Abstract
Tumors of the head and neck represent a molecularly diverse set of human cancers, but relatively few proteins have actually been shown to drive the disease at the molecular level. To identify new targets for individualized diagnosis or therapeutic intervention, we performed a kinase centric chemical proteomics screen and quantified 146 kinases across 34 head and neck squamous cell carcinoma (HNSCC) cell lines using intensity-based label-free mass spectrometry. Statistical analysis of the profiles revealed significant intercell line differences for 42 kinases (p < 0.05), and loss of function experiments using siRNA in high and low expressing cell lines identified kinases including EGFR, NEK9, LYN, JAK1, WEE1, and EPHA2 involved in cell survival and proliferation. EGFR inhibition by the small molecule inhibitors lapatinib, gefitinib, and erlotinib as well as siRNA led to strong reduction of viability in high but not low expressing lines, confirming EGFR as a drug target in 10–20% of HNSCC cell lines. Similarly, high, but not low EPHA2-expressing cells showed strongly reduced viability concomitant with down-regulation of AKT and ERK signaling following EPHA2 siRNA treatment or EPHA1-Fc ligand exposure, suggesting that EPHA2 is a novel drug target in HNSCC. This notion is underscored by immunohistochemical analyses showing that high EPHA2 expression is detected in a subset of HNSCC tissues and is associated with poor prognosis. Given that the approved pan-SRC family kinase inhibitor dasatinib is also a very potent inhibitor of EPHA2, our findings may lead to new therapeutic options for HNSCC patients. Importantly, the strategy employed in this study is generic and therefore also of more general utility for the identification of novel drug targets and molecular pathway markers in tumors. This may ultimately lead to a more rational approach to individualized cancer diagnosis and therapy.
Head and neck squamous cell carcinoma (HNSCC)1 is the sixth most common form of cancer with ∼600,000 new cases worldwide every year. The prognosis for this disease is poor (40–50% survival rate over 5 years) and has not markedly improved over the past decades (1). At the molecular level, HNSCC is a very heterogeneous disease that can roughly be divided into human papillomavirus-positive (20%) and human papillomavirus-negative cases (80%) (1). Genome instability and aneuploidity are frequently observed, and genetic alterations have been identified in both tumor suppressors (e.g., PTEN and TP53) (2–5) and oncogenes (e.g. PIK3CA and EGFR) (6–8), but there are also many cases in which the genome appears to be rather normal (1). Relatively few signaling pathways have so far been shown to be involved in the pathogenesis of HNSCC. Among these are the transforming growth factor-β/SMAD (9–11) and EGFR/phosphatidylinositol 3-kinase/AKT pathways (12). The latter offers a number of possible therapeutic intervention points particularly at the level of EGFR itself (13), which is amplified and/or overexpressed in many HNSCC cases (7). Anti-EGFR monoclonal antibody (14) or tyrosine kinase inhibitor (15) therapies have shown some clinical benefits, notably for combined antibody and radiation therapy (16). In comparison, tyrosine kinase inhibitors have shown rather low response rates, the reasons for which are currently not clear (15). Despite the success of EGFR-targeted therapy, there is a great need to identify new molecular targets whose activity may drive this cancer in the many individuals for whom EGFR does not play a major role. Recently, several other kinase-centric molecular mechanisms have been investigated. These include aurora kinase A (AURKA) (17), polo-like kinase 1 (PLK1) (18), and c-MET (19), indicating that the observed molecular heterogeneity of the disease may be rooted in multiple kinase signaling pathways and underscoring the need for potential biomarkers and/or therapeutic targets for an individualized approach to the management of HNSCC.
Signaling pathways are best studied at the protein level, and quantitative proteomics methods are increasingly used to address signaling in a systematic fashion (20, 21). We have recently developed a chemical proteomics screening method that allows the interrogation of many signaling pathways in parallel (22). The approach comprises two main elements. The first element is an affinity purification matrix termed Kinobeads, which consists of seven immobilized nonselective kinase inhibitors. It allows the purification and identification of several hundred kinases and other ATP-binding proteins from cell lines or tissues (22, 23). The second element is intensity-based label-free quantitative mass spectrometry that enables the identification and relative quantification of the purified proteins across many biological samples (24). Although the Kinobeads approach was initially developed to profile the selectivity of small molecule kinase inhibitors, it also lends itself to profiling the expression of kinases in cells or tissues (22, 25). In this study, we utilized the Kinobeads approach to identify systematically kinases from HNSCC cell lines that might represent novel candidate targets for individualized therapeutic intervention and/or candidate biomarkers. Quantitative profiling and statistical analysis of 146 protein kinases across 34 HNSCC cell lines revealed that 42 kinases showed highly significant differential protein expression. These included disease associated kinases such as EGFR, c-MET, and AURKA but also novel candidates. Loss of function experiments using siRNA and small molecule kinase inhibitors showed that EGFR, EPHA2, NEK9, RIPK2, WEE1, and JAK1 are involved in cell survival, and the validation data assembled thus far suggest EPHA2 as a novel target for HNSCC therapy.
EXPERIMENTAL PROCEDURES
Cell Culture and Harvesting
All the 34 cell lines used in this study represent HNSCC of the tongue, and further information on the cell lines (including relatively sparse clinical information) can be found supplemental Table S1 and several publications (26–42). With the exception of the four cell lines HSC-3, HSC-4, OSC-19, and OSC-20, all were originally obtained from primary tumors. We note that these lines are not primary cells but are cell lines adapted to grow in culture over an extended period of time. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with high glucose and glutamine (PAA, Pasching, Austria) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS; PAA), 1× nonessential amino acids (NEAA; PAA) at 37 °C in humidified air with 10% CO2. To standardize conditions, all of the cell lines were grown without specific additives such as hydrocortisone. In the case of cell lines originally maintained with hydrocortisone (BICR16 and BICR56), the culture medium was depleted of hydrocortisone, and the cells were then grown without it for at least seven further passages without observing major effects on growth rate or cell morphology. Prior to harvesting, the cells were cultured in medium for 48 h. The cells were grown to 90–100% confluence while avoiding substantial overgrowth. Before lysis, the cells were washed three times with ice-cold PBS. Radioimmunoprecipitation assay 100 buffer (mixed micelle buffer with 20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.5% deoxycholic acid, 0.1% SDS) with freshly added protease (2× protease inhibitor mixture; Roche Applied Science, Mannheim, Germany) and phosphatase inhibitors (5× phosphatase inhibitor cocktail, Sigma-Aldrich; 5× phosphatase inhibitor mixture 2, Sigma-Aldrich; 1 mm sodium orthovanadate and 1 mm sodium molybdate) was used for cell lysis. One ml of ice-cold lysis buffer/175-cm2 cell culture flask was then added to the cells. The cells were scraped immediately, collected in a precooled microcentrifuge tube and incubated on an orbital mixer for 30 min at 4 °C. Lysates were then centrifuged at 2 °C for 30 min at 20,000 × g, and the supernatants were collected, aliquoted, frozen in liquid nitrogen, and stored at −80 °C until further use. Protein concentration in lysates was determined by the Bradford assay.
Affinity Purification and Protein Digestion
Kinobead pulldowns were performed as described previously (22). Briefly, cell lysates were diluted with equal volumes of 1× compound pulldown buffer (50 mm Tris/HCl, pH 7.5, 5% glycerol, 1.5 mm MgCl2, 150 mm NaCl, 20 mm NaF, 1 mm sodium orthovanadate, 1 mm DTT, 5 mm calyculin A, and protease inhibitors). Lysates were further diluted if necessary to a final protein concentration of 5 mg/ml using 1× compound pulldown buffer supplemented with 0.4% Nonidet P-40. Kinobeads (100 μl suspension) were incubated with lysates (total of 5 mg of protein) at 4 °C for 4 h. Subsequently, beads were washed with 1× compound pulldown buffer and collected by centrifugation. The bound proteins were eluted with 2× NuPAGE® LDS sample buffer (Invitrogen), and the eluates were reduced and alkylated by 10 mm DTT and 55 mm iodoacetamide. The samples were then run into a 4–12% NuPAGE gel (Invitrogen) for about 1 cm to concentrate the sample prior to in-gel trypsin digestion. In-gel trypsin digestion was performed according to standard procedures.
