Abstract
Patients with liver metastases from colon carcinoma show highly variable responses to chemotherapy and tumor recurrence is frequently observed. Therapy-resistant cancer stem cells have been implicated in drug resistance and tumor recurrence. However, the factors determining therapy resistance and tumor recurrence are poorly understood. The aim of this study was to gain insight into these mechanisms by comparing the proteomes of patient-derived cancer stem cell cultures and their differentiated isogenic offspring.
We established colonosphere cultures derived from resection specimens of liver metastases in patients with colon cancer. These colonospheres, enriched for colon cancer stem cells, were used to establish isogenic cultures of stably differentiated nontumorigenic progeny. Proteomics based on one-dimensional gel electrophoresis coupled to nano liquid chromatography tandem MS was used to identify proteome differences between three of these paired cultures. The resulting data were analyzed using Ingenuity Pathway Software.
Out of a total data set of 3048 identified proteins, 32 proteins were at least twofold up-regulated in the colon cancer stem cells when compared with the differentiated cells. Pathway analysis showed that “cell death ” regulation is strikingly different between the two cell types. Interestingly, one of the top-up-regulated proteins was BIRC6, which belongs to the class of Inhibitor of Apoptosis Proteins. Knockdown of BIRC6 sensitized colon cancer stem cells against the chemotherapeutic drugs oxaliplatin and cisplatin.
This study reveals that differentiation of colon cancer stem cells is accompanied by altered regulation of cell death pathways. We identified BIRC6 as an important mediator of cancer stem cell resistance against cisplatin and oxaliplatin. Targeting BIRC6, or other Inhibitors of Apoptosis Proteins, may help eradicating colon cancer stem cells.
Treatment of colorectal cancer patients with chemotherapy is characterized by highly divergent tumor responses, but tumor recurrence is almost always observed. Therefore, chemotherapy is not considered to be a curative modality in the treatment of colorectal cancer (1). Tumor recurrence may be because of the presence of therapy-resistant, genetically distinct tumor subclones. These subclones may either be pre-existent or may be generated as a direct result of the chemotherapy itself. More recently, it has been suggested that therapy resistance and subsequent tumor recurrence could be mediated by the “cancer stem cell ” fraction of colorectal tumors. Cancer stem cells make up only a few percent of the total tumor cell mass, but are uniquely endowed with tumor-initiating capacity (2–5).
Interestingly, normal intestinal stem cells have been identified as the cell-of-origin of intestinal tumors (6, 7). Cancer stem cells may therefore be transformed descendants of normal tissue stem cells. Although normal stem cells give rise to differentiated cells lacking tissue-regenerating capacity, cancer stem cells give rise to differentiated tumor cells lacking tumor-regenerating capacity (8, 9).
Normal colon stem cells are exposed to toxins and drugs for an entire lifetime. To cope with this continuous challenge, stem cells must possess intrinsic resistance mechanisms that protect their DNA from being mutated and that allow prolonged survival. Inheritance of these resistance mechanisms by cancer stem cells may protect them from the cytotoxic action of chemotherapeutic drugs. If cancer stem cells are indeed the major driving force behind tumor recurrence, novel strategies are required to target this subset of cancer cells. Indeed, several studies have shown that residual tumor tissue after chemotherapy is enriched for cancer stem cell-like cells (10, 11). However, the relationship between chemo-resistance and tumor-initiating potential and the mechanisms underlying cancer stem cell selective drug resistance are currently poorly understood (12–15).
Here we set out to address the relationship between colorectal cancer stem cells and drug resistance. To this end, we have generated cancer stem cell enriched human colonosphere cultures from colorectal liver metastases. In addition, we have generated colonosphere-derived stably differentiated progeny. These isogenic cell pairs were then used to identify proteome differences using mass spectrometry. Analysis of the data revealed that proteins governing cell survival are overrepresented in the cancer stem cell cultures. The most prominently overexpressed survival protein, BIRC6/BRUCE/Apollon, was identified as a key mediator of cancer stem cell resistance to cisplatin and oxaliplatin.
EXPERIMENTAL PROCEDURES
Colorectal Cancer Stem Cells and Differentiated Tumor Cell Cultures
Collection of tumor specimens, isolation and expansion of colorectal cancer stem cell and differentiated cell cultures was performed as described in Emmink et al. (16). Human colorectal tumor specimens were obtained from patients undergoing a liver resection for metastatic adenocarcinoma, in accordance with the ethical committee on human experimentation. Informed consent was obtained from all patients. All tumors were diagnosed as colorectal adenocarcinomas. Liver metastases were excised from segment VII (L145), segment IV (L146), and segment II-IV (L167). Differentiation status was not determined.
Isolation and Expansion of Colorectal Cancer Stem Cell Cultures
The obtained tissue fragments were washed extensively with phosphate-buffered saline and were mechanically dissociated using scalpels and vigorous trituration to yield small fragments (<1 mm3) and single cells. Enzymatic digestion was performed using thermolysin 0.05% (Sigma, Type X) in Dulbecco's modified Eagle's medium/F12 containing 5 mm Hepes (Invitrogen, Carlsbad, CA) for 2 h at 37 °C. The suspension was then filtered through a 40-μm-pore size nylon cell strainer (BD Falcon) to separate the tissue fragments from the single cells. The single cell suspension was cultured in advanced Dulbecco's modified Eagle's medium/F12 (Invitrogen) supplemented with 0.6% glucose (BDH Lab. Supplies), 2 mm l-glutamine (Biowhittaker, Rockland, ME), 9.6 μg/ml putrescin (Sigma), 6.3 ng/ml progesterone (Sigma), 5.2 ng/ml sodium selenite (Sigma), 25 μg/ml insulin (Sigma), 100 μg/ml apotransferrin (Sigma), 5 mm hepes (Invitrogen), 0,005 μg/ml trace element A (Cellgro), 0.01 μg/ml trace element B (Cellgro), 0.01 μg/ml trace element C (Cellgro), 100 μm β-mercaptoethanol (Merck), 10 ml antibiotic-antimycotic (Invitrogen), 4 μg/ml gentamicine (Invitrogen), 0.002% lipid mixture (Sigma), 5 μg/ml glutathione (Roche), and 4 μg/ml Heparin (Sigma). Growth factors (20 ng/ml EGF (Invitrogen) and 10 ng/ml b-FGF (Abcam, Cambridge, UK)) were added to the cell culture medium freshly each week. All cell culture was carried out in nontissue culture treated flasks (BD Falcon) at 37 °C in a 5% CO2 humidified incubator. In vitro differentiation was induced by culturing colon colonospheres for 3 weeks on collagen-coated dishes in Dulbecco's modified Eagle's medium/F12 (GIBCO) supplemented with 20% fetal bovine serum. Passage numbers of the clones used (n = 3) spheroid cultures and the accompanying differentiated tumor cell cultures were all below 10.
