. Furthermore, human sera from two healthy European individuals, i.e. from nonendemic area of strongyloidiasis and from ten individuals living in West Africa, i.e. an area endemic for the genetically closely related S. stercoralis and other intestinal helminths were examined. The ELISA revealed graduated IgG reactivities of sera from both Strongyloides-exposed rats and humans, with ESP from pF (high), iL3 (mid), and flS (low) (Figs. 4A, 4B; significances varied between p < 0.05 and p < 0.001).
Fig. 4.
Demonstration of IgG antibody reactivity with proteins in E/S products from iL3, pF and flS using sera from rats (A) and humans (B). ELISA titers—shown as box plots and quartiles—obtained (A) for 10 sera before (pre) and 32 days after (post) infection with S. ratti and (B) for two nonexposed Europeans not exposed to S. stercoralis and 10 Africans exposed to or infected with S. stercoralis. The antibody titers differed significantly (Mann-Whitney U test: p < 0.05 - p < 0.001) within the two respective groups of rat sera (for iL3: p < 0.001, pF: p < 0.001 and for fls: p < 0.05) and human sera for E/S products from iL3 and pF and flS (p < 0.05 for all).
Identification of Sr-prolyl Serine Carboxypeptidase (PSC-1) Full-Length Gene Sequence
In the pF E/S specimens, we identified several high-scoring pF-specific peptides that were assigned to two ESTs, both showing significant similarity to prolyl oligopeptidase (PSC-1; SR01641 and SR03191; see Table IIB). The S. ratti EST database contained a third cluster, SR03122, that had an overlapping N-terminal region with SR03191. Aligning SR01641, SR03191, and SR03122 resulted in a fragment with 454 amino acids (supplemental Fig. S3). The 5′ and 3′ RACE amplification was used to get the full-length Sr-PSC-1 cDNA sequence, which was confirmed by cloning and subsequent sequencing. Combining the previously known EST sequences and newly obtained 5′- and 3′ sequences resulted in the first full-length S. ratti prolyl oligopeptidase (prolyl serine carboxypeptidase; E.C. 3.4.21.26). The S. ratti prolyl oligopeptidase (PSC-1) contains an open reading frame of 2364 nucleotides encoding for a protein of 797 amino acids length and corresponding to a molecular weight of 91 kDa (supplemental Fig. S4; GenBank Accession number FJ011551.1). The protein sequence was added to the search database, and the ProteinPilot searches were repeated. The search identified 25 peptides with a confidence score of 99% and sequence coverage of 32%, resulting in an unused protein score of 65.71. Accordingly, Sr-PSC-1 was the highest-scoring pF-specific protein (with the caveat, that PSC-1 was one of the few full-length S. ratti proteins in the database used for the protein identification searches). In accordance, Sr-PSC-1 was found extremely up-regulated in pF applying the qRT-PCR (Fig. 3).
Sequence Analysis of Sr-PSC-1
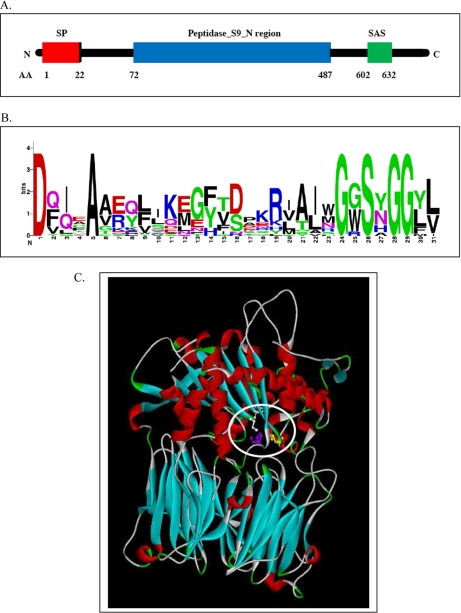
Supplemental Fig. S4 shows the full-length nucleotide and amino acid sequence of Sr-PSC-1. The yellow nucleotide sequences were the previously unknown residues of Sr-PSC-1. Using Prosite, the following features were identified: Sr-PSC-1 contained a signal peptide for secretion with the cleavage site between position 22 and 23 (Fig. 5A); the main domains of Sr-PSC-1 were the Peptidase_S9_N region ranging from amino acid residue 72 to 487 and the serine-active site (SAS) between amino acid residue 602 and 632 (supplemental Fig. S4, Fig. 5A). Aligning and comparing the sequences of 24 different prolyl oligopeptidase up- and downstream of the active-site serine residue identified six conserved amino acid residues (Fig. 5B): an aspartic acid residue in the –26 position, an alanine residue in the –21 postion, and glycine residues in the –2, +2, and +3 positions. The corresponding sequence logo can be found in Fig. 5B. The active-site serine (Ser627 in Sr-PSC-1) is part of the catalytic triad essential for serine proteases, which also includes Asp713 and His753 (39). The residues of this catalytic triad (Ser, Asp, His) are shown in red in supplemental Fig. S4.
Fig. 5.