LC-MS/MS Analysis
Nanoflow LC-MS/MS was performed by coupling an Eksigent nanoLC-Ultra 1D+ (Eksigent, Dublin, CA) to a LTQ-Orbitrap XL electron transfer dissociation (Thermo Scientific, Bremen, Germany). Tryptic peptides were dissolved in 20 μl of 0.1% formic acid, and 10 μl were injected for each analysis. Peptides were delivered to a trap column (100 μm inner diameter × 2 cm, packed with 5 μm C18 resin, Reprosil PUR AQ; Dr. Maisch, Ammerbuch, Germany) at a flow rate of 5 μl/min in 100% buffer A (0.1% FA in HPLC grade water). After 10 min of loading and washing, the peptides were transferred to an analytical column (75-μm × 40-cm C18 column Reprosil PUR AQ, 3 μm; Dr. Maisch, Ammerbuch, Germany) and separated using a 210-min gradient from 2 to 35% of buffer B (0.1% FA in acetonitrile) at 300 nl/min flow rate. The LTQ-Orbitrap was operated in data-dependent mode, automatically switching between MS and MS2. Full scan MS spectra were acquired in the Orbitrap at 60,000 resolution. Internal calibration was performed using the ion signal (Si(CH3)2O)6H+ at m/z 445.120025 present in ambient laboratory air. Tandem mass spectra were generated for up to eight peptide precursors in the linear ion trap for fragmentation by using collision-induced dissociation.
Peptide and Protein Quantification and Identification
Progenesis software (version 3.1; Nonlinear Dynamics, Newcastle, UK) was used for intensity-based label-free quantification. Briefly, after selecting one sample as a reference, the retention times of all eluting precursor m/z values in all other samples within the experiment were aligned creating a large list of “features” representing the same peptide in each sample. Features with two to six charges were included for further analysis. Features with two or less isotopes were excluded. After alignment and feature filtering, replicate samples were grouped together, and raw abundances of all features were normalized to determine a global scaling factor for correcting experimental variation such as differences in the quantity of protein loaded into the instrument. Briefly, for each sample, one unique factor is calculated and used to correct all features in the sample for experimental variation according to Ref. 43. Given that multiple MS/MS spectra are frequently collected for the same feature (precursor ion) across all the samples, the precursor intensities were ranked, and the MS/MS spectra of the five most intense precursors for each feature were transformed into peak lists and exported to generate Mascot generic files. The Mascot generic files were searched against the protein sequence database IPI human (v. 3.68, 87,061 sequences) using Mascot (v.2.2, Matrix Science, London, UK). Search parameters were as follows: fixed modification of carbamidomethylation of cysteine residues, variable modification of S, T, and Y phosphorylation and M oxidation, trypsin as proteolytic enzyme with up to two miss cleavages, precursor ion mass tolerance of 5 ppm, fragment ion mass tolerance of 0.6 Da, decoy search enabled. Search results for spectrum to peptide matches were exported in .xml format and then imported into Progenesis software to enable the combination of peptide quantification and identification. Peptides with mascot ion scores <33 (p < 0.05 identity threshold) were filtered out, and only unique peptides for corresponding proteins were used for identification and quantification. Single peptide identifications were removed except for identified kinases. Tandem mass spectra for single peptide identifications were annotated and are documented in supplemental Fig. S1. For protein quantification, the feature intensities of all unique peptides of a protein were summed up. The results of Progenesis analysis (raw data and normalized data) are tabulated in supplemental Tables S2 and S3.
Statistical Analysis
To investigate the data distribution and ensure the appropriate application of statistical tools, frequency histograms and quantile-quantile plots were created for all non-normalized kinase intensities provided by Progenesis analysis (see above and supplemental Fig. S2). The data were then normalized by variance stabilization normalization (VSN) (44), which addresses the error structure in the data and stabilizes the variance across the entire intensity range. VSN has previously been shown to be applicable and useful for MS-based quantification (45). The power of the VSN methodology is greatest in situations where hypothesis tests are used to detect changes in protein expression. The underlying assumptions of these hypothesis tests are data normality and homogeneity of variance, which tend to be more appropriate represented by VSN-transformed rather than non-normalized data (supplemental Fig. S2). To detect differential kinase protein expression between multiple cell lines, a moderated linear model was applied using the limma package (46) in Bioconductor (47). Briefly, a complete pairwise comparison was performed as follows: Let ykc be the expression values for kinases k = 1 … K and cell lines c = 1…C, preprocessed and normalized (supplemental Fig. S1), and then the systematic effect for each kinase can be described by a linear model E(yk) = Xβk, where yk = (yk1…ykC)T is the vector of expression values for kinase k, X is a known design matrix with full column rank R, and βk = (βk1,..,βkR)T is a kinase-specific vector of regression coefficients. Regression coefficients represent comparisons of interest between cell lines in the experiment. These coefficients were estimated with the least squares linear model fitting procedure and tested for differential expression (testing any particular βkR equal to 0) by moderated Student's t statistics via the empirical Bayesian statistics described in the limma package (46). The null hypothesis was accepted or rejected on the basis of p values computed for the omnibus F-statistic via limma as described above, at a specified significance level. The p values were adjusted for multiple testing to control the false discover rate at 5% using the algorithm of Benjamini and Hochberg (48). Adjusted p values allowed us to identify differentially expressed kinases. The results of the statistical analysis are tabulated in supplemental Table S4.
Kinase Inhibitor Treatments
The EGFR inhibitors gefetinib and erlotinib, the dual EGFR/HER2 inhibitor lapatinib, and the pan SRC family inhibitor dasatinib were purchased from LC Laboratories (Woburn, MA) and the c-MET inhibitor PHA665752 was purchased from Tocris Bioscience (Bristol, UK). All of the inhibitors were dissolved as 10 mm stock solutions in DMSO and kept at −20 °C. HNSCC cells were seeded in 96-well plates at 3 × 10 3 cells/well and grown in DMEM supplemented with 10% (v/v) FBS and 1× NEAA for 24 h prior to experimental treatments. Next, 100 μl of fresh medium supplemented with different concentrations of kinase inhibitors (range of 40 nm to 10 μm in 0.1% DMSO depending on the experiment; control cells were treated with 0.1% DMSO as a vehicle control) were added to the cells. Cell viability was monitored after 96 h of treatment using the XTT cell proliferation kit II (Roche Applied Science).
Immunoblot Analysis
Anti- EGFR, p-EGFR, AKT, p-AKT, ERK, p-ERK, and FAK antibodies were purchased from Cell Signaling Technology (provided by New England Biolabs Inc., Ipswich, MA), and anti-p-FAK, EPHA2, and α-tubulin antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). For immunoblot analysis, the cells were washed with precooled PBS and lysed in radioimmunoprecipitation assay buffer. Protein concentration was determined by the Bradford assay. Fifty μg of lysate from control and treated cells were mixed with equal volumes of 2× NuPAGE® LDS sample buffer containing 10 mm DTT and heated at 95 °C for 5 min. The proteins were subsequently separated by 4–12% NuPAGE gel and transferred onto to PVDF membranes (Invitrogen). The membranes were blocked for 1 h in blocking solution (2% BSA in 1× Tris-buffered saline, 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 0.1% Tween 20) at room temperature and probed overnight at 4 °C with the respective primary antibody. Immunoreactivity was detected using IgG-conjugated peroxidase (GE Healthcare, Little Chalfont, UK) and visualized by an ImageQuant LAS 4000 mini (GE Healthcare). For phosphorylation analysis, the membranes were first probed with the respective phospho-specific antibody and then stripped with 62.5 mm Tris-HCl, pH 6.8, 100 mm β-mercaptoethanol, 2% SDS for 30 min at room temperature to allow probing the same blot for the respective total protein. The EGFR and EPHA2 antibodies used for Western blotting were validated by single band detection on Western blots of full HNSCC cell lines (supplemental Fig. S3).
Protein Knockdown by siRNA
Small interfering RNA (siRNA) reagents were purchased from Qiagen. The concentrations used in the described experiments were twice the values recommended by the manufacturer (see supplemental Table S5). The cells serving as negative controls were treated with transfection reagent only; positive controls were transfected with 1 nm AllStars Hs cell death control siRNA (Qiagen). 7500 cells were seeded in 96-well plates and incubated in DMEM containing 10% (v/v) FBS and 1× NEAA at 37 °C 1 day before transfection. siRNA duplexes were diluted in 50 μl of DMEM without serum, followed by the addition of 1 μl of INTERFERinTM (Polyplus Transfection, Illkirch, France) and immediate homogenization for 10 s. The mixture was then incubated for 10 min at room temperature to allow formation of the transfection complex between siRNA duplexes and INTERFERinTM. After exchanging the medium with 125 μl of fresh prewarmed complete medium, the transfection mixture was added to the cells and mixed gently by swirling. Cell viability was assessed by the XTT assay after 4 days as described above.