Cell lysis and SDS-PAGE
Paired colonospheres and differentiated tumor cell cultures were seeded in 10 cm2 diameter tissue culture plates and cultured for 24 h in serum-free stem cell medium. Cells were subsequently washed twice with phosphate-buffered saline, centrifuged and washed with water to get rid of the excess salts. Lysis buffer (20 mm HEPES pH7.4, 1% Nonidet P-40, 150 mm NaCl, 5 mm MgCl2, 10% glycerol) containing proteinase inhibitor was used to lyse cells. Equal amounts of protein (50 μg) were separated on NuPAGE Novex Bis-Tris Mini Gels (Invitrogen). Gels were stained with Coomassie brilliant blue G-250 (Pierce, Rockford, IL), washed and each lane was sliced into ten bands using a band pattern to guide the slicing. The gel slicing and in-gel digesting was performed in a laminar flow under keratin-free conditions.
In-gel Digestion
Before MS analysis, separated proteins were in-gel digested as described (17). Gel lanes corresponding to the different protein samples were sliced into ten bands. The bands were washed and dehydrated three times in 50 mm ammonium bicarbonate pH 7.9 + 50% acetonitrile. Subsequently, cysteine bonds were reduced with 10 mm dithiotreitol for 1 h at 56 °C and alkylated with 50 mm iodoacetamide for 45 min at room temperature in the dark. After two subsequent wash and dehydration cycles the bands were dried 10 min in a vacuum centrifuge and incubated overnight with 0.06 μg/μl trypsin at 25 °C. Peptides were extracted once in 1% formic acid and subsequently two times in 50% acetonitrile in 5% formic acid. The volume was reduced to 50 μl in a vacuum centrifuge prior to liquid chromatography tandem MS (LC-MS/MS) analysis (17).
NanoLC-MS/MS Analysis
Peptides were separated by an Ultimate 3000 nanoLC-MS/MS system (Dionex LC-Packings, Amsterdam, The Netherlands) equipped with a 20 cm × 75 μm ID fused silica column custom packed with 3 μm 120 Å ReproSil Pur C18 aqua (Dr Maisch GMBH, Ammerbuch-Entringen, Germany). After injection, peptides were trapped at 30 μl/min on a 5 mm × 300 μm ID Pepmap C18 cartridge (Dionex LC-Packings) at 2% buffer B (buffer A: 0.05% formic acid in MQ; buffer B: 80% acetonitrile + 0.05% formic acid in MQ) and separated at 300 nl/min in a 10–40% buffer B gradient in 60 min. Eluting peptides were ionized at 1.7 kV in a Nanomate Triversa Chip-based nanospray source using a Triversa LC coupler (Advion, Ithaca, NY). Intact peptide mass spectra and fragmentation spectra were acquired on a LTQ-FT hybrid mass spectrometer (Thermo Fisher, Bremen, Germany). Intact masses were measured at resolution 50.000 in the ICR cell using a target value of 1 × 106 charges. In parallel, after an FT prescan, the top 5 peptide signals (charge-states 2+ and higher) were submitted to MS/MS in the linear ion trap (3 amu isolation width, 30 ms activation, 35% normalized activation energy, Q value of 0.25 and a threshold of 5000 counts). Dynamic exclusion was applied with a repeat count of 1 and an exclusion time of 30s.
Label-free protein quantitation by spectral counting has emerged as a powerful alternative to labeling-based strategies, with each approach having its pros and cons. Notably, label-free analysis is less accurate than approaches in which proteins are labeled, in particular when small differences in protein levels across samples are quantified. However, with a good parallel workflow, label-free quantitation is simple, cheap, and allows for quantitation at a relative large dynamic range.
Database Searching, Statistics, and Ingenuity Pathway Analysis
MS/MS spectra were searched against the human International Protein Index database 3.31 (67,511 entries) using Sequest (version 27, rev 12), which is part of the BioWorks 3.3 data analysis package (Thermo Fisher, San Jose, CA). MS/MS spectra were searched with a maximum allowed deviation of 10 ppm for the precursor mass and 1 amu for fragment masses. Methionine oxidation and cysteine carboxamidomethylation were allowed as variable modifications, two missed cleavages were allowed and the minimum number of tryptic termini was 1. After database searching the DTA and OUT files were imported into Scaffold 2.01.01 (Proteome software, Portland, OR). Scaffold was used to organize the gel-band data and to validate peptide identifications using the Peptide Prophet algorithm (18). Only peptide identifications with a probability >95% were retained. Subsequently, the ProteinProphet algorithm (19) was applied and protein identifications with a probability of >99% with two peptides or more in at least one of the samples were retained. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped. For each protein identified, the number of spectral counts (identified MS/MS spectra) in each sample (10 gel bands) was exported to Excel. For each sample the spectral counts per protein were normalized on the sum of the spectral counts for that sample. The list of differentially expressed proteins, including fold changes was imported in the online software package Ingenuity (Ingenuity IPA, version 7.6) and pathway and network analysis was performed with only direct relationships. Furthermore the list of differentially expressed proteins was imported in the online STRING database of known and predicted protein interactions. The interactions include direct (physical) and indirect (functional) associations between proteins.
Lentiviral Constructs and Transduction
The lentiviral short-hairpin RNA (shRNA) constructs targeting baculoviral inhibitor of apoptosis protein repeat containing 6 (BIRC6)1 were obtained from the TRC-Mm1.0 library (Sigma Aldrich). The target set used for BIRC6 (NM_016252) included TRCN0000004157, TRCN0000004158, TRCN00000059, TRCN0000004160, and TRCN0000004161. Of these constructs, transduction of cells with 58 and 59 produced the best knock down. As the control vector, we used the same vector containing a sequence targeting luciferase, TGACCAGGCATTCACAGAAAT.
Western Blotting and Antibodies
Lysates of colonospheres and their differentiated offspring were prepared in lysisbuffer (20 mm HEPES pH7.4, 1% Nonidet P-40, 150 mm NaCl, 5 mm MgCl2, 10% glycerol). Equal amounts of protein were run out on NuPAGE Novex Tris-Acetate Mini Gel (Invitrogen) and were analyzed by Western blotting using antibodies directed against BIRC6 (Ab19609, Abcam) and β-Actin (AC-15, Novus Biologicals, Littleton, CO) was used.