A, Domain structure of the Sr-PSC-1. AA, amino acid; SP, signal peptide; SAS, serine active site. B, Sequence logo of the serine active site generated from multiple sequence alignments of 24 different prolyl oligopeptidase active sites. The total height of a logo position depends on the degree of conservation in the corresponding multiple sequence alignment column. Highly conserved alignment columns produce high logo positions. The tallest letters of six amino acids (D, A, G, S, G, G) correspond to the most conserved amino acids which are underlined in the sequence shown in the supplemental Fig. S4 (source http://us.expasy.org/cgi-bin/prosite). C, Ribbon diagram of Sr-PSC-1. The peptidase domain is located in the upper region, the β-propeller domain in the lower region. a) light blue - β-propeller structures, b) red - helical structures, c) white - Ser627, d) violet - His753, e) yellow - Asp713.
Using Swiss Model as protein structure homology-modeling service (template number: 1e5tA) we generated a model for the three-dimensional structure of Sr-PSC-1 (Fig. 5C) (40). Based on this three-dimensional model, the enzyme is cylindrical and consists of two domains, a peptidase domain and a seven-bladed beta-propeller. The catalytic triad is located in a large cavity at the interface of the two domains. The serine 627 (white) is found at the tip of a sharp turn and directly next to histidine 753 (violet), containing the catalytic imidazole group. The spatially adjacent aspartic acid 713 (yellow) and the histidine 753 are in contact via hydrogen bonds between one of the two oxygen atoms of the carboxylate group and the NH-group of the imidazole ring.
Inhibition of Sr-PSC-1 Enzyme Activity
Based on the finding that Sr-PSC-1 is (1) highly expressed (Fig. 3), (2) efficiently secreted, and (3) specific for the parasitic female stage, Sr-PSC-1 is potentially very important for drug development as the inhibition of this serine protease might affect relevant proteolytic activities. Prolyl oligopeptidases have recently gained pharma-ceutical interest, because prolyl oligopeptidase inhibitors have been shown to have anti-amnesic properties in rats, increase the brain levels of several neuropeptides (41) and improve cognition (42). We investigated the effect of prolyl oligopeptidase inhibitors on parasitic nematodes, which has never been done before. To this end, we added four different pyrrolidine derivatives (1A, B and 2A, B) (supplemental Fig. S1) (29) to in vitro cultures of parasitic females and observed their motility as proxy for their health status. All treated worms showed a dose-dependent decrease in motility when compared with the nontreated control group within 30 to 60 min of adding the prolyl oligopeptidase inhibitors (Fig. 6). The inhibitor compounds 2A and 2B were much more effective than the compounds 1A and 1B (29). The prolyl oligopeptidase inhibitor-induced change in motility was not reversible as replacing the culture medium with fresh medium without the inhibitors 18 h after the treatment did not result in recurrence of the motility confirming that motility is a good proxy for mortality.
Fig. 6.
Effect of different prolyl oligopeptidase inhibitors during in vitro culture of S. ratti parasitic female worms. Single graphs show different concentrations of compounds 1 (blue line, diamonds), 2 (red line, circles), 3 (green line, triangles), and 4 (purple line, x's). The mortality represents the percentage of worms that do not show any movements at a certain time point. The inhibitors included are: (1A) isophthalic acid 2(S)-(cyclopentanecarbonyl)pyrrolidine-l-prolyl-2(S)-cyanopyrrolidine amide; (1B) iso-phthalic acid 2(S)-(cyclopentanecarbonyl) pyrrolidine-l-prolyl-2(S)-(hydroxy-acetyl)-pyrrolidine amide; (2A) 4-phenylbutanoyl-l-prolyl-pyrrolidine; (2B) 4-phenyl-butanoyl-l-prolyl-2(S)-cyanopyrrolidine (56).
DISCUSSION
E/S products (ESPs) secreted by cells and organisms play pivotal biological roles across a wide range of parasitic organisms. Representing the primary interface between the parasite and the host, the E/S components include proteins involved in biological processes like cell migration, cell adhesion, cell-cell communication, proliferation, differentiation, morphogenesis, and the regulation of immune responses (43). We have undertaken the proteomic study of S. ratti as model organism to investigate the parasite-host interactions in parasitic nematode infections. Accordingly, we aimed to provide insights into the E/S proteome of S. ratti and the interplay of the excretome/secretome with the development of S. ratti which has a complex life cycle with free-living stages, each having unique roles in host-pathogen interactions. In fact, for the entire genus Strongyloides, only a single proteomic study has been published (44). To this end, we isolated E/S specimens from three distinct developmental stages: iL3, pF, and flS.
Prior to starting this endeavor we showed that the proteins present in the culture supernatant are indeed caused by secretion and not because of leakage of dead or damaged cells. Interestingly, the pattern of protein bands differed strongly between ESPs and somatic crude extracts of the stages (supplemental Fig. S2). Furthermore, a rank-order correlation of the identified ESP and extract proteins indicated little to no rank-order correlation, thus providing further evidence in support of secretion and against worm lysis. We applied cycloheximide, a potent inhibitor of protein translation, in the iL3 cultures. In addition, we used azide treatment and exposure to unphysiological conditions at 4 °C and 70 °C. The almost complete reduction of proteins in the supernatant and also the disappearance of the astacin metalloproteinase, which was the most prominent proteolytic component in the gelatin gels, confirmed bona fide excretion/secretion and excluded leakage and/or random lysis as reason for observing the identified proteins. This validation experiment was important, as our analysis identified numerous proteins normally considered as cytosolic or nuclear. However, recent studies have shown that many proteins, including peroxidoxin, galectins, heat shock proteins, and high-mobility group box 1, were indeed excreted and secreted proteins (45–47).