Sequence Determination of EGFR Transcripts from Different Cell Lines
Total mRNA was prepared from cell lines Cal27, UPCI:SCC056, UTSCC87, UTSCC14, and UTSCC40 using the magnetic mRNA isolation kit (New England Biolabs Inc., Ipswich, MA). The purified mRNA served as template for cDNA synthesis with anchored poly(dT) primers employing the ProtoScript Moloney murine leukemia virus Taq reverse transcription-PCR kit (New England Biolabs Inc., Ipswich, MA). EGFR encoding transcripts were selectively amplified by touchdown PCR. Because of the limited reading length of the sequencing reaction, eight consecutive, overlapping segments were generated by designing eight corresponding PCR primer pairs to cover the full-length EGFR encoding sequence. Individual PCRs were prepared in 50-μl aliquots by mixing 2 μl of cDNA (at varying dilutions), 0.2 μm primers (biomers.net, Ulm, Germany), 1 unit of Phusion high fidelity DNA polymerase (New England Biolabs Inc.), 10 μl of 5× reaction buffer, and 200 μm dNTP mix. Touchdown PCR was initiated in a Primus25 thermocycler (PEQlab, Erlangen, Germany) at 98 °C for 3 min, followed by 10 touchdown cycles with 30 s of denaturation at 98 °C, 60 °C annealing decreased by 1 °C per cycle for 15 s, and 72 °C synthesis for 45 s. After 25 additional cycles at a constant annealing temperature of 55 °C, the reaction was completed by a final extension step at 72 °C for 5 min. The PCR products were purified by agarose gel electrophoresis employing the QIAquick gel extraction kit (Qiagen). The DNA sequence of individual PCR amplicon was determined using the ABI Prism 3730 automatic sequencer (Applied Biosystems) and the primers employed for transcript isolation by PCR.
EphrinA1-Fc Treatment
For cell viability assays, 3 × 10 3 cells/well were seeded in 96-well plate with 100 μl of DMEM containing 10% (v/v) FBS and 1× NEAA and allowed to attach for 24 h. Then medium with the respective concentrations of EphrinA1-Fc (or 1 μg/ml Fc for control cells; R & D Systems, Minneapolis, MN) was added to the cells, and cell viability was assessed by the XTT assay after 4 days as described above. For Western blots, 105 cells were seeded in 6-well plates in 1 ml of DMEM containing 10% (v/v) FBS and 1× NEAA. After 24 h, the cells were starved by culturing in serum-free DMEM for another 24 h. Subsequently, the cells were stimulated either with 1 μg/ml EphrinA1-Fc or 1 μg/ml Fc as a control for 30 min. The cells were subsequently washed with PBS and lysed for Western blot analysis as described above.
Immunohistochemistry
The levels of EGFR and EphA2 were analyzed by immunohistochemistry in tissue microarrays (TMAs). All of the tissue samples were obtained from patients who were diagnosed with primary HNSCC and underwent a surgical resection. In total, 92 primary formalin-fixed and paraffin-embedded samples from different anatomic subsites were obtained from the archives of the Institute of Pathology, Technische Universität München. All of the tumor tissue specimens were procured from patients giving written informed consent according to the requirements of the ethics committee of the Technische Universität München. TMAs were generated from these samples in house by sampling one tumor tissue core (1.0 mm in diameter) from each paraffin-embedded tissue block using the technique pioneered by Kononen et al. (78). The TMAs were constructed solely for the purpose of profiling protein expression in HNSCC patients. Therefore, no further clinical data (such as human papillomavirus status, smoking habit, alcohol abuse) are available for this patient material. Immunohistochemical staining for EGFR and EphA2 were carried out using an automated stainer (Ventana Discovery, Tuscon, AZ) and the DAB Map kit (Ventana) as described elsewhere (49). The tissues were incubated with either an EGFR antibody (Dako EGFR pharmDXTM kit) or an EphA2 polyclonal antibody (sc-924; Santa Cruz) in a dilution of 1:250. The EGFR antibody used for TMAs was validated by the vendor. The EPHA2 antibody used for TMAs was validated by single band detection on Western blots against a number of human tissues in the Human Protein Atlas project (http://www.proteinatlas.org/) and by single band detection on Western blots against complete cell lysates of HNSCC cell lines in our laboratory (supplemental Fig. S3).
Data Availability
The protein identification data from this study are available from the following ftp site: ftp://ftp.lrz.de/transfer/proteomics/.
RESULTS
Strategy and Identification of Differential Kinase Protein Expression in HNSCC Cell Lines
The strategy employed for the identification of differential kinase protein expression from HNSCC cell lines is illustrated in Fig. 1. Briefly, cell lines originally derived from primary HNSCC tumors (30 of 34) or metastases (4 of 34; supplemental Table S1) were grown in vitro and subsequently lysed in the presence of high concentrations of phosphatase inhibitors. Kinases were partially purified using Kinobeads, which comprise a mixed Sepharose matrix of seven immobilized kinase inhibitors. The enriched proteins were subjected to trypsin digestion and nanoscale LC-MS/MS and identified by database searching of the tandem mass spectra. Label-free quantification was performed using the m/z and retention time aligned precursor ion intensities of the identified peptides integrated across the chromatographic peak. Moderated F-statistics was employed to identify kinases that are significantly differentially expressed across the 34 cell lines.
Fig. 1.
Quantitative chemical proteomics strategy for the identification of kinase drug targets or signaling pathway biomarkers.
Suitability of Precursor Intensity-based Label-free Relative Quantification
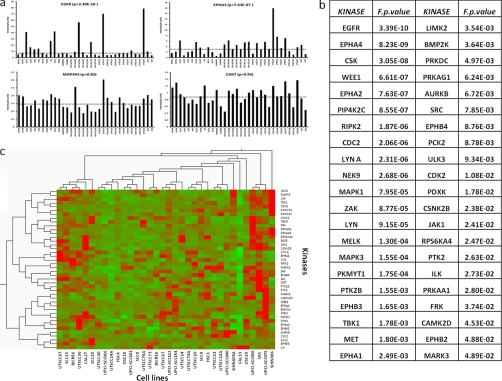
In this study, 34 cell lines were profiled for kinase expression in duplicate generating 68 samples, each of which was subjected to a 4-h LC-MS/MS experiment for label-free protein quantification and identification (2 × 34 × 4 h = 272 h LC-MS/MS time). In total, 17,873 precursor ion and retention time features led to successful peptide identifications. This corresponds to 665 unique proteins including 146 kinases with the latter contributing ∼50% of the entire signal intensity (supplemental Tables S2 and S3). Obviously, intensity-based label-free quantification requires an intensity response that is directly proportional to the amount of analyte. LC-MS experiments using a dilution series of the Universal Proteomics Standard (Sigma), show that the response of the system is linear over more than 2 orders of magnitude (supplemental Fig. S4). For our kinobeads experiments, we do not know the absolute quantities of any of the proteins we purify. It should therefore be noted that all of our quantification data are relative (i.e. comparing the relative quantities of one protein across the different experiment is possible, but the quantities of different proteins cannot be compared with each other). Other prerequisites for successful quantification using MS intensity data are consistent sample processing and a stable LC-MS system to avoid bias in the analysis. To address this, we processed and analyzed experiments in two blocks of 34 samples (i.e. cell lines). This minimized variance in LC-MS conditions so that the data are comparable within each replicate. Variation between replicates can then be easily addressed by standard normalization methods. Supplemental Fig. S5 shows that the retention time distribution in the data is very narrow, and the median CV of all identified LC-MS features (i.e. peptides) is 1.1% (42% of all features show retention time CVs <1, and 93% show CVs <2%). The median precursor mass error of identified peptides was 0.8 ppm, and 95% of all identified peptides were measured within 1.4 ppm mass error. This demonstrates that the employed LC-MS conditions were very stable and thus well suited for quantification purposes. For two proteins (EGFR and MET), we also assessed how the MS intensity data compare to Western blot staining across both batches of 34 cell lines (supplemental Figs. S6 and S7). The correlation of the two quantification methods was found to be very good (R2 = 0.85 for EGFR), proving that label-free mass spectrometry is a valid quantification approach in this study. To evaluate the technical merits of the overall approach, we performed three biological and technical repeats using the cell line Cal27 (supplemental Fig. S8). The overall variance in the biological (technical) replicates after normalization is a relatively low CV of 0.08 (0.04). As one would expect, the variance in the quantification data decreases as the number of peptides measured per protein increases. Still, even for proteins quantified by just a single peptide, the variance between biological replicates is rarely larger than 20%. Because Kinobead affinity pulldowns represent relatively noncomplex proteomes, the reproducibility of protein identification is also very high. In the above replicates, all of the proteins with four or more peptides are identified in all of the replicates, and 80% of all proteins reproduce in all replicates if two peptides are found for a protein. In our analytical set-up, the reproducibility of protein identification is further improved by the alignment of the data. As long as a protein is robustly identified in at least one of the samples, the alignment of the retention times together with the accurate mass of the peptide precursor will lead to the identification of all peptides in all samples (as long as they are present above noise) (50). As a side note, the data from this controlled sample set highlight a distinct advantage of MS intensity-based quantification over spectrum counting. The percentage of missing data points (i.e. identified/quantified proteins across the three biological replicates) is 0% for MS intensity and 6.9% for spectrum counting. For the screen of 34 cell lines (in duplicate), the respective numbers are 5.6% missing values for MS intensity and 76.6% for spectrum counting. From the above, we conclude that the experimental approach generates robust qualitative and quantitative data for subsequent data analysis. 146 kinases were identified and quantified across all 34 cell lines, and Fig. 2 and supplemental Tables S2–S4 summarize these data. Most kinases were similarly expressed across the panel of cell lines (e.g. MAP4K5 and CDK7; Fig. 2a, bottom panel), but some kinases showed marked differences in expression (e.g. EGFR and EPHA2; Fig. 2a, upper panel). Moderated F-statistics based on the normalized MS intensity data of all quantified kinases resulted in 42 kinases showing highly significant differential expression between cell lines (p < 0.05; Fig. 2b). These include receptor tyrosine kinases such as EGFR, EPH receptor family members, DDR1, and c-MET as well as a diverse set of cytoplasmic kinases of the NEK, SRC, and other families. Unsupervised clustering of the differential kinases, and 34 cell lines (Fig. 2c) failed to show strong grouping of either the kinases or the cell lines. Attempts to cluster the samples or kinases by criteria such as (the sparsely available) clinical information, kinase family membership, or member of signaling pathways did also not result in a significant grouping, indicating that the kinase profiles and underlying properties of the respective cancer cells are probably indeed quite different.