Flow Cytometry and Cell Sorting
Dead cells were excluded using viability marker 7-aminoactinomycin D (7-AAD) (R&D, Detroit, MI) and cell doublets and clumps were excluded using doublet discrimination gating. Aldefluor®-positive cells were analyzed according to the manufacturer's protocol by using Aldefluor® and DEAB (STEMCELL Technologies). The cell sorting experiments were conducted with DAKO-Cytomation MoFlo High Speed Sorter.
Cisplatin and Oxaliplatin Sensitivity Assay
Control and BIRC6 knockdown colonospheres and differentiated tumor cells were cultured in the presence of oxaliplatin (Pharmachemie BV, Haarlem) or cisplatin (Pharmachemie BV, Haarlem) at the indicated concentrations for 5 days. Mitochondrial activity was evaluated using CellTiter 96® AQueous Nonradioactive Cell Proliferation Assay (MTS) (Promega, Charbonnières, France). All absorbance values are expressed as percentages of vehicle-treated control wells.
RESULTS
Proteome Differences Between Colon Cancer Stem Cells and Their Differentiated Progeny
To compare the proteome of colon cancer stem cells to their differentiated progeny, we analyzed three isogenic pairs of colonospheres and differentiated tumor cells derived from freshly resected liver metastases. All colonosphere cultures were enriched for cancer stem cells based on their high clone- and tumor-forming potential (16).
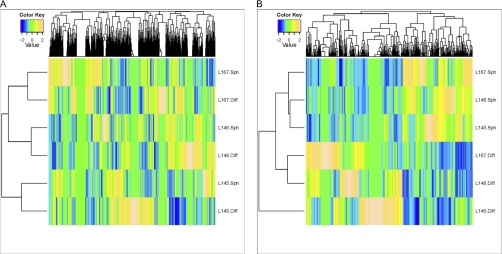
The protein lysates of these cultures were fractionated on an SDS-PAGE gel (Fig. 1A), followed by in-gel tryptic digestion. Analysis of the extracted peptides was performed by Nano-LC-MS/MS, followed by database searching. In total, 3048 proteins were identified in all sets of cells together, with an average of 2269 proteins per isogenic couple (Table I). The number of proteins that was at least twofold up-regulated in the colonospheres of each isogenic pair varied from 377 to 491. Of these >twofold up-regulated proteins, 32 proteins were up-regulated in the colonospheres of all 3 isogenic pairs (Fig. 1B). The unsupervised heat map of global clustering shows that the similarity between isogenic pairs of colonosphere cells and differentiated tumor cells is greater than the similarity among the different colonospheres cultures (Fig. 2A). In contrast, supervised clustering including all significantly up- and down-regulated proteins shows that all colonosphere cultures and differentiated cultures now cluster together (Fig. 2B). This suggests that it may be possible to identify a cancer stem cell protein signature in colorectal cancer.
Fig. 1.
NanoLC-MS/MS/based analysis of proteome differences between colonospheres and isogenic differentiated tumor cells. A, Coomassie-stained protein gradient gel loaded with protein samples from colonospheres (Sph) and differentiated tumor cells (Diff) of the indicated tumors. This gel was used for MS analysis. B, Venn diagram of all >twofold up-regulated proteins in colonospheres.
Table I. Proteome differences between colonospheres and isogenic differentiated cells. The numbers refer to all identified up-, down- and non-regulated proteins (top row), all up-regulated proteins (middle row), and all >twofold up-regulated proteins. The number of proteins in all three categories that were identified in 2/3 and in 3/3 pairs are shown in the last two columns.
| L145 | L146 | L167 | Overlap |
||
|---|---|---|---|---|---|
| 2/3 pairs | 3/3 pairs | ||||
| All proteins | 2242 | 2290 | 2376 | 1739 | 516 |
| Total upregulated proteins | 1078 | 1001 | 1093 | 676 | 229 |
| >2 fold upregulated | 491 | 377 | 437 | 275 | 32 |
Fig. 2.
Separation of colonospheres from differentiated tumor cells by cluster analysis. A, Heat map of unsupervised cluster analysis of all identified proteins. B, Heat map of supervised cluster analysis of all significantly up- and down-regulated proteins (p < 0.05 according to the Fisher exact test per pair.) The selected list of regulated proteins separates colonospheres from differentiated tumor cells.
Proteins Associated with Survival are Up-regulated in Colon Cancer Stem Cells
Next, we divided the up-regulated proteins in three categories: (1) Top up-regulated, ≥ twofold up-regulated in three out of three isogenic pairs, (2) Subtop up-regulated, ≥1.5-fold up-regulated in three out of three isogenic pairs, and (3) Rest up-regulated, ≥1.5-fold up-regulated in two out of three isogenic pairs. A total amount of 119 proteins was found to be enriched in colonospheres (32 in category I; 22 in category II, and 65 in category III) (Table II, supplemental Table S1). Interestingly, among the top up-regulated proteins is ALDH1A1 (aldehyde dehydrogenase 1A1), which was recently identified by us and others as a bona fide colon (cancer) stem cell marker (11, 16, 20). The identification of ALDH1A1 demonstrates the validity of our proteomics approach to identify up-regulated factors in cancer stem cells.
Table II. All proteins up-regulated in colonospheres in comparison to differentiated tumor cells. The table shows all proteins that are up-regulated in colonospheres when compared to differentiated tumor cells according to the following criteria: (1) top up-regulated, ≥2-fold up-regulated in three out of three isogenic pairs (32) and (2) subtop up-regulated, ≥1.5-fold up-regulated in three out of three isogenic pairs (22). Sph = colonosphere culture enriched for cancer stem cells, Diff = isogenic differentiated progeny of colonosphere culture.