Once we confirmed that excretion and secretion was the cause for observing the proteins in the supernatant, we used SDS-PAGE in combination with LC/MS (GeLC/MS) for a comprehensive analysis of the different excretomes/secretomes during S. ratti development. In total 586 ESPs were identified. Our study is the first proteomic study of S. ratti-derived specimens and furthermore is the largest proteomic study in the genus Strongyloides by far. Previously, only a single proteomic study of Strongyloides-derived samples been published and this study identified merely 26 proteins (44).
Any proteomic study in Strongyloides (or many other parasitic nematodes) is hampered by the limited availability of protein sequence information. However, the use of available protein and EST sequences from S. ratti and S. stercoralis (combined with other nematodes) and new technologies can partially overcome the problems associated with lack of organism-specific sequence information. For instance, new search engines, such as ProteinPilot, allow for database-wide substitutions during the protein identification search. Thus, despite this lack of sequence information we did generate the second largest nematode E/S proteome map to date. The only larger nematode E/S study was recently published by Bennuru et al. (13) who studied the B. malayi E/S proteome and profited from the availability of the complete genome sequence (48).
Proteins from a wide range of biological processes and functions were identified in our study including cell migration, cell adhesion, cell-cell communication, proliferation, differentiation, morphogenesis, and the regulation of immune responses. In addition, numerous proteins of unknown function and hypothetical proteins were found. For the subsequent analysis of stage-specific E/S proteins, we did not attempt any detailed quantitative analysis of the identified proteins. Instead, we followed a stringent binary observed/not observed approach.
Recently, a microarray with 2,227 putative genes was used to identify genes likely to play a key role in the parasitic life of S. ratti (49). In this report, the microarray was probed with cDNA prepared from parasites subjected to low or high immune pressures, i.e. harvested 6 and 15 days postinfection, respectively. Comparison of these transcript expression data with our proteomic data identified several proteins which were specific for a particular stage or were expressed in two or three studied stages. For example, the cluster SR00984, which relates to a small heat-shock protein (HSP), is only observed as an ESP in pF-derived specimens and as an actively transcribed gene. Of note, the only stage-specific excreted and secreted heat shock proteins were found in the pF-derived specimens (50). In addition, numerous other nonstage specific HSPs were identified, including the abundant Sr-HSP-10 and Sr-HSP-60 proteins, which were recently partially characterized (17).
The stage-specific transcripts from the free-living and parasitic stages, which were also observed in the qRT-PCR of selected S. ratti genes, confirmed the stage-specific expression found in the proteomic data. One notable exception was PhRT, which was identified only in the pF sample using proteomics, but showed equal transcript levels throughout S. ratti developmental stages. A possible explanation for this observation is that although the majority of the stage-specific ESPs are controlled at the transcriptional level, the excretion and secretion of PhRT is controlled post-transcriptionally, i.e. by timed translation or by post-translational events, such as stage-specific post-translational modifications. The remarkable diversity of the expression levels of the various genes in the iL3, pF, and flF, as determined in our study, reflects changes associated with the transition to parasitic lifestyle and the adaptation to the host. These include the iL3-specific Sr astacin (Sr-AST), which is homologous to a promising vaccine candidate against the hookworm infections (51). The astacin-like metalloprotease Ac-MTP-1 (32) is specifically secreted by the iL3 of the hookworm, showing the same stage specificity as Sr-AST (SR11111) in our study. Work from our laboratory had characterized a homologous astacin in the filaria O. volvulus (19). The Sr-AST identified in the iL3 specimens corresponded to a full-length astacin metalloproteinase sequence that was identified in our laboratory based on a homologous S. stercoralis sequence (19). It was interesting to note that this metalloprotease showed elevated expression in iL3 stages of both S. ratti and S. stercoralis (32), thus underlining its putative role to facilitate skin penetration at the initiation of infection. Besides the iL3-specific astacin, two other EST clusters (SR03587 and SR02663) were identified that showed similarities to astacins. However, these two astacins were pF-specific, thus indicating that different astacins were expressed during different developmental stages. These results show protease activities tailored to the needs of each life cycle stage.
S. ratti prolyl oligopeptidase Sr-PSC-1 is another example of a stage-specific protease that has been exclusively identified in parasitic females. This protein was only partially covered by the EST clusters used for the protein identification. For further characterization of this protein, we cloned the full-length gene; its characteristics are shown in supplemental Fig. S4 and in Fig. 5. Sr-PSC-1 represents a typical serine protease characterized by the conserved catalytic triad (aaSer, aaAsp and aaHis). A homolog of Sr-PSC-1 with 37% identity with the whole protein was found in B. malayi, but shared only 52% identity with the C-terminal region that contained the catalytic domain (www.nematodes.org/downloads/databases/). In contrast, no significant similarity on a primary sequence level was found with the nonparasitic nematode C. elegans, which was unusual because a majority of proteins were commonly shared between C. elegans and other nematode species. This may suggest that this protease played a role in parasite establishment within the host, a notion that was supported by the observation that a similar stage-specific gene expression for PSC-1 had been reported before in Teladorsagia circumcincta (52). The stage-specificity of PSC-1 in S. ratti was supported by the fact that the ESTs encoding PSC-1 originated only from the EST libraries generated from pF. In addition, the comparative PCR analysis (Fig. 3) only showed a positive result for parasitic female cDNA. Interestingly, the EST cluster SR04440, which in the present study occurred in the extracts of pF (data not shown), was also found in a microarray analysis in samples from pF under high-immune pressure (49). The Strongyloides PSC-1 represents a novel abundant stage-specific protein that might have relevance for the containment of parasitism.