Fig. 2.
Identification of differential kinase protein expression across 34 HNSCC cell lines. a, examples for protein expression levels across all cell lines utilizing retention time aligned precursor ion intensities. Horizontal lines mark the median expression values. b, results of moderated F-statistics reveal kinases that are differentially expressed between cell lines (only proteins with F p values of <0.05 are listed). c, unsupervised clustering does not reveal any obvious grouping of differentially expressed kinases or cell lines, indicating strong differences between individual tumor biologies.
Some but Not All HNSCC Cell Lines Are Dependent on EGFR for Survival
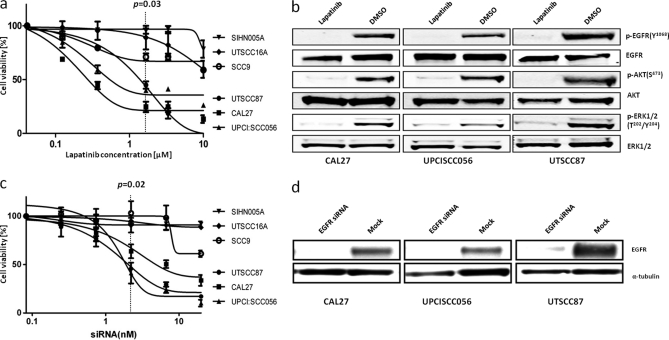
EGFR has been shown to be overexpressed in the majority of HNSCC patients (14), and it is the most differentially expressed kinase in our cell line panel (p = 3.39E-10). Interestingly, the difference in EGFR expression between the lowest and highest expressing cell lines is a factor of 25 (supplemental Table S2), and 7 of the 34 cell lines investigated (21%) show very high expression levels (>3-fold over median; Fig. 2a). This is in very good agreement with immunohistochemical data collected from HNSCC tumor samples, where 6 of 25 patients (24%) showed very strong EGFR staining (supplemental Fig. S9). We hypothesized that our approach would enable us to highlight abnormally expressed kinases as drivers for the proliferation of a subpopulation of HNSCC cells. To verify this hypothesis, we compared cell viability in three high EGFR expressing cell lines (Cal27, UPCI:SCC056, and UTSCC87) with three low EGFR-expressing cell lines (SIHN005A, SCC9, and UTSCC16A) in response to EGFR small molecule inhibitors and RNA interference. Fig. 3a shows that submicromolar concentrations of lapatinib, a highly selective EGFR/HER inhibitor, kills high but not low EGFR-expressing cells with a concomitant down-regulation of the AKT and ERK signaling pathways (Fig. 3b). The same differential drug response was observed for the EGFR inhibitors gefitinib and erlotinib (supplemental Fig. S10). Similarly, knocking down EGFR protein expression in cells by siRNA led to a strong reduction in cell viability of high but not low EGFR-expressing cells (Fig. 3, c and d). Because pharmacological inhibition and genetic knockdown show the same viability phenotypes and because we did not detect any mutations in EGFR by cDNA sequencing (supplemental Fig. S11), we conclude that EGFR overexpression is a major driver of survival in some (20–25%) but not all HNSCC cell lines.
Fig. 3.
Some but not all HNSCC cell lines are growth-dependent on EGFR overexpression and activity. a, high EGFR-expressing cells respond more strongly to lapatinib treatment than low EGFR-expressing cells. b, signaling pathway analysis using phospho-specific antibodies reveals that lapatinib treatment inhibits the EGFR/AKT and EGFR/ERK pathways. c, high EGFR-expressing cells respond more strongly to siRNA-mediated EGFR knockdown than low EGFR-expressing cells. d, Western blot analysis documenting EGFR knockdown efficiency in CAL27, UPCISCC056, and UTSCC87 cells.
Loss of Function Screening by siRNA Highlights New Candidate Drug Targets
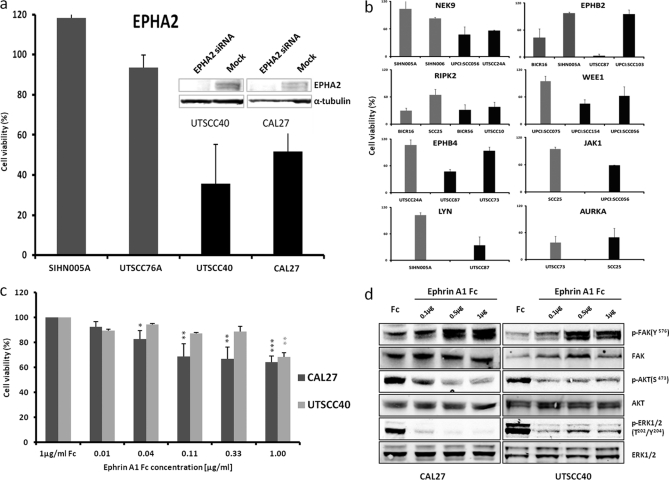
To elucidate whether other differentially expressed kinases are also involved in the proliferation of some of the cell lines, we chose a further 22 proteins from the list of differentially expressed kinases (p < 0.05) (Fig. 2b), and six kinases for which no strong evidence for differential behavior was obtained from the statistical analysis. Each of these was subjected to siRNA knockdown in the respective high and low expressing cell lines (supplemental Table S5). The data show that for many proteins, loss of expression did not significantly alter cell viability. However, nine kinases (AURKA, EPHA2, EPHB2, EPHB4, JAK1, LYN, NEK9, RIPK2, and WEE1) showed strong (>40%) reduction of cell viability 96 h post-siRNA transfection, suggesting that these kinases play an important role in promoting and maintaining the survival of cells. Interestingly, whereas EPHA2, NEK9, LYN, WEE1, and JAK1 knockdown killed cells with high but not low expression, AURKA and RIPK2 knockdown killed both high and low expressing cells (Fig. 4, a and b). These data suggest that at least EPHA2, NEK9, WEE1, LYN, and JAK1 might constitute novel targets with therapeutic potential in HNSCC, whereas the other proteins are more generally important for cell survival and/or proliferation.
Fig. 4.
EPHA2 target validation. a, cell viability of low (gray) and high (black) EPHA2-expressing cell lines following EPHA2 protein knockdown by siRNA. EPHA2 protein levels are shown in the inset. b, example results from a siRNA screen of 29 differentially expressed kinases in the respective low (gray) and high (black) expressing cell lines. c, cell viability decreases in high EPHA2-expressing cell lines following treatment with Fc-conjugated Ephrin A1. d, signaling pathway analysis using phospho-specific antibodies reveals that Ephrin A1 treatment inhibits ERK and AKT and induces FAK phosphorylation.