| Protein | Gene | Accession number | L145 |
L146 |
L167 |
Fold change | Location | Function | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Diff | Sph | Diff | Sph | Diff | Sph | ||||||
| >2-fold up-regulated proteins in 3/3 combinations | |||||||||||
| Glutathione peroxidase 2 (gastrointestinal) | GPX2 | IPI00298176 | 0 | 13 | 0 | 23 | 0 | 12 | ∞ | Cytoplasm | Enzyme |
| Glutathione peroxidase 1 | GPX1 | IPI00293975 | 0 | 10 | 0 | 8 | 0 | 12 | ∞ | Cytoplasm | Enzyme |
| Ferritin, light polypeptide | FTL | IPI00852596 | 0 | 4 | 0 | 4 | 0 | 7 | ∞ | Cytoplasm | Other |
| Neuroblastoma amplified sequence | NBAS | IPI00333913 | 0 | 5 | 0 | 6 | 0 | 3 | ∞ | Unknown | Other |
| Baculoviral IAP repeat-containing 6 | BIRC6 | IPI00299635 | 0 | 4 | 0 | 6 | 0 | 3 | ∞ | Cytoplasm | Enzyme |
| Microsomal glutathione S-transferase 2 | MGST2 | IPI00017767 | 0 | 3 | 0 | 4 | 0 | 3 | ∞ | Cytoplasm | Enzyme |
| Glutathione peroxidase 4 (phospholipid hydroperoxidase) | GPX4 | IPI00304814 | 0 | 2 | 0 | 5 | 0 | 3 | ∞ | Cytoplasm | Enzyme |
| Eukaryotic translation initiation factor 3, subunit H | EIF3H | IPI00647650 | 0 | 3 | 0 | 3 | 0 | 3 | ∞ | Cytoplasm | Translation regulator |
| CDGSH iron sulfur domain 3 | CISD3 | IPI00783359 | 0 | 2 | 0 | 4 | 0 | 3 | ∞ | Unknown | Other |
| Endosulfine alpha | ENSA | IPI00220797 | 0 | 3 | 0 | 3 | 0 | 2 | ∞ | Unknown | Transporter |
| Family with sequence similarity 98, member A | FAM98A | IPI00174442 | 0 | 2 | 0 | 2 | 0 | 3 | ∞ | Unknown | Other |
| RAN binding protein 3 | RANBP3 | IPI00456728 | 0 | 2 | 0 | 2 | 0 | 3 | ∞ | Nucleus | Other |
| Chromosome 11 open reading frame 31 | C11ORF31 | IPI00218054 | 0 | 2 | 0 | 2 | 0 | 3 | ∞ | Nucleus | Other |
| Mitochondrial ribosomal protein S18B | MRPS18B | IPI00022316 | 0 | 2 | 0 | 3 | 0 | 2 | ∞ | Cytoplasm | Other |
| RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) | RRS1 | IPI00014253 | 0 | 2 | 0 | 3 | 0 | 2 | ∞ | Nucleus | Other |
| DEAH (Asp-Glu-Ala-His) box polypeptide 38 | DHX38 | IPI00294211 | 0 | 2 | 0 | 2 | 0 | 2 | ∞ | Nucleus | Enzyme |
| C-terminal binding protein 1 | CTBP1 | IPI00012835 | 0 | 2 | 0 | 2 | 0 | 2 | ∞ | Nucleus | Enzyme |
| SIN3 homolog A, transcription regulator (yeast) | SIN3A | IPI00170596 | 0 | 2 | 0 | 2 | 0 | 2 | ∞ | Nucleus | Transcription regulator |
| Succinate dehydrogenase complex, subunit B, iron sulfur (Ip) | SDHB | IPI00294911 | 2 | 5 | 0 | 6 | 0 | 8 | 9.2 | Cytoplasm | Enzyme |
| RNA binding motif protein 25 | RBM25 | IPI00004273 | 0 | 6 | 0 | 5 | 2 | 7 | 8.7 | Nucleus | Other |
| E1A binding protein p400 | EP400 | IPI00064931 | 2 | 5 | 0 | 7 | 0 | 5 | 8.2 | Nucleus | Other |
| Myosin, heavy chain 10, non-muscle | MYH10 | IPI00397526 | 0 | 3 | 0 | 4 | 4 | 26 | 8.1 | Cytoplasm | Other |
| Leucine rich repeat containing 16A | LRRC16A | IPI00014843 | 0 | 17 | 4 | 10 | 0 | 4 | 7.4 | Unknown | Enzyme |
| CDC42 binding protein kinase beta (DMPK-like) | CDC42BPB | IPI00477763 | 2 | 6 | 0 | 4 | 0 | 3 | 6.2 | Cytoplasm | Kinase |
| Eukaryotic translation initiation factor 3, subunit J | EIF3J | IPI00290461 | 2 | 6 | 0 | 2 | 0 | 3 | 5.2 | Cytoplasm | Translation regulator |
| TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa | TAF15 | IPI00020194 | 0 | 3 | 0 | 2 | 2 | 5 | 4.9 | Nucleus | Transcription regulator |
| PTK2 protein tyrosine kinase 2 | PTK2 | IPI00012885 | 0 | 3 | 0 | 3 | 2 | 4 | 4.8 | Cytoplasm | Kinase |
| Protein arginine methyltransferase 5 | PRMT5 | IPI00441473 | 0 | 4 | 0 | 2 | 3 | 7 | 4.2 | Cytoplasm | Enzyme |
| Activating signal cointegrator 1 complex subunit 3 | ASCC3 | IPI00430472 | 2 | 6 | 0 | 5 | 2 | 6 | 4.1 | Nucleus | Enzyme |
| Aldehyde dehydrogenase 1 family, member A1 | ALDH1A1 | IPI00218914 | 11 | 76 | 26 | 102 | 45 | 113 | 3.4 | Cytoplasm | Enzyme |
| Mitochondrial ribosomal protein S27 | MRPS27 | IPI00022002 | 0 | 2 | 2 | 6 | 2 | 6 | 3.4 | Cytoplasm | Other |
| Vacuolar protein sorting 13 homolog C (S. cerevisiae) | VPS13C | IPI00465428 | 9 | 19 | 5 | 13 | 5 | 13 | 2.3 | Unknown | Other |
| >1.5-fold up-regulated proteins in 3/3 combinations | |||||||||||
| Adenylosuccinate lyase | ADSL | IPI00026904 | 0 | 5 | 3 | 5 | 0 | 5 | 4.8 | Cytoplasm | Enzyme |
| Transmembrane protein 205 | TMEM205 | IPI00063130 | 0 | 5 | 0 | 3 | 3 | 5 | 4.2 | Unknown | Other |
| Transformation/transcription domain-associated protein | TRRAP | IPI00069084 | 5 | 8 | 0 | 7 | 0 | 6 | 4.0 | Nucleus | Transcription regulator |
| Nuclear receptor co-repressor 1 | NCOR1 | IPI00289344 | 0 | 3 | 0 | 4 | 3 | 5 | 3.9 | Nucleus | Transcription regulator |
| ATP-binding cassette, sub-family B (MDR/TAP), member 1 | ABCB1 | IPI00027481 | 0 | 7 | 0 | 11 | 9 | 17 | 3.8 | Plasma Membrane | Transporter |