The serine proteases of the prolyl oligopeptidase family were previously reported in the protozoan parasites Trypanosoma brucei and Leishmania major (29). In these parasites, the prolyl oligopeptidase was likely to be involved in host cell invasion and hydrolysis of host proteins (53, 54). Prolyl oligopeptidase had also been studied as a potential therapeutic agent for the treatment of celiac sprue, an inflammatory disease of the small intestine (55). In vertebrates, prolyl oligopeptidase activity had been found throughout the body, with a highest concentration within the brain (56). The prolyl oligopeptidase family S9 was comprised of beta-hydrolase enzymes also sharing the classical catalytic triad. The 80-kDa prolyl oligopeptidase were able to hydrolyze the peptide bond on the carboxyl side of internal proline residues. Evaluation of the sequence data from parasitic S. ratti females revealed that the culture supernatants contained sequences matching EST clusters SR01641 and SR03191 that showed homology to prolyl oligopeptidase (22).
Prolyl oligopeptidase had been studied in human and mouse in the context of their neurological roles. The high occurrence of prolyl oligopeptidase in the brain suggested that it was involved in the maturation and degradation of peptide hormones and neuropeptides, such as substance P, oxytocin, and angiotensin. Published data describe different prolyl oligopeptidase inhibitors, that are active in vitro and in vivo in humans and rodents and increase levels of several neuropeptides in the brain (57). When testing for the effects of several pyrrolidine-derived prolyl oligopeptidase inhibitors on S. ratti parasitic females, we observed a clear concentration- and time-dependent reduction in motility (Fig. 6), which was used in the present study as proxy for vitality status of S. ratti. This observation might lead to novel treatment options for strongyloidasis.
A variety of proteins that we have identified in the S. ratti ESP are homologous to proteins from other nematode species that may have been involved in either the containment of parasitism or the suppression or induction of host-immune responses. These included anti-oxidative proteins (thioredoxin peroxidase, glutathione peroxidase, superoxide dismutase), various proteinases, serpin, galectins, small HSPs, and macrophage migration inhibitory factor (50,58). Rats infected by S. ratti form a marked immunity against a challenge infection when immunized with ESP fraction from adult worms (59). Thus, ESP from pF can influence the host defense system. S. ratti infection was shown to induce transient nematode-specific Th2 response characterized by the generation of interleukin-4, -5, and -13 that foster eosinophilic granulocytes and mast cells and induce IgG4 and IgE antibody isotype production, which were involved in effector responses (60). Interestingly, ESP from pF were found to be strongly recognized by IgG in sera from S. ratti-infected rats and Strongyloides-exposed persons living in Africa, whereas ESP from iL3 showed a lower reactivity and ESP from flS were hardly recognized (Fig. 4).
Although the main focus of our study was the identification of stage-specific ESPs, analyzing the list of commonly found ESPs was also very revealing. Proteins abundantly observed in all three investigated developmental stages were—among others—HSPs, galectins, and proteins involved in oxidative phosphorylation, carbohydrate synthesis and metabolism, biosynthesis, developmental processes, sugar- and fatty acid-binding (supplemental Table S2). In addition, similar to the findings in previous studies of the excretomes/secretomes of parasitic nematodes, we identified several homologous structural proteins such as actin, profiling, or myosin (1, 13, 43, 61–63). Furthermore, we identified tropomyosin, a fibrillar protein involved in the contraction of muscle cells, in the stage independent E/S product. The identification of tropomyosin as an E/S product was not unprecedented, as proteins from the tropomyosin family were also detected in the secretions of adult B. malayi stages (61) and S. mansoni cercariae (64). Similarly, Hartmann et al. (65) showed that birds are partially protected after infection with A. viteae infective larvae (iL3) infection when immunized with recombinant tropomyosin. Thus, it can be concluded that the tropomyosin is a widely occurring allergen excreted/secreted by numerous tissue- and intestine-dwelling nematodes (66).
In the present study, almost 600 proteins were identified in the E/S products derived from different developmental stages of S. ratti. Half of the E/S products were found in two stages, the largest number (>33%) in iL3 and 13% in pF. Individual parasitic stage-specific proteins were expected to exhibit important biological functions in the general, as well as stage-specific parasite-host-interaction and may reveal immunomodulatory activities, which are of relevance for both the parasite in its mucosal habitat and for the infested host (67). A biological role for the iL3-secreted astacin, the Nie antigen and a fatty acid retinoid binding protein, parasitic female-released prolyl oligopeptidase, small heat shock proteins, and a secreted acidic and rich in cysteine-related protein may be anticipated. These identified proteins include putative immunemodulators that will strengthen research aimed to develop novel intervention tools to control the parasite and prevent the diseases.