EPHA2 Target Validation
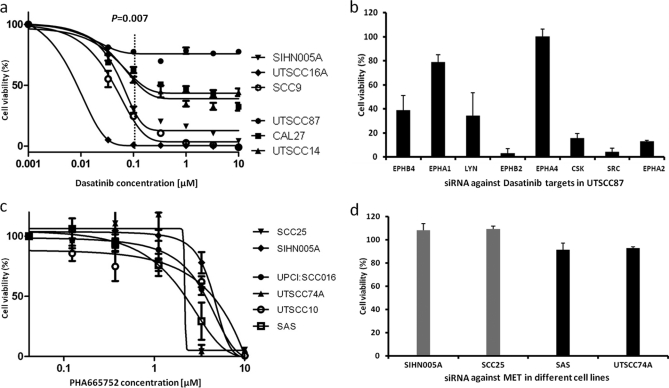
Analysis of EPHA2 protein expression by immunohistochemistry in a HNSCC tissue array (n = 92 patients; supplemental Fig. S12) revealed that ∼15% of all patients express very high levels of EPHA2 (55% show moderate expression, and 30% of stains are weak or absent), which correlates well with data obtained by MS intensity profiling (Fig. 1). We next addressed whether EPHA2 overexpression had any functional link to cancer cell viability. As described above, siRNA of EPHA2 killed high (Cal27 and UTSCC40) but not low (SIHN005A and UTSCC76A) expressing cells (Fig. 4a). In addition, we treated high EPHA2-expressing cells (Cal27 and UTSSC40) with Fc-conjugated Ephrin A1 (a negative regulator of EPHA2 activity), which led to a modest (30–40%) but statistically highly significant and dose-dependent reduction in cell viability relative to control Fc-treated cells (Fig. 4c). This is in line with such data reports for other cell lines in the literature (51). Western blot analysis showed that downstream AKT and ERK phosphorylation were reduced, and FAK phosphorylation (an EPHA2 proximal signaling molecule) was increased by EPHA1-Fc treatment (Fig. 4d), suggesting that EPHA2 signaling is functional in the cell lines studied. Further evidence for a role of EPHA2 in cell survival comes from double siRNA knockdown experiments. Cal27 cells express high levels of EPHA2 as well as EGFR. The viability of Cal 27 cells was strongly reduced by siRNA-mediated knockdown of either EGFR (30%, p = 0.01) or EPHA2 (58%, p = 6E-4; supplemental Fig. S13). A simultaneous knockdown of both proteins led to an additive reduction in cell viability (75%, p = 3E-5), suggesting that both receptors act independently. The abrogation of ERK and AKT phosphorylation, however, suggests that the pathways intersect or converge at some point. EPHA2 signaling is not well studied in general and much less so in HNSCC. However, from mining the literature of the Ephrin receptor family and EGFR using the Ingenuity Pathway Analysis software, it is possible that intersections of the pathways occur at the kinase FAK or the adaptor protein SHC. Both FAK and SHC are in fact interaction partners of EPHA2, and SHC is also a described interaction partner of EGFR (52). These connections would link both EPHA2 and EGFR to the observed phosphorylation effects on FAK, AKT, and ERK (see supplemental Fig. S14 for a possible pathway model). However, future work will have to address whether these axes are also present in HNSCC cells. Pharmacological inhibition is an important complement to siRNA knockdown for target validation. Unfortunately, no selective EPHA2 inhibitor has yet been described. However, it is known that EPHA2 is a low nanomolar target of the small molecule dasatinib (a potent pan-SRC family kinase inhibitor) (53). We observed that not only the EGFR inhibitors used earlier but also dasatinib discriminates between high expressing (UTSCC87, CAL27, and UTSCC14) and low (SIHN005A, UTSCC16A, and SCC9) EGFR-expressing cells (p = 0.007 at 100 nm dasatinib dose; Fig. 5a). This effect cannot be attributed to EGFR inhibition because dasatinib has no activity against EGFR at the concentration employed. To investigate which proteins might be responsible for the observed effect, we knocked down known dasatinib targets in the highly sensitive cell line UTSCC87 (EC50 for dasatinib <10 nm; Fig. 5b). Interestingly, the loss of SRC, CSK, EPHA2, or EPHB2 expression each led to a >80% reduction in cell viability, which may be taken as circumstantial evidence that a part of the cell killing induced by dasatinib might be mediated via EPHA2. Formal proof of this hypothesis would require a selective EPHA2 inhibitor that, as mentioned above, is not available yet.
Fig. 5.
Role of SRC family kinases and c-MET in the survival of HNSCC cell lines. a, high EGFR-expressing cells respond more strongly to Dasatinib treatment than low expressing cells. b, siRNA-mediated protein knockdown of known Dasatinib targets in the highly Dasatinib-sensitive cell line UTSCC87 shows that differential killing observed in a can be attributed to inhibition of several proteins including EPHA2. c, high and low c-MET-expressing cell lines respond similarly to the selective c-MET inhibitor PHA665752. d, siRNA-mediated c-MET knockdown of low (gray) and high (black) c-MET-expressing cell lines shows no significant effect on cell viability.
Evaluation of c-MET as a Target in HNSCC
It has been suggested that c-MET is a target in HNSCC (19). Our label-free MS protein expression profiling and Western blot profiling data (supplemental Fig. S7) show that c-MET is moderately differentially expressed across the panel of 34 cell lines. We therefore treated four relatively high (UPCI:SCC016, UTSCC74A, UTSCC10, and SAS) and two relatively low (SCC25 and SIHN005A) c-MET-expressing cell lines with the potent and selective c-MET inhibitor PHA665752 (biochemical kinase assay IC50 of 9 nm; Fig. 5c). None of the cell lines responded to the drug below the cytotoxic dose of 2 μm, and siRNA-mediated c-MET knockdown also showed no significant effect on cell viability in any of the four cell lines tested (Fig. 5d). Thus, from our data, we currently do not have any evidence supporting c-MET inhibition as an effective cancer cell killing mechanism for treating HNSCC.
DISCUSSION
An increasing appreciation of the role of protein kinases in oncogenesis and tumor progression has led to many being evaluated as therapeutic targets using small molecules or biological agents (54, 55). At present, the majority of molecularly targeted drugs used to treat cancer patients are directed against kinases. However, clinical evidence often shows that only a certain subset of patients significantly benefit from such approaches (56). This is because the underlying molecular tumor biology of a particular cancerous disease can vary greatly between patients. Hence, there is a need both for the identification of new drug targets as well as for the identification of biomarkers that can be used to stratify patients for treatment (avoiding exposure to ineffective drugs) and to measure their responses (generating evidence for efficacy of therapeutic regimens) (57).
In this study, we have used a kinase-focused chemical proteomics approach in conjunction with intensity-based label-free quantitative mass spectrometry to profile the expression of 146 kinases across 34 HNSCC cell lines of the tongue to identify novel potential drug targets and/or signaling pathway biomarker candidates. The analysis revealed a great molecular heterogeneity within this group of cell lines and identified a number of proteins previously implicated in the disease (EGFR, c-MET, etc.) as well as novel candidates indicative of individual tumor biology that open new avenues for the development of cancer drugs and companion diagnostics (EPHA2, NEK9, etc.).
These discoveries were enabled in part by the use of the Kinobeads approach, which has been shown to allow the purification of ∼60% of all human protein kinases from cell lines and tissues (22). HNSCC has also been studied by mRNA profiling. For example, the Gene Expression Atlas at the European Bioinformatics Institute (http://www.ebi.ac.uk/gxa) contains mRNA expression data on 152 protein kinases for the Cal27 cell line, which was also used in our study. The kinobeads proteomic data contain 146 kinase from Cal27 cells, and the two data sources together provide evidence for 249 kinases, suggesting that the two techniques are complementary and that kinobeads purify ∼60% of the total expressed kinome of Cal 27 cells. The Gene Expression Atlas mRNA resource lists a total of 350 protein kinases in the context of HNSCC (regardless of cell line, cell type or cancer site), and the proteomic data add a further 63 kinases not covered by the mRNA data. Kinobeads thus allow the profiling of a significant portion of kinome at the protein level, which cannot be feasibly done by classical protein-based methods such as Western blots. Further below, we discuss how the mRNA and protein profiles of the target/marker candidates compare. It has to be borne in mind, however, that the relationship between mRNA and the protein level is often not simple and that the two data types represent quite fundamentally different types of biological information.
A further enabling factor was the use of the MS intensity-based label-free quantification method, which allowed comparison of many (here 34) HNSCC cell lines without requiring a reference sample for pair-wise comparisons as is typically done in stable isotope labeling strategies. The goal to identify proteins representing individual tumor pathologies or subgroups of patient populations necessitates the analysis of larger panels of samples. This in turn requires an analytical platform that can deliver stable operation across extended periods of time. With retention time variations of only ∼1% (enabled by ultra high pressure LC) and peptide mass errors of ∼1 ppm (enabled by Orbitrap detection), the variation in the quantitative data is <10%, showing that MS intensity-based label-free quantification can deliver data quality comparable with stable isotope labeling (24).
Kinobeads were originally developed for selectivity profiling of small molecule kinase inhibitors (22), but we and others have also shown that kinobeads can be employed to identify kinases from human tissue including tumor cells isolated from patients (25). However, direct proteomic analysis of patient samples, although desirable, is challenging because of the often limited available sample quantity, varying ratios of tumor versus stroma versus vasculature, extent of necrosis and hypoxia, infiltration of inflammatory cells, etc., as well as significant issues with candidate validation options. This is why we decided to perform the target discovery and initial validation experiments using cell lines representing one precise cancer site and then to translate the laboratory findings into patient material using immunohistochemistry in human cancer tissue. All of the cell lines studied were originally derived from primary and metastatic tumors of patients with HNSCC of the tongue. We acknowledge that tumor-derived cell lines may not always be representative of the original cancer, but they are much more feasible models to study molecular mechanisms that cannot be addressed experimentally in the tumor itself.