| CDGSH iron sulfur domain 1 | CISD1 | IPI00020510 | 0 | 3 | 0 | 8 | 6 | 10 | 3.4 | Cytoplasm | Other |
| Methionine adenosyltransferase II, beta | MAT2B | IPI00002324 | 5 | 8 | 0 | 5 | 0 | 5 | 3.4 | Cytoplasm | enzyme |
| NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa | NDUFA11 | IPI00329301 | 2 | 5 | 0 | 6 | 4 | 6 | 2.7 | Cytoplasm | Enzyme |
| Succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | SDHA | IPI00305166 | 5 | 18 | 12 | 21 | 6 | 23 | 2.6 | Cytoplasm | Enzyme |
| 15 kDa selenoprotein | SEP15 | IPI00030877 | 3 | 8 | 5 | 9 | 2 | 7 | 2.3 | Cytoplasm | Enzyme |
| Aldehyde dehydrogenase 1 family, member B1 | ALDH1B1 | IPI00103467 | 8 | 17 | 15 | 46 | 14 | 22 | 2.2 | Cytoplasm | Enzyme |
| Acetyl-CoA acyltransferase 1 | ACAA1 | IPI00012828 | 2 | 11 | 7 | 11 | 9 | 15 | 2.0 | Cytoplasm | Enzyme |
| Eukaryotic translation initiation factor 5B | EIF5B | IPI00299254 | 6 | 12 | 6 | 11 | 3 | 8 | 2.0 | Cytoplasm | Translation regulator |
| Heterogeneous nuclear ribonucleoprotein A0 | HNRNPA0 | IPI00011913 | 3 | 10 | 4 | 6 | 5 | 9 | 2.0 | Nucleus | Other |
| UDP-glucose 6-dehydrogenase | UGDH | IPI00031420 | 4 | 22 | 30 | 45 | 19 | 37 | 1.9 | Nucleus | Enzyme |
| Valyl-tRNA synthetase | VARS | IPI00000873 | 9 | 24 | 15 | 22 | 14 | 23 | 1.8 | Cytoplasm | Enzyme |
| Tubulin tyrosine ligase-like family, member 12 | TTLL12 | IPI00029048 | 6 | 10 | 12 | 27 | 8 | 12 | 1.8 | Unknown | Other |
| Interleukin enhancer binding factor 3, 90kDa | ILF3 | IPI00298788 | 20 | 32 | 17 | 28 | 13 | 27 | 1.7 | Nucleus | Transcription regulator |
| Carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase | CAD | IPI00301263 | 24 | 36 | 33 | 58 | 19 | 33 | 1.6 | Cytoplasm | Enzyme |
| RAN binding protein 2 | RANBP2 | IPI00221325 | 28 | 45 | 22 | 35 | 28 | 42 | 1.5 | Nucleus | Enzyme |
Next, we used the Ingenuity Pathway Knowledge Base tool to identify biological functions and canonical pathways that distinguish colonospheres from differentiated cells. For this analysis all 119 colonosphere-enriched proteins (Table II, supplemental Table S1) were included. When classified according to function, proteins regulating “Cell Death ” (20) and proteins regulating “Post-Translational Modification ” (12) were most frequently identified. “Cell cycle control of chromosomal replication,” “aryl hydrocarbon receptor signaling,” “estrogen receptor signaling,” and “mitochondrial dysfunction” were identified as the canonical pathways that are overrepresented in colonosphere cultures (Table III).
Table III. Ingenuity analysis of colonosphere-enriched proteins. All 119 up-regulated proteins were analyzed by Ingenuity. The main Molecular and Cellular functions represented by this protein group are “Cell Death” (20) and “Post Translational Modifications” (12). The main canonical pathways represented by this protein group are also shown.
| Top functions | Associated molecules | Focus molecules |
|---|---|---|
| Molecular and cellular functions | ||
| Cell death | ABCB1, ALDH1A1, ARMC10, BAX, BIRC6, CTBP1, EIF3H, EP400, GPX2, HSPA4, HTT, MCM2, MDC1, PRKDC, PTK2, RBM25, SDHA, SDHB, TIMM50, TRAP1 | 20 |
| Post-translational modification | BAX, CTBP1, FTL, HSPA4, HTT, NEDD8, PRKDC, PRMT5, SEP15, ST13, TIMM50, TTN, ABCB1, ACO2, ADSL, AKR1C3, ALDH1A1, BAX, BDH2, GPX4, HSPA4, HTT, MAT2B, SDHA, TST, UGDH | 12 |
| Top functions | Associated molecules | Number of identified molecules | Number of molecules in pathway |
|---|---|---|---|
| Canonical pathways | |||
| Cell cycle control of chromosomal replication | MCM2, MCM3, MCM5, MCM7 | 4 | 30 |
| Aryl hydrocarbon receptor signaling | ALDH1A1, ALDH1B1, BAX, MCM7, MGST2, NEDD8 | 6 | 141 |
| Estrogen receptor signaling | CTBP1, NCOR1, PRKDC, TAF15, TRRAP | 5 | 134 |
| Mitochondrial dysfunction | GPX4, NDUFA11, NDUFB11, SDHA, SDHB | 5 | 133 |
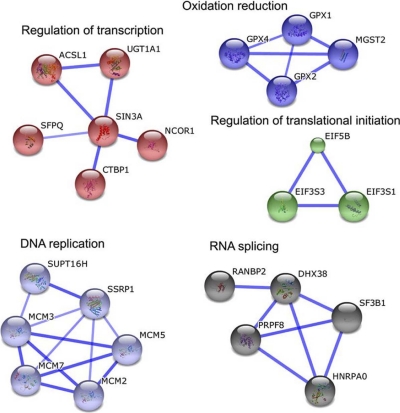
Next, the STRING database was used to identify known and predicted protein interactions among the up-regulated proteins. STRING analysis of all up-regulated proteins showed five major clusters involved in redox regulation, transcription control, RNA splicing, DNA replication, and regulation of translation initiation (Fig. 3).
Fig. 3.
Colonopshere-enriched proteins display functional interactions. All colonosphere-enriched proteins (119) were analyzed against the STRING database for functional protein association networks. The strength of the associations is represented by line thickness. Networks with three or more protein interactions are shown. Required confidence (score) of protein association was 0.700 (high confidence).