Acknowledgments
We thank Dr. G. Pluschke (Department Medical Parasitology and Infection Biology, Swiss Tropical and Public Health Institute, Basel, Switzerland) for providing the S. ratti cycle. We thank Dr. Elina M. Jarho (Department of Pharmaceutical Chemistry, University of Kuopio, Finland) for providing the prolyl oligopeptidase inhibitors. The expert assistance of Mrs. K. Krausz and M. Lintzel (Bernhard Nocht Institute), Dr. Christina M. Taylor (The Genome Institute, Washington University School of Medicine, St. Louis), and Zachary Waldon, Peter Warren and Yin Yin Lin (The Proteomics Center at Children's Hospital Boston) is appreciated. We acknowledge the support of Dr. Peter U. Fischer (Department of Internal Medicine, Infectious Diseases Division, Washington University School of Medicine, St. Louis, USA) in the data analysis and helpful discussion. We thank Dr. J. Paulo (The Proteomics Center at Children's Hospital Boston) for critical reading the manuscript. Data of this work form major parts of the doctoral theses by Soblik H and Younis, A. E. in the Faculty of Natural Sciences of the University of Hamburg, Germany.
Footnotes
* This work was supported by the Leibniz Association and the Vereinigung der Freunde des Tropeninstituts Hamburg, the Boehringer Ingelheim Fonds (H. S.), the Egyptian Ministry of Higher Education (A. E. Y.) and the NIH-NIAID support (M. M.).
 This article contains supplemental Figs. S1 to S4 and Tables S1 to S5.
This article contains supplemental Figs. S1 to S4 and Tables S1 to S5.
1 The abbreviations used are:
- E/S
- excretory/secretory
- ESP
- excretory/secretory proteins
- iL3
- infective larvae
- pF
- parasitic female
- flS
- free-living stages
- flF
- free-living females
- PSC
- prolyl serine carboxypeptidase
- EST
- expressed sequence tag.
REFERENCES
- 1. Nagaraj S. H., Gasser R. B., Ranganathan S. (2008) Needles in the EST haystack: large-scale identification and analysis of excretory-secretory (ES) proteins in parasitic nematodes using expressed sequence tags (ESTs). PLoS Negl. Trop. Dis. 2, e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hewitson J. P., Grainger J. R., Maizels R. M. (2009) Helminth immuno regulation: the role of parasite secreted proteins in modulating host immunity. Mol. Biochem. Parasitol. 167, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Woolhouse M. E., Webster J. P., Domingo E., Charlesworth B., Levin B. R. (2002) Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 32, 569–577 [DOI] [PubMed] [Google Scholar]
- 4. Rook G. A. (2007) The hygiene hypothesis and the increasing prevalence of chronic inflammatory disorders. Trans. R. Soc. Trop. Med. Hyg. 101, 1072–1074 [DOI] [PubMed] [Google Scholar]
- 5. Viney M. E. (2006) The biology and genomics of Strongyloides. Med. Microbiol. Immunol. 195, 1–6 [DOI] [PubMed] [Google Scholar]
- 6. Grove D. I. (1996) Human strongyloidiasis. Adv. Parasitol. 38, 251–309 [DOI] [PubMed] [Google Scholar]
- 7. Keiser P. B., Nutman T. B. (2004) Strongyloides stercoralis in the immuno compromised population. Clin. Microbiol. Rev. 17, 208–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olsen A., van Lieshout L., Marti H., Polderman T., Polman K., Steinmann P., Stothard R., Thybo S., Verweij J. J., Magnussen P. (2009) Strongyloidiasis – the most neglected of the neglected tropical diseases? Trans. R. Soc. Trop. Med. Hyg. 103, 967–972 [DOI] [PubMed] [Google Scholar]
- 9. Marcos L. A., Terashima A., Dupont H. L., Gotuzzo E. (2008) Strongyloides hyperinfection syndrome: an emerging global infectious disease. Trans. R. Soc. Trop. Med. Hyg. 102, 314–318 [DOI] [PubMed] [Google Scholar]
- 10. Montes M., Sawhney C., Barros N. (2010) Strongyloides stercoralis: there but not seen. Curr. Opin. Infect. Dis. 23, 500–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kramme S., Nissen N., Soblik H., Erttmann K., Tannich E., Fleischer B., Panning M., Brattig N. W. (2010) Novel real-time PCR for the universal detection of Strongyloides spp. J. Med. Microbiol. 60, 454–458 [DOI] [PubMed] [Google Scholar]