The most differentially expressed protein identified across the panel of cell lines was the EGF receptor. This protein has long been implicated in HNSCC, and several studies have shown at least some level of overexpression in 80–90% of the cases but with a wide range in interindividual variability (14, 58). EGFR overexpression is also clearly correlated with therapeutic response and overall survival (1, 14, 58). Our proteomic data faithfully reproduce these earlier findings in that 7 of 34 (21%) cell lines display very high expression levels, which is also in line with our immunohistochemistry data on primary HNSCC tissue microarrays (supplemental Fig. S9). This also correlates with an impressive response of high but not low EGFR-expressing cells to specific EGFR inhibitors and siRNA treatment (Fig. 3 and supplemental Fig. S10). This behavior can be interpreted using the well established “target addiction” model (59), which states that continued very high expression of a protein is required for maintaining cell survival and the malignant phenotype. In turn, the model also explains why only those patients whose tumors are addicted to EGFR overexpression respond to EGFR-targeted therapy.
The statistical analysis of the proteomic data revealed 42 kinases with varying degrees of differential expression across the panel of cell lines (p < 0.05; Fig. 2b). One might expect that several of these kinases may represent other examples of oncogene addiction akin to the EGFR case, which would immediately qualify these proteins as potential drug targets. Altered protein expression may also result from genome aberrations or other possibly complex molecular phenotypes with or without direct impact on cell survival or proliferation but possibly with value as marker proteins. Thirteen of the cell lines used in this study have also been investigated for copy number variations and resulting mRNA expression (Ref. 29 and supplemental Table S7). For eight of the differential proteins in Fig. 2b (MAPK1, RIPK2, NEK9, PRKDC, ULK3, MELK, TBK1, and RPS6KA4), changes in gene copy number and mRNA levels have been detected. The differential protein expression observed in our study may therefore in part be rooted in genomic aberrations. For a further 11 proteins, significant gene copy number and mRNA changes have been detected, but no statistically significant change in the protein levels across the 34 cell lines were found. Interestingly, no gene copy number changes were observed for EGFR in the above study, and our cDNA sequencing data on EGFR in five cell lines (supplemental Fig. S11) also indicate that the EGFR protein is wild type. However, EGFR is the most differentially expressed protein in our cell line panel, confirming earlier studies that the relationships between mRNA and protein levels can be very complex.
Given that the impact of differential kinases expression on cell viability could not be deduced from the proteomic (and transcriptomic) data alone, we sought to validate and prioritize the candidate list by loss of function experiments using siRNA. In particular, we asked whether cell viability would be differentially affected by knocking down a candidate protein in high versus low expressing cells because such proteins would represent possible targets for individualized therapeutic intervention. The results revealed that there are indeed some proteins that fulfill this criterion, notably EPHA2, JAK1, LYN, NEK9, and WEE1 (Fig. 4 and supplemental Table S4). At the same time, the majority of the differential kinases do not show any noticeable cell viability phenotype in response to siRNA knockdown, indicating that these proteins on their own have no decisive role in cell survival or proliferation (but of course possibly in other biological functions) or that the respective cells can tolerate or compensate for the effects of varying expression levels within the observed range. Knockdown of two kinases (AURKA and RIPK2) led to cell death regardless of expression level differences, indicating central roles of the proteins for cell survival (60, 61).
Clearly, the above exemplifies that merely quantifying differential protein expression behavior by proteomics (or any other omics for that matter) does not suffice to qualify such proteins as potential therapeutic targets or signaling biomarkers. Some level of validation is required to prioritize the list of candidates. It was beyond the scope of this work to evaluate systematically the siRNA-mediated cell viability phenotype of all kinases in all cell lines, but the experiments conducted thus far clearly highlight a small number of proteins in addition to EGFR that merit further investigation. EPHA2 is a receptor tyrosine kinase that has been functionally implicated in a number of solid tumors (62–64) and, more recently, also in HNSCC with potential roles in tumor metastasis and angiogenesis (18, 65–67). However, the details of these possible functions are still unclear (68). EPHA2 mRNA levels have been found to be up-regulated in some but not all HNSCC studies (69), but a clinically interesting but preliminary observation is that HNSCC patients with high EPHA2 protein expression levels (as measured by immunohistochemistry, n = 14) have a poorer survival prognosis than those with low EPHA2 expression (65, 67). Our immunohistochemistry data for EPHA2 protein expression (n = 92 patients) agree well with the earlier study and, together with the siRNA treatment, natural ligand stimulation and kinase inhibition data, suggest that EPHA2 may well be a new drug target in addition to being a potential molecular biomarker for a subpopulation (10–20%) of HNSCC patients. Compelling further evidence would come from the use of selective inhibitors of EPHA2 but unfortunately such molecules have not yet been reported in the literature. It was beyond the scope of this study to follow up the other potential targets highlighted in our analysis in more detail (JAK1, LYN, NEK9, and WEE1). However, JAK1 and LYN are established drug targets in a number of cancers (70, 71) and may thus also represent genuine targets in HNSCC. JAK1 (but not LYN) mRNA levels have been found to be up-regulated in some HNSCC cases (Gene Expression Atlas, #109). The validity of NEK9 and WEE1 as cancer targets is currently less established, but both are very plausible candidates. NEK9 is important in the regulation of mitosis and gene copy number, and associated mRNA changes have been detected in some HNSCC cell lines (29). WEE1 is an important regulator of the G2/M cell cycle checkpoint (72), and again, there is evidence of increased mRNA levels in some cases of HNSCC (Gene Expression Atlas, #109). It has recently been shown that WEE1 inhibition leads to an increase in the sensitivity of osteosarcoma cells to radiation (73). This finding is particularly relevant in the context of our work on the molecular basis for HNSCC, because surgery and radiation are the mainstay therapies in this disease, and radiation sensitization of cells via WEE1 inhibition might therefore further increase the effectiveness of this therapeutic regimen (74).
Dasatinib treatments of HNSCC cell lines resulted in another interesting observation. Dasatinib is a potent pan-SRC family kinase inhibitor but does not inhibit EGFR. Nevertheless, cells expressing high levels of EGFR responded better to the drug than cells expressing low levels (Fig. 5a). This may be explained by the fact that several SRC family members are activated downstream of EGFR in HNSCC (75). If the EGFR-SRC family axis is a dominant survival or proliferation signal, either treatment using an EGFR or SRC inhibitor would lead to cell death. Given that dasatinib is an approved drug, this observation suggests that dasatinib may be an effective treatment for HNSCC cases that are driven by EGFR, SRC family kinases, EPHA2, or a combination thereof. It may thus increase the response rate of patients that would normally only be treated with an EGFR inhibitor alone (or in combination with standard therapy) (76).
Based on mRNA expression profiling and pharmacological inhibition studies, it has been suggested before that c-MET may be a potential molecular target for HNSCC therapies (19). The Kinobead profiling data show that c-MET is also moderately differentially expressed in our cell line panel (Fig. 2b and supplemental Fig. S7). However, neither siRNA knockdown nor pharmacological inhibition using appropriate doses of a selective c-MET inhibitor reduced the viability of c-MET-overexpressing cells in our experiments (Fig. 5, c and d). The earlier studies required very high doses (2–5 μm) of the c-MET inhibitor SU11274 to kill HNSCC cells. This is more than 10 times the reported effective dose for inhibition of cellular c-MET phosphorylation (19) and ∼200 higher than the reported in vitro dose for inhibition of kinase activity. We used a different selective c-MET inhibitor (PHA665752), which has a similar potency and more than 50-fold selectivity across a panel of diverse tyrosine and serine-threonine kinases (77). With this compound, we also observed killing of HNSCC cells when using doses of 2–5 μm. However, our interpretation of these data is that the observed reduction in cell viability is more likely because of off-target effects and nonspecific cytotoxicity rather than the result of selective c-MET inhibition. We therefore argue that potential therapeutic value of c-MET inhibition in HNSCC needs further investigation. Clearly, c-MET may still play important roles in regulating other aspects such as motility and invasiveness of tumor cells in many HNSCCs, but this was not investigated in this study.
CONCLUSIONS
Using a kinase-focused chemical proteomics approach, we were able to profile simultaneously the protein expression of ∼150 kinases across 34 cell lines derived from head and neck cancer of the tongue. This study identified known and novel protein kinases that are important for the survival and proliferation of HNSCC cells, including several ephrin receptors that may represent targets for therapeutic intervention or act as biomarkers indicative of active signaling pathways. One logical extension of this work would be to perform retrospective or prospective studies on HNSCC patients that correlate the expression of the candidates with clinical parameters such as disease progression and survival. On the molecular level, an attractive future avenue is to extend the analysis to the phosphoproteome to address the molecular consequences of differential kinase expression in individual tumor pathologies. Such future work may shed further light on details of individual tumor biologies and may thus provide new signaling biomarkers that can be used to prioritize pathways to target by small molecule drugs and to monitor the response to therapeutic intervention at the molecular level. Last but not least, our work also again highlights the need for more selective pharmacological tools that allow investigation of some of the drug target candidates, particularly from the EPH receptor family.