Given our interest in drug resistance we further focused on proteins regulating cell death and survival (Table III). An extensive literature search revealed that the majority of overrepresented “Cell Death ” regulators (15/20) have anti-apoptotic activity (Table IV). Of these, Baculoviral IAP repeat-containing 6 (BIRC6; also known as BRUCE or Apollon), an Inhibitor of Apoptosis Protein (IAP), is a key regulator of the intrinsic apoptosis pathway and has previously been implicated in drug resistance (21, 22).
Table IV. Functional annotation of colonosphere-enriched “Cell Death” proteins. The 20 colonosphere-enriched cell death regulators that were identified by Ingenuity were classified according to their pro-and anti-apoptotic function, based on literature.
| Protein | Gene | Accession number | Fold change | Anti-apoptotic | Pre-apoptotic |
|---|---|---|---|---|---|
| Glutathione peroxidase 2 (gastrointestinal) | GPX2 | IPI00298176 | ∞ | + | |
| Baculoviral IAP repeat-containing 6 | BIRC6 | IPI00299635 | ∞ | + | |
| Eukaryotic translation initiation factor 3, subunit H | EIF3H | IPI00647650 | ∞ | + | |
| C-terminal binding protein 1 | CTBP1 | IPI00012835 | ∞ | + | |
| Armadillo repeat containing 10 | ARMC10 | IPI00217968 | ∞ | + | |
| Succinate dehydrogenase complex, subunit B, iron sulfur | SDHB | IPI00294911 | 9.2 | + | + |
| RNA binding motif protein 25 | RBM25 | IPI00004273 | 8.7 | + | + |
| E1A binding protein p400 | EP400 | IPI00064931 | 8.2 | + | |
| PTK2 protein tyrosine kinase 2 | PTK2 | IPI00012885 | 4.8 | + | |
| Aldehyde dehydrogenase 1 family, member A1 | ALDH1A1 | IPI00218914 | 3.4 | + | |
| ATP-binding cassette, sub-family B (MDR), member 1 | ABCB1 | IPI00027481 | 3.8 | + | |
| BCL2-associated X protein | BAX | IPI00071059 | 3.0 | + | |
| Succinate dehydrogenase complex, subunit A | SDHA | IPI00305166 | 2.6 | + | |
| Huntingtin | HTT | IPI00002335 | ∞ | + | + |
| Translocase of inner mitochondrial membrane 50 | TIMM50 | IPI00418497 | 3.1 | + | |
| Mediator of DNA-damage checkpoint 1 | MDC1 | IPI00470805 | 2.9 | + | + |
| Minichromosome maintenance complex component 2 | MCM2 | IPI00184330 | 1.7 | + | |
| TNF receptor-associated protein 1 | TRAP1 | IPI00030275 | 1.6 | + | |
| Protein kinase, DNA-activated, catalytic polypeptide | PRKDC | IPI00296337 | 1.4 | + | |
| Heat shock 70kDa protein 4 | HSPA4 | IPI00002966 | 1.3 | + |
BIRC6 Mediates Resistance of Colorectal Cancer Stem Cells to Platinum Compounds
Next, we tested whether BIRC6 plays a role in mediating chemotherapy resistance in colorectal cancer stem cells. First, we analyzed BIRC6 expression in the three sets of isogenic cell pairs by Western blotting. In line with the proteomics data, BIRC6 was highly up-regulated in colonospheres when compared with differentiated cells (Fig. 4A). Western blot analysis of other top up-regulated proteins (GPX2, GPX1, and ALDH1) also confirmed the proteomics data (data not shown, and see (16)). Our previous results have shown that ALDH activity (as measured by the fluorescent substrate Aldefluor®) defines the tumorigenic and clonogenic cancer stem cell population within colonospheres (16). Therefore, we used FACS sorting to separate Aldefluor®high from Aldefluor®low cells (Fig. 4B) and analyzed BIRC6 expression in both tumor cell fractions. Fig. 4C shows that BIRC6 is highly expressed in tumorigenic Aldefluor®high cells, but not in nontumorigenic Aldefluor®low cells.
Fig. 4.
BIRC6 is expressed in the tumorigenic Aldefluorhigh fraction of colonosphere cells. A, Western blot analysis of BIRC6 levels in colonospheres and differentiated tumor cells in all pairs. B, Single cell cultures of L145 colonospheres were separated into Aldefluor®high and Aldefluor®low cell populations by FACS sorting. C, Western blot analysis of Aldefluor®high and Aldefluor®low cell populations for expression of BIRC6.
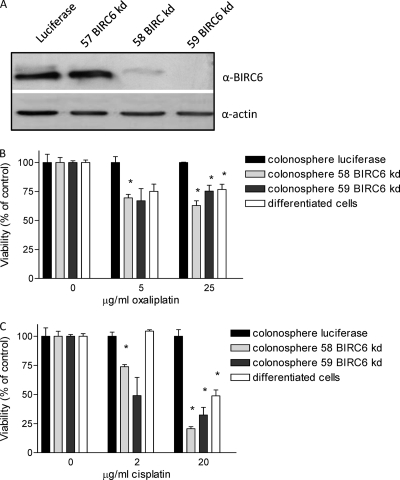
Next, we assessed the importance of BIRC6 in mediating colonosphere resistance to oxaliplatin and cisplatin, two frequently used chemotherapeutic drugs. To this end, expression of BIRC6 was suppressed in colonospheres by using a set of lentiviral RNA interference (RNAi) vectors. Two vectors (58 and 59) were found to suppress BIRC6 expression very efficiently (Fig. 5A).
Fig. 5.
BIRC6 confers resistance against cisplatin and oxaliplatin. A, Western blot analysis of BIRC6 expression in L145 colonospheres transduced with control (luciferase, 57) and BIRC6 knockdown vectors (58, 59). B, Control and BIRC6 knockdown L145 colonospheres were treated with oxaliplatin or cisplatin for 3 days using the indicated concentrations. Cell viability was then assessed by MTS assays for mitochondrial activity. Absorbance values (in triplicate) are expressed as percentage of vehicle-treated control wells. *Statistical significance (unpaired, 2-tailed t test: p < 0.05).
Control and BIRC6 knockdown colonopsheres and differentiated tumor cells were treated with increasing concentrations of oxaliplatin or cisplatin and cell viability was measured by standard MTS assays. Knockdown of BIRC6 resulted in a significantly higher response of colonosphere cells to oxaliplatin and cisplatin (Fig. 5B and 5C). The sensitivity of BIRC6 knockdown cells to both drugs was comparable to that of the differentiated tumor cells. These results identify BIRC6 as an important mediator of resistance to oxaliplatin and cisplatin and suggest that high BIRC6 expression may selectively protect the cancer stem cell fraction against these drugs.