- 12. Lok J. B. (2007) Strongyloides stercoralis: a model for translational research on parasitic nematode biology. WormBook. 17, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bennuru S., Semnani R., Meng Z., Ribeiro J. M., Veenstra T. D., Nutman T. B. (2009) Brugia malayi excreted/secreted proteins at the host/parasite interface: stage- and gender-specific proteomic profiling. PLoS Negl. Trop. Dis. 3, e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Keiser J., Thiemann K., Endriss Y., Utzinger J. (2008) Strongyloides ratti: in vitro and in vivo activity of tribendimidine. PLoS Negl. Trop. Dis. 2, e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Viney M. E., Lok J. B. (2007) Strongyloides spp. WormBook. 23, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Whitehead A. G., Hemming J. R. (1965) A comparison of some quantitative methods of extracting small vermiform nematodes from soil. Ann. Appl. Biol. 55, 25–38 [Google Scholar]
- 17. Tazir Y., Steisslinger V., Soblik H., Younis A. E., Beckmann S., Grevelding C. G., Steen H., Brattig N. W., Erttmann K. D. (2009) Molecular and functional characterisation of the heat shock protein 10 of Strongyloides ratti. Mol. Biochem. Parasitol. 168, 149–157 [DOI] [PubMed] [Google Scholar]
- 18. McKerrow J. H., Pino-Heiss S., Lindquist R., Werb Z. (1985) Purification and characterization of an elastinolytic proteinase seceted by cercariae of Schistosoma mansoni. J. Biol. Chem. 260, 3703–3707 [PubMed] [Google Scholar]
- 19. Borchert N., Becker-Pauly C., Wagner A., Fischer P., Stöcker W., Brattig N. W. (2007) Identification and characterization of onchoastacin, an astacin-like metalloproteinase from the filaria Onchocerca volvulus. Microbes Infect. 9, 498–506 [DOI] [PubMed] [Google Scholar]
- 20. Renard B. Y., Kirchner M., Monigatti F., Ivanov A. R., Rappsilber J., Winter D., Steen J. A., Hamprecht F. A., Steen H. (2009) When less can yield more - Computational preprocessing of MS/MS spectra for peptide identification. Proteomics 9, 4978–4984 [DOI] [PubMed] [Google Scholar]
- 21. Renard B. Y., Timm W., Kirchner M., Steen J. A., Hamprecht F. A., Steen H. (2010) Estimating the confidence of peptide identifications without decoy databases. Anal Chem. 82, 4314–4318 [DOI] [PubMed] [Google Scholar]
- 22. Mitreva M., McCarter J. P., Martin J., Dante M., Wylie T., Chiapelli B., Pape D., Clifton S. W., Nutman T. B., Waterston R. H. (2004) Comparative genomics of gene expression in the parasitic and free-living nematodes Strongyloides stercoralis and Caenorhabditis elegans. Genome Res. 14, 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin J., Abubucker S., Wylie T., Yin Y., Wang Z., Mitreva M. (2009) Nematode.net update 2008: improvements enabling more efficient data mining and comparative nematode genomics. Nucleic Acids Res. 37, D571–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 25. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using realtime quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 27. Mpagi J. L., Büttner D. W., Tischendorf F. W., Erttmann K. D., Brattig N. W. (2000) Humoral responses to a secretory Onchocerca volvulus protein: differences in the pattern of antibody isotypes to recombinant Ov20/OvS1 in generalized and hyperreactive onchocerciasis. Parasite Immunol. 22, 455–460 [DOI] [PubMed] [Google Scholar]
- 28. Tischendorf F. W., Brattig N. W., Büttner D. W., Pieper A., Lintzel M. (1996) Serum levels of eosinophil cationic protein, eosinophil-derived neurotoxin and myeloperoxidase in infections with filariae and schistosomes. Acta Trop. 62, 171–182 [DOI] [PubMed] [Google Scholar]
- 29. Venäläinen J. I., Juvonen R. O., Männistö P. T. (2004) Evolutionary relationships of the prolyl oligopeptidase family enzymes. Eur. J. Biochem. 271, 2705–2715 [DOI] [PubMed] [Google Scholar]
- 30. Jacob J., Vanholme B., Haegeman A., Gheysen G. (2007) Four transthyretin-like genes of the migratory plant-parasitic nematode Radopholus similis: members of an extensive nematode-specific family. Gene 402, 9–19 [DOI] [PubMed] [Google Scholar]
- 31. Mpagi J. L., Erttmann K. D., Brattig N. W. (2000) The secretory Onchocerca volvulus protein OvS1/Ov20 exhibits the capacity to compete with serum albumin for the host's long-chain fatty acids. Mol. Biochem. Parasitol. 105, 273–279 [DOI] [PubMed] [Google Scholar]
- 32. Gomez Gallego S., Loukas A., Slade R. W., Neva F. A., Varatharajalu R., Nutman T. B., Brindley P. J. (2005) Identification of an astacin-like metalloproteinase transcript from the iL3 of Strongyloides stercoralis. Parasitol. Int. 54, 123–133 [DOI] [PubMed] [Google Scholar]
- 33. Williamson A. L., Lustigman S., Oksov Y., Deumic V., Plieskatt J., Mendez S., Zhan B., Bottazzi M. E., Hotez P. J., Loukas A. (2006) Ancylostoma caninum MTP-1, an astacin-like metalloprotease secreted by infective hookworm larvae is involved in tissue migration. Infect. Immun. 7 4, 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ravi V., Ramachandran S., Thompson R. W., Andersen J. F., Neva F. A. (2002) Characterization of a recombinant immunodiagnostic antigen (NIE) from Strongyloides stercoralis L3-stage larvae. Mol. Biochem. Parasitol. 125, 73–81 [DOI] [PubMed] [Google Scholar]