Footnotes
* This work was supported by grants from the China Scholarship Council (to Z. W.) and the Heads Up cancer charity (to S. F.).
 This article contains supplemental Tables S1–S5 and Figs. S1–S14.
This article contains supplemental Tables S1–S5 and Figs. S1–S14.
1 The abbreviations used are:
- HNSCC
- head and neck squamous cell carcinoma
- DMEM
- Dulbecco's modified eagle medium
- FA
- Formic acid
- FBS
- fetal bovine serum
- NEAA
- nonessential amino acids
- VSN
- variance stabilization normalization
- EGFR
- epidermal growth factor receptor
- ERK
- extracellular signal-regulated kinase
- AURKA
- aurora kinase A
- siRNA
- small interfering RNA
- TMA
- tissue microarray.
REFERENCES
- 1. Leemans C. R., Braakhuis B. J., Brakenhoff R. H. (2011) The molecular biology of head and neck cancer. Nat. Rev. Cancer 11, 9–22 [DOI] [PubMed] [Google Scholar]
- 2. Brennan J. A., Boyle J. O., Koch W. M., Goodman S. N., Hruban R. H., Eby Y. J., Couch M. J., Forastiere A. A., Sidransky D. (1995) Association between cigarette smoking and mutation of the p53 gene in squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 332, 712–717 [DOI] [PubMed] [Google Scholar]
- 3. Okami K., Wu L., Riggins G., Cairns P., Goggins M., Evron E., Halachmi N., Ahrendt S. A., Reed A. L., Hilgers W., Kern S. E., Koch W. M., Sidransky D., Jen J. (1998) Analysis of PTEN/MMAC1 alterations in aerodigestive tract tumors. Cancer Res. 58, 509–511 [PubMed] [Google Scholar]
- 4. Smeets S. J., van der Plas M., Schaaij-Visser T. B., van Veen E. A., van Meerloo J., Braakhuis B. J., Steenbergen R. D., Brakenhoff R. H. (2011) Immortalization of oral keratinocytes by functional inactivation of the p53 and pRb pathways. Int. J. Cancer 128, 1596–1605 [DOI] [PubMed] [Google Scholar]
- 5. Somers K. D., Merrick M. A., Lopez M. E., Incognito L. S., Schechter G. L., Casey G. (1992) Frequent p53 mutations in head and neck cancer. Cancer Res. 52, 5997–6000 [PubMed] [Google Scholar]
- 6. Redon R., Muller D., Caulee K., Wanherdrick K., Abecassis J., du Manoir S. (2001) A simple specific pattern of chromosomal aberrations at early stages of head and neck squamous cell carcinomas: PIK3CA but not p63 gene as a likely target of 3q26-qter gains. Cancer Res. 61, 4122–4129 [PubMed] [Google Scholar]
- 7. Sheu J. J., Hua C. H., Wan L., Lin Y. J., Lai M. T., Tseng H. C., Jinawath N., Tsai M. H., Chang N. W., Lin C. F., Lin C. C., Hsieh L. J., Wang T. L., Shih I., Tsai F. J. (2009) Functional genomic analysis identified epidermal growth factor receptor activation as the most common genetic event in oral squamous cell carcinoma. Cancer Res. 69, 2568–2576 [DOI] [PubMed] [Google Scholar]
- 8. Murugan A. K., Hong N. T., Fukui Y., Munirajan A. K., Tsuchida N. (2008) Oncogenic mutations of the PIK3CA gene in head and neck squamous cell carcinomas. Int. J. Oncol. 32, 101–111 [PubMed] [Google Scholar]
- 9. Bornstein S., White R., Malkoski S., Oka M., Han G., Cleaver T., Reh D., Andersen P., Gross N., Olson S., Deng C., Lu S. L., Wang X. J. (2009) Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J. Clin. Invest. 119, 3408–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hague A., Eveson J. W., MacFarlane M., Huntley S., Janghra N., Thavaraj S. (2004) Caspase-3 expression is reduced, in the absence of cleavage, in terminally differentiated normal oral epithelium but is increased in oral squamous cell carcinomas and correlates with tumour stage. J. Pathol. 204, 175–182 [DOI] [PubMed] [Google Scholar]
- 11. Wang D., Song H., Evans J. A., Lang J. C., Schuller D. E., Weghorst C. M. (1997) Mutation and downregulation of the transforming growth factor beta type II receptor gene in primary squamous cell carcinomas of the head and neck. Carcinogenesis 18, 2285–2290 [DOI] [PubMed] [Google Scholar]
- 12. Freudlsperger C., Burnett J. R., Friedman J. A., Kannabiran V. R., Chen Z., Van Waes C. (2011) EGFR-PI3K-AKT-mTOR signaling in head and neck squamous cell carcinomas: Attractive targets for molecular-oriented therapy. Expert Opin. Ther. Targets 15, 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharafinski M. E., Ferris R. L., Ferrone S., Grandis J. R. (2010) Epidermal growth factor receptor targeted therapy of squamous cell carcinoma of the head and neck. Head Neck 32, 1412–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zimmermann M., Zouhair A., Azria D., Ozsahin M. (2006) The epidermal growth factor receptor (EGFR) in head and neck cancer: Its role and treatment implications. Radiat. Oncol. 1, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Astsaturov I., Cohen R. B., Harari P. M. (2008) Clinical application of EGFR inhibitors in head and neck squamous cell cancer. Cancer Treat. Res. 139, 135–152 [PubMed] [Google Scholar]
- 16. Huang S. M., Harari P. M. (2000) Modulation of radiation response after epidermal growth factor receptor blockade in squamous cell carcinomas: Inhibition of damage repair, cell cycle kinetics, and tumor angiogenesis. Clin. Cancer Res. 6, 2166–2174 [PubMed] [Google Scholar]
- 17. Mazumdar A., Henderson Y. C., El-Naggar A. K., Sen S., Clayman G. L. (2009) Aurora kinase A inhibition and paclitaxel as targeted combination therapy for head and neck squamous cell carcinoma. Head Neck 31, 625–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerster K., Shi W., Ng B., Yue S., Ito E., Waldron J., Gilbert R., Liu F. F. (2010) Targeting polo-like kinase 1 enhances radiation efficacy for head-and-neck squamous cell carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 77, 253–260 [DOI] [PubMed] [Google Scholar]
- 19. Seiwert T. Y., Jagadeeswaran R., Faoro L., Janamanchi V., Nallasura V., El Dinali M., Yala S., Kanteti R., Cohen E. E., Lingen M. W., Martin L., Krishnaswamy S., Klein-Szanto A., Christensen J. G., Vokes E. E., Salgia R. (2009) The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma. Cancer Res. 69, 3021–3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: A critical review. Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 21. Mallick P., Kuster B. (2010) Proteomics: A pragmatic perspective. Nat. Biotechnol. 28, 695–709 [DOI] [PubMed] [Google Scholar]
- 22. Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S., Mathieson T., Perrin J., Raida M., Rau C., Reader V., Sweetman G., Bauer A., Bouwmeester T., Hopf C., Kruse U., Neubauer G., Ramsden N., Rick J., Kuster B., Drewes G. (2007) Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 23. Bantscheff M., Hopf C., Kruse U., Drewes G. (2007) Proteomics-based strategies in kinase drug discovery. Ernst Schering Found. Symp. Proc. 2007, 1–28 [DOI] [PubMed] [Google Scholar]
- 24. Patel V. J., Thalassinos K., Slade S. E., Connolly J. B., Crombie A., Murrell J. C., Scrivens J. H. (2009) A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J. Proteome Res. 8, 3752–3759 [DOI] [PubMed] [Google Scholar]
- 25. Kruse U., Pallasch C. P., Bantscheff M., Eberhard D., Frenzel L., Ghidelli S., Maier S. K., Werner T., Wendtner C. M., Drewes G. (2011) Chemoproteomics-based kinome profiling and target deconvolution of clinical multi-kinase inhibitors in primary chronic lymphocytic leukemia cells. Leukemia 25, 89–100 [DOI] [PubMed] [Google Scholar]
- 26. Edington K. G., Loughran O. P., Berry I. J., Parkinson E. K. (1995) Cellular immortality: A late event in the progression of human squamous cell carcinoma of the head and neck associated with p53 alteration and a high frequency of allele loss. Mol. Carcinog. 13, 254–265 [DOI] [PubMed] [Google Scholar]