DISCUSSION
In the present study we have used a proteomics approach to identify potential regulators of the cancer stem cell phenotype in colorectal tumors. We identified known (ALDH1A1) and novel factors enriched in cancer stem cell cultures (colonospheres) when compared with stably differentiated tumor cells. Interestingly, STRING analysis revealed that distinct protein complexes involved in transcriptional repression, DNA replication, RNA splicing, translation initiation and redox control are significantly enriched in colonospheres when compared with differentiated tumor cells. Future work should reveal the function of these complexes in the maintenance of colorectal cancer stem cells.
Importantly, cancer stem cells were also characterized by high expression of a set of survival proteins, the most prominent of which was BIRC6. BIRC6 deletion is associated with sensitization to chemotherapy in in vivo and in vitro studies (21, 22). Furthermore, BIRC6 deletion promotes p53 stabilization and caspase 3 activation (23).
Our data show that specifically colorectal cancer stem cell cultures display increased resistance to oxaliplatin and to cisplatin and that BIRC6 is an important mediator of resistance. Previously, it was shown by gene expression profiling that BIRC1 and BIRC6 are up-regulated in colorectal tumors when compared with normal intestinal issue (24). Our results suggest that it is predominantly the cancer stem cell-fraction in colorectal tumors that expresses this survival protein.
BIRC6, also known as Apollon or Bruce, belongs to the family of IAP proteins. IAP's are major regulators of apoptosis due, at least in part, to their ability to inhibit caspase activation (25, 26). Human IAP family members include X-chromosome-linked IAP (XIAP, also known as BIRC4), cellular IAP 1 (c-IAP1 also known as BIRC2), c-IAP2 (also known as BIRC3), neuronal apoptosis inhibitory protein (also known as BIRC1), and survivin (also known BIRC5). IAP proteins contain one to three baculovirus IAP repeat (BIR) domains that are required for their anti-apoptotic activity (25). Our results are in line with previous studies showing that cancer stem cells express high levels of anti-apoptotic proteins and resist apoptotic stimuli (27, 28). Recently it was demonstrated that IL4-stimulated expression of survivin (BIRC5) protects colorectal cancer stem cells against apoptosis (10, 28, 29). The proteomics approach described here did not identify BIRC5 as a cancer stem cell-enriched protein. Possibly, different tumors resist apoptotic stimuli by increasing the expression of distinct IAP family members.
Because IAP's play an important role in tumor maintenance and therapy resistance they represent attractive targets for targeted therapy. Furthermore, IAP's are highly expressed in several cancer tissues (30). Several small molecule IAP inhibitors have been developed, including Smac-based peptides and Smac mimetics targeting a broad spectrum of IAP's (31, 32). Preclinical studies in mice carrying xenograft tumors have shown promising antitumor efficacy in the treatment of malignant glioma, breast cancer, nonsmall cell lung cancer, and multiple myeloma. However, most of these preclinical studies have focused on BIRC4 and surviving (BIRC5), rather than on BIRC6 (33, 34). Several IAP inhibitors are being tested for their safety and anti-tumor efficacy in clinical trials either in combination with irradiation or with chemotherapy (35, 36). It is not yet established whether these compounds also target BIRC6. Second mitochondrial-derived activator of caspases (SMAC) mimetics bind to and inhibit the BIR domains in IAP's. Because SMAC also binds to BIRC6 (37), it is not unlikely that SMAC mimetics will inhibit BIRC6.
Based on the data presented here we propose that BIRC6 protects the cancer stem cell fraction of colorectal tumors against oxaliplatin and cisplatin. Targeting BIRC6 by SMAC mimetics or by novel specific BIRC6 inhibitors may therefore be effective in combination with platinum-based anticancer drugs. This may help eradicating the cancer stem cell fraction in colorectal tumors.
Footnotes
* This work has been supported by the WvH: Dutch Foundation for medical scientific research foundation; BLE: Dutch Cancer Society (KWF). We acknowledge the VUmc-Cancer Center Amsterdam for financial support of the proteomics infrastructure, TVP and CRJ.
 This article contains supplemental Table S1.
This article contains supplemental Table S1.
1 The abbreviations used are:
- BIRC
- baculoviral inhibitor of apoptosis protein repeat containing
- ALDH1A1
- aldehyde dehydrogenase 1A1
- IAP
- inhibitor of apoptosis protein
- SMAC
- second mitochondrial-derived activator of caspases.
REFERENCES
- 1. Chau I., Cunningham D. (2009) Treatment in advanced colorectal cancer: what, when and how? Br. J. Cancer 100, 1704–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ricci-Vitiani L., Lombardi D. G., Pilozzi E., Biffoni M., Todaro M., Peschle C., De Maria R. (2007) Identification and expansion of human colon-cancer-initiating cells. Nature 445, 111–115 [DOI] [PubMed] [Google Scholar]
- 3. O'Brien C. A., Pollett A., Gallinger S., Dick J. E. (2007) A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445, 106–110 [DOI] [PubMed] [Google Scholar]
- 4. Todaro M., Francipane M. G., Medema J. P., Stassi G. (2010) Colon cancer stem cells: promise of targeted therapy. Gastroenterology 138, 2151–2162 [DOI] [PubMed] [Google Scholar]
- 5. Vermeulen L., Todaro M., de Sousa M. F., Sprick M. R., Kemper K., Perez A. M., Richel D. J., Stassi G., Medema J. P. (2008) Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc. Natl. Acad. Sci. U.S.A. 105, 13427–13432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barker N., Ridgway R. A., van Es J. H., van de W. M., Begthel H., van den B. M., Danenberg E., Clarke A. R., Sansom O. J., Clevers H. (2009) Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 [DOI] [PubMed] [Google Scholar]