- 35. Schwarzbauer J. E., Musset-Bilal F., Ryan C. S. (1994) Extracellular calcium-binding protein SPARC/osteonectin in Caenorhabditis elegans. Methods Enzymol. 245, 257–270 [DOI] [PubMed] [Google Scholar]
- 36. Ko R. C., Fan L. (1996) Heat shock response of Trichinella spiralis and T. pseudospiralis. Parasitology 112, 89–95 [DOI] [PubMed] [Google Scholar]
- 37. Feng J., Zhan B., Liu Y., Liu S., Williamson A., Goud G., Loukas A., Hotez P. (2007) Molecular cloning and characterization of Ac-MTP-2, an astacin-like metalloprotease released by adult Ancylostoma caninum. Mol. Biochem. Parasitol. 152, 132–138 [DOI] [PubMed] [Google Scholar]
- 38. Jiang L. I., Sternberg P. W. (1999) An HMG1-like protein facilitates Wnt signaling in Caenorhabditis elegans. Genes Dev. 13, 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Szeltner Z., Polgár L. (2008) Structure, function and biological relevance of prolyl oligopeptidase. Curr. Protein. Pept. Sci. 9, 96–107 [DOI] [PubMed] [Google Scholar]
- 40. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 41. Atack J. R., Suman-Chauhan N., Dawson G., Kulagowski J. J. (1991) In vitro and in vivo inhibition of prolyl endopeptidase. Eur. J. Pharmacol. 205, 157–163 [DOI] [PubMed] [Google Scholar]
- 42. Schneider J. S., Giardiniere M., Morain P. (2002) Effects of the prolyl endopeptidase inhibitor S 17092 on cognitive deficits in chronic low dose MPTP-treated monkeys. Neuropsychopharmacol 26, 176–182 [DOI] [PubMed] [Google Scholar]
- 43. Maizels R. M., Yazdanbakhsh M. (2003) Immune regulation by helminth parasites: cellular and molecular mechanisms. Nat. Rev. Immunol. 3, 733–744 [DOI] [PubMed] [Google Scholar]
- 44. Marcilla A., Sotillo J., Pérez-Garcia A., Igual-Adell R., Valero M. L., Sánchez-Pino M. M., Bernal D., Muñoz-Antolí C., Trelis M., Toledo R., Esteban J. G. (2010) Proteomic analysis of Strongyloides stercoralis L3 larvae. Parasitology 137, 1577–1583 [DOI] [PubMed] [Google Scholar]
- 45. Rubartelli A., Lotze M. T. (2007) Inside, outside, upside down: damage-associated molecular-pattern molecules (DAMPs) and redox. Trends Immunol. 28, 429–436 [DOI] [PubMed] [Google Scholar]
- 46. Robinson M. W., Hutchinson A. T., Donnelly S., Dalton J. P. (2010) Worm secretory molecules are causing alarm. Trends Parasitol. 26, 371–372 [DOI] [PubMed] [Google Scholar]
- 47. Robinson M. W., Hutchinson A. T., Dalton J. P., Donnelly S. (2010) Peroxiredoxin: a central player in immune modulation. Parasite Immunol. 32, 305–313 [DOI] [PubMed] [Google Scholar]
- 48. Ghedin E., Wang S., Spiro D., Caler E., Zhao Q., Crabtree J., Allen J. E., Delcher A. L., Guiliano D. B., Miranda-Saavedra D., Angiuoli S. V., Creasy T., Amedeo P., Haas B., El-Sayed N. M., Wortman J. R., Feldblyum T., Tallon L., Schatz M., Shumway M., Koo H., Salzberg S. L., Schobel S., Pertea M., Pop M., White O., Barton G. J., Carlow C. K., Crawford M. J., Daub J., Dimmic M. W., Estes C. F., Foster J. M., Ganatra M., Gregory W. F., Johnson N. M., Jin J., Komuniecki R., Korf I., Kumar S., Laney S., Li B. W., Li W., Lindblom T. H., Lustigman S., Ma D., Maina C. V., Martin D. M., McCarter J. P., McReynolds L., Mitreva M., Nutman T. B., Parkinson J., Peregrin-Alvarez J. M., Poole C., Ren Q., Saunders L., Sluder A. E., Smith K., Stanke M., Unnasch T. R., Ware J., Wei A. D., Weil G., Williams D. J., Zhang Y., Williams S. A., Fraser-Liggett C., Slatko B., Blaxter M. L., Scott A. L. (2007) Draft genome of the filarial nematode parasite Brugia malayi. Science 317, 1756–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thompson F. J., Barker G. L., Hughes L., Viney M. E. (2008) Genes important in the parasitic life of the nematode Strongyloides ratti. Mol. Biochem. Parasitol. 158, 112–119 [DOI] [PubMed] [Google Scholar]
- 50. Younis A. E., Geisinger F., Ajonina-Ekoti I., Soblik H., Steen H., Mitreva M., Erttmann K. D., Perbandt M., Liebau E., Brattig N. W. (2011) Stage-specific excretory-secretory small heat shock proteins from the parasitic nematode Strongyloides ratti – putative links to host's intestinal mucosal defense system. FEBS J. 278, 3319–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hotez P. J., Ashcom J., Zhan B., Bethony J., Loukas A., Hawdon J., Wang Y., Jin Q., Jones K. C., Dobardzic A., Dobardzic R., Bolden J., Essiet I., Brandt W., Russell P. K., Zook B. C., Howard B., Chacon M. (2003) Effect of vaccination with a recombinant fusion protein encoding an astacinlike metalloprotease (MTP-1) secreted by host-stimulated Ancylostoma caninum third-stage iL3. J. Parasitol. 89, 853–855 [DOI] [PubMed] [Google Scholar]
- 52. Nisbet A. J., Redmond D. L., Matthews J. B., Watkins C., Yaga R., Jones J. T., Nath M., Knox D. P. (2008) Stage-specific gene expression in Teladorsagia circumcincta (Nematoda: Strongylida) iL3 and early parasitic stages. Int. J. Parasitol. 38, 829–838 [DOI] [PubMed] [Google Scholar]
- 53. Grellier P., Vendeville S., Joyeau R., Bastos I. M., Drobecq H., Frappier F., Teixeira A. R., Schrével J., Davioud-Charvet E., Sergheraert C., Santana J. M. (2001) Trypanosoma cruzi prolyl oligopeptidase Tc80 is involved in nonphagocytic mamalian cell invasion by trypomastigotes. J. Biol. Chem. 276, 47078–47086 [DOI] [PubMed] [Google Scholar]
- 54. Bastos I. M., Motta F. N., Charneau S., Santana J. M., Dubost L., Augustyns K., Grellier P. (2010) Prolyl oligopeptidase of Trypanosoma brucei hydrolyzes native collagen, peptide hormones and is active in the plasma of infected mice. Microbes Infect. 12, 457–466 [DOI] [PubMed] [Google Scholar]
- 55. Ehren J., Govindarajan S., Morón B., Minshull J., Khosla C. (2008) Protein engineering of improved prolyl endopeptidases for celiac sprue therapy. Protein Eng. Des. Sel. 21, 699–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Irazusta J., Larrinaga G., Gonzáles-Maesto J., Gil J., Meana J. J., Casis L. (2002) Distribution of prolyl endopeptidase activities in rat and human brain. Neurochem. Int. 40, 337–345 [DOI] [PubMed] [Google Scholar]
- 57. Venäläinen J. I., Garcia-Horsman J. A., Forsberg M. M., Jalkanen A., Wallén E. A. A., Jarho E. M., Christiaans J. A., Gynther J., Männistö P. T. (2006) Binding kinetics and duration of in vivo action of novel prolyl oligopeptidase inhibitors. Biochem. Pharmacol. 71, 683–692 [DOI] [PubMed] [Google Scholar]
- 58. Younis A. E., Soblik H., Ajonina-Ekoti I., Erttmann K. D., Luersen K., Liebau E., Brattig N. W. (2011) Characterization of a secreted macrophage migration inhibitory factor homologue of the parasitic nematode Strongyloides acting at the parasite-host cell interface. Microbes Infect. October 17 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 59. Mimori T., Tanaka M., Tada I. (1987) Strongyloides ratti: Formation of protection in rats by excretory/secretory products of adult worms. Exp. Parasitol., 64, 342–346 [DOI] [PubMed] [Google Scholar]
- 60. Eschbach M. L., Klemm U., Kolbaum J., Blankenhaus B., Brattig N., Breloer M. (2010) Strongyloides ratti infection induces transient nematode-specific Th2 response and reciprocal suppression of IFN-gamma production in mice. Parasite Immunol. 32, 370–383 [DOI] [PubMed] [Google Scholar]
- 61. Hewitson J. P., Harcus Y. M., Curwen R. S., Dowle A. A., Atmadja A. K., Ashton P. D., Wilson A., Maizels R. M. (2008) The secretome of the filarial parasite, Brugia malayi: proteomic profile of adult excretory-secretory products. Mol. Biochem. Parasitol. 160, 8–21 [DOI] [PubMed] [Google Scholar]
- 62. Mulvenna J., Hamilton B., Nagaraj S. H., Smyth D., Loukas A., Gorman J. J. (2009) Proteomics analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum. Mol. Cell. Proteomics. 8, 109–121 [DOI] [PubMed] [Google Scholar]
- 63. Robinson M. W., Menon R., Donnelly S. M., Dalton J. P., Ranganathan S. (2009) An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica: proteins associated with invasion and infection of the mammalian host. Mol. Cell. Proteomics. 8, 1891–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Knudsen G. M., Medzihradszky K. F., Lim K. C., Hansell E., McKerrow J. H. (2005) Proteomic analysis of Schistosoma mansoni cercarial secretions. Mol. Cell. Proteomics 4, 1862–1875 [DOI] [PubMed] [Google Scholar]
- 65. Hartmann S., Sereda M. J., Sollwedel A., Kalinna B., Lucius R. (2006) A nematode allergen elicits protection against challenge infection under specific conditions. Vaccine 24, 3581–3590 [DOI] [PubMed] [Google Scholar]
- 66. Sereda M. J., Hartmann S., Büttner D. W., Volkmer R., Hovestädt M., Brattig N., Lucius R. (2010) Characterization of the allergen filarial tropomyosin with an invertebrate specific monoclonal antibody. Acta Trop. 116, 61–67 [DOI] [PubMed] [Google Scholar]
- 67. Johnston M. J., MacDonald J. A., McKay D. M. (2009) Parasitic helminths: a pharmacopeia of anti-inflammatory molecules. Parasitology 136, 125–147 [DOI] [PubMed] [Google Scholar]