- 7. Zhu L., Gibson P., Currle D. S., Tong Y., Richardson R. J., Bayazitov I. T., Poppleton H., Zakharenko S., Ellison D. W., Gilbertson R. J. (2009) Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 457, 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clarke M. F., Dick J. E., Dirks P. B., Eaves C. J., Jamieson C. H., Jones D. L., Visvader J., Weissman I. L., Wahl G. M. (2006) Cancer stem cells–perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 66, 9339–9344 [DOI] [PubMed] [Google Scholar]
- 9. Hamburger A. W., Salmon S. E. (1977) Primary bioassay of human tumor stem cells. Science 197, 461–463 [DOI] [PubMed] [Google Scholar]
- 10. Todaro M., Alea M. P., Di Stefano A. B., Cammareri P., Vermeulen L., Iovino F., Tripodo C., Russo A., Gulotta G., Medema J. P., Stassi G. (2007) Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 1, 389–402 [DOI] [PubMed] [Google Scholar]
- 11. Dylla S. J., Beviglia L., Park I. K., Chartier C., Raval J., Ngan L., Pickell K., Aguilar J., Lazetic S., Smith-Berdan S., Clarke M. F., Hoey T., Lewicki J., Gurney A. L. (2008) Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS. ONE. 3, e2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dean M., Fojo T., Bates S. (2005) Tumour stem cells and drug resistance. Nat. Rev. Cancer 5, 275–284 [DOI] [PubMed] [Google Scholar]
- 13. Dean M. (2009) ABC transporters, drug resistance, and cancer stem cells. J. Mammary. Gland. Biol. Neoplasia. 14, 3–9 [DOI] [PubMed] [Google Scholar]
- 14. Liu G., Yuan X., Zeng Z., Tunici P., Ng H., Abdulkadir I. R., Lu L., Irvin D., Black K. L., Yu J. S. (2006) Analysis of gene expressio! and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer 5, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hermann P. C., Huber S. L., Herrler T., Aicher A., Ellwart J. W., Guba M., Bruns C. J., Heeschen C. (2007) Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 1, 313–323 [DOI] [PubMed] [Google Scholar]
- 16. Emmink B. L., van Houdt W. J., Vries R. G., Hoogwater F. J. H., Nijkamp M. W., Govaert. K. M., Verheem A., Steller E. J. A., Jimenez C. R., Clevers H., Borel Rinkes I. H. M., Kranenburg O. (2011)Differentiated colorectal cancer cells protect tumor-initiating cells from irinotecan. Gastroenterology 141, 269–278 [DOI] [PubMed] [Google Scholar]
- 17. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 18. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 19. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 20. Huang E. H., Hynes M. J., Zhang T., Ginestier C., Dontu G., Appelman H., Fields J. Z., Wicha M. S., Boman B. M. (2009) Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 69, 3382–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chu L., Gu J., Sun L., Qian Q., Qian C., Liu X. (2008) Oncolytic adenovirus-mediated shRNA against Apollon inhibits tumor cell growth and enhances antitumor effect of 5-fluorouracil. Gene Ther. 15, 484–494 [DOI] [PubMed] [Google Scholar]
- 22. Chen Z., Naito M., Hori S., Mashima T., Yamori T., Tsuruo T. (1999) A human IAP-family gene, apollon, expressed in human brain cancer cells. Biochem. Biophys. Res. Commun. 264, 847–854 [DOI] [PubMed] [Google Scholar]
- 23. Lopergolo A., Pennati M., Gandellini P., Orlotti N. I., Poma P., Daidone M. G., Folini M., Zaffaroni N. (2009) Apollon gene silencing induces apoptosis in breast cancer cells through p53 stabilisation and caspase-3 activation. Br. J. Cancer 100, 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bianchini M., Levy E., Zucchini C., Pinski V., Macagno C., De S. P., Valvassori L., Carinci P., Mordoh J. (2006)Comparative study of gene expression by cDNA microarray in human colorectal cancer tissues and normal mucosa. Int. J. Oncol. 29, 83–94 [PubMed] [Google Scholar]
- 25. Salvesen G. S., Duckett C. S. (2002) IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 3, 401–410 [DOI] [PubMed] [Google Scholar]
- 26. Deveraux Q. L., Stennicke H. R., Salvesen G. S., Reed J. C. (1999) Endogenous inhibitors of caspases. J. Clin. Immunol. 19, 388–398 [DOI] [PubMed] [Google Scholar]
- 27. Eramo A., Ricci-Vitiani L., Zeuner A., Pallini R., Lotti F., Sette G., Pilozzi E., Larocca L. M., Peschle C., De M. R. (2006) Chemotherapy resistance of glioblastoma stem cells. Cell Death. Differ. 13, 1238–1241 [DOI] [PubMed] [Google Scholar]
- 28. Signore M., Ricci-Vitiani L., De M. R. (2011) Targeting apoptosis pathways in cancer stem cells. Cancer Lett [DOI] [PubMed] [Google Scholar]
- 29. Di Stefano A. B., Iovino F., Lombardo Y., Eterno V., Hoger T., Dieli F., Stassi G., Todaro M. (2010) Survivin is regulated by interleukin-4 in colon cancer stem cells. J. Cell. Physiol. 225, 555–561 [DOI] [PubMed] [Google Scholar]
- 30. Yang L., Cao Z., Yan H., Wood W. C. (2003) Coexistence of high levels of apoptotic signaling and inhibitor of apoptosis proteins in human tumor cells: implication for cancer specific therapy. Cancer Res. 63, 6815–6824 [PubMed] [Google Scholar]
- 31. Chen D. J., Huerta S. (2009) Smac mimetics as new cancer therapeutics. Anticancer Drugs 20, 646–658 [DOI] [PubMed] [Google Scholar]
- 32. Flygare J. A., Fairbrother W. J. (2010) Small-molecule pan-IAP antagonists: a patent review. Expert. Opin. Ther. Pat 20, 251–267 [DOI] [PubMed] [Google Scholar]
- 33. Oost T. K., Sun C., Armstrong R. C., Al-Assaad A. S., Betz S. F., Deckwerth T. L., Ding H., Elmore S. W., Meadows R. P., Olejniczak E. T., Oleksijew A., Oltersdorf T., Rosenberg S. H., Shoemaker A. R., Tomaselli K. J., Zou H., Fesik S. W. (2004) Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer. J. Med. Chem. 47, 4417–4426 [DOI] [PubMed] [Google Scholar]
- 34. Fulda S., Wick W., Weller M., Debatin K. M. (2002) Smac agonists sensitize for Apo2L/T. Nat. Med. 8, 808–815 [DOI] [PubMed] [Google Scholar]
- 35. Gyrd-Hansen M., Meier P. (2010) IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat. Rev. Cancer 10, 561–574 [DOI] [PubMed] [Google Scholar]
- 36. Fulda S. (2008) Targeting inhibitor of apoptosis proteins (IAPs) for cancer therapy. Anticancer Agents Med. Chem. 8, 533–539 [DOI] [PubMed] [Google Scholar]
- 37. Chen D. J., Huerta S. (2009) Smac mimetics as new cancer therapeutics. Anticancer Drugs 20, 646–658 [DOI] [PubMed] [Google Scholar]