- 27. Eicheler W., Zips D., Dörfler A., Grénman R., Baumann M. (2002) Splicing mutations in TP53 in human squamous cell carcinoma lines influence immunohistochemical detection. J. Histochem. Cytochem. 50, 197–204 [DOI] [PubMed] [Google Scholar]
- 28. Gioanni J., Fischel J. L., Lambert J. C., Demard F., Mazeau C., Zanghellini E., Ettore F., Formento P., Chauvel P., Lalanne C. M., Courdi A. (1988) Two new human tumor cell lines derived from squamous cell carcinomas of the tongue: Establishment, characterization and response to cytotoxic treatment. Eur. J. Cancer Clin. Oncol. 24, 1445–1455 [DOI] [PubMed] [Google Scholar]
- 29. Järvinen A. K., Autio R., Kilpinen S., Saarela M., Leivo I., Grénman R., Mäkitie A. A., Monni O. (2008) High-resolution copy number and gene expression microarray analyses of head and neck squamous cell carcinoma cell lines of tongue and larynx. Genes Chromosomes Cancer 47, 500–509 [DOI] [PubMed] [Google Scholar]
- 30. Kawashiri S., Kumagai S., Kojima K., Harada H., Yamamoto E. (1995) Development of a new invasion and metastasis model of human oral squamous cell carcinomas. Eur. J. Cancer B Oral. Oncol. 31B, 216–221 [DOI] [PubMed] [Google Scholar]
- 31. Lansford C. D., Grenman R., Bier H., Somers K. D., Kim S. Y., Whiteside T. L., Clayman G. L., Welkoborsky H. J., Carey T. E. (1999) Human Cell Culture, Cancer Cell Lines. In Head and Neck Cancers (Masters J., Palsson B. eds.) pp. 185–255, Kluwer Academic Press, Dordrecht, The Netherlands [Google Scholar]
- 32. Lin C. J., Grandis J. R., Carey T. E., Gollin S. M., Whiteside T. L., Koch W. M., Ferris R. L., Lai S. Y. (2007) Head and neck squamous cell carcinoma cell lines: Established models and rationale for selection. Head Neck 29, 163–188 [DOI] [PubMed] [Google Scholar]
- 33. Momose F., Araida T., Negishi A., Ichijo H., Shioda S., Sasaki S. (1989) Variant sublines with different metastatic potentials selected in nude mice from human oral squamous cell carcinomas. J. Oral Pathol. Med. 18, 391–395 [DOI] [PubMed] [Google Scholar]
- 34. O-charoenrat P., Modjtahedi H., Rhys-Evans P., Court W. J., Box G. M., Eccles S. A. (2000) Epidermal growth factor-like ligands differentially up-regulate matrix metalloproteinase 9 in head and neck squamous carcinoma cells. Cancer Res. 60, 1121–1128 [PubMed] [Google Scholar]
- 35. Pekkola-Heino K., Joensuu H., Klemi P., Grenman R. (1994) Relation of DNA ploidy and proliferation rate to radiation sensitivity in squamous carcinoma cell lines. Arch. Otolaryngol. Head Neck Surg. 120, 750–754 [DOI] [PubMed] [Google Scholar]
- 36. Pekkola-Heino K., Kulmala J., Klemi P., Lakkala T., Aitasalo K., Joensuu H., Grenman R. (1991) Effects of radiation fractionation on four squamous cell carcinoma lines with dissimilar inherent radiation sensitivity. J. Cancer Res. Clin. Oncol. 117, 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dos Reis P. P., Bharadwaj R. R., Machado J., Macmillan C., Pintilie M., Sukhai M. A., Perez-Ordonez B., Gullane P., Irish J., Kamel-Reid S. (2008) Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer 113, 3169–3180 [DOI] [PubMed] [Google Scholar]
- 38. Rheinwald J. G., Beckett M. A. (1981) Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 41, 1657–1663 [PubMed] [Google Scholar]
- 39. White J. S., Weissfeld J. L., Ragin C. C., Rossie K. M., Martin C. L., Shuster M., Ishwad C. S., Law J. C., Myers E. N., Johnson J. T., Gollin S. M. (2007) The influence of clinical and demographic risk factors on the establishment of head and neck squamous cell carcinoma cell lines. Oral Oncol. 43, 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yoshida H., Hasegawa S., Kuromi H., Inagaki N., Seino S., Takahashi K., Sato K. (1993) A variant form of laminin is responsible for the neurite outgrowth-promoting activity in conditioned medium from a squamous carcinoma cell line. Connect. Tissue Res. 30, 23–35 [DOI] [PubMed] [Google Scholar]
- 41. Zhang N., Erjala K., Kulmala J., Qiu X., Sundvall M., Elenius K., Grénman R. (2009) Concurrent cetuximab, cisplatin, and radiation for squamous cell carcinoma of the head and neck in vitro. Radiother. Oncol. 92, 388–392 [DOI] [PubMed] [Google Scholar]
- 42. Eng C., Thiele H., Zhou X. P., Gorlin R. J., Hennekam R. C., Winter R. M. (2001) PTEN mutations and proteus syndrome. Lancet 358, 2079–2080 [DOI] [PubMed] [Google Scholar]
- 43. Hauck S. M., Dietter J., Kramer R. L., Hofmaier F., Zipplies J. K., Amann B., Feuchtinger A., Deeg C. A., Ueffing M. (2010) Deciphering membrane-associated molecular processes in target tissue of autoimmune uveitis by label-free quantitative mass spectrometry. Mol. Cell Proteomics 9, 2292–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huber W., von Heydebreck A., Sultmann H., Poustka A., Vingron M. (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 1, (Suppl. 18) S96–S104 [DOI] [PubMed] [Google Scholar]
- 45. Karp N. A., Huber W., Sadowski P. G., Charles P. D., Hester S. V., Lilley K. S. (2010) Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell Proteomics 9, 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smyth G. K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 47. Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. (2004) Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 [Google Scholar]
- 49. Rauser S., Langer R., Tschernitz S., Gais P., Jütting U., Feith M., Höfler H., Walch A. (2010) High number of CD45RO+ tumor infiltrating lymphocytes is an independent prognostic factor in non-metastasized (stage I–IIA) esophageal adenocarcinoma. BMC Cancer 10, 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fang R., Elias D. A., Monroe M. E., Shen Y., McIntosh M., Wang P., Goddard C. D., Callister S. J., Moore R. J., Gorby Y. A., Adkins J. N., Fredrickson J. K., Lipton M. S., Smith R. D. (2006) Differential label-free quantitative proteomic analysis of Shewanella oneidensis cultured under aerobic and suboxic conditions by accurate mass and time tag approach. Mol. Cell Proteomics 5, 714–725 [DOI] [PubMed] [Google Scholar]
- 51. Wykosky J., Gibo D. M., Debinski W. (2007) A novel, potent, and specific ephrinA1-based cytotoxin against EphA2 receptor expressing tumor cells. Mol. Cancer Ther. 6, 3208–3218 [DOI] [PubMed] [Google Scholar]
- 52. Pratt R. L., Kinch M. S. (2002) Activation of the EphA2 tyrosine kinase stimulates the MAP/ERK kinase signaling cascade. Oncogene 21, 7690–7699 [DOI] [PubMed] [Google Scholar]
- 53. Lombardo L. J., Lee F. Y., Chen P., Norris D., Barrish J. C., Behnia K., Castaneda S., Cornelius L. A., Das J., Doweyko A. M., Fairchild C., Hunt J. T., Inigo I., Johnston K., Kamath A., Kan D., Klei H., Marathe P., Pang S., Peterson R., Pitt S., Schieven G. L., Schmidt R. J., Tokarski J., Wen M. L., Wityak J., Borzilleri R. M. (2004) Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J. Med. Chem. 47, 6658–6661 [DOI] [PubMed] [Google Scholar]
- 54. Pearson M. A., Fabbro D. (2004) Targeting protein kinases in cancer therapy: A success? Expert Rev. Anticancer Ther. 4, 1113–1124 [DOI] [PubMed] [Google Scholar]
- 55. Cruz J. J., Ocaña A., Del Barco E., Pandiella A. (2007) Targeting receptor tyrosine kinases and their signal transduction routes in head and neck cancer. Ann. Oncol. 18, 421–430 [DOI] [PubMed] [Google Scholar]
- 56. McDermott U., Settleman J. (2009) Personalized cancer therapy with selective kinase inhibitors: An emerging paradigm in medical oncology. J. Clin. Oncol. 27, 5650–5659 [DOI] [PubMed] [Google Scholar]
- 57. Fishman M. C., Porter J. A. (2005) Pharmaceuticals: A new grammar for drug discovery. Nature 437, 491–493 [DOI] [PubMed] [Google Scholar]
- 58. Maurizi M., Almadori G., Ferrandina G., Distefano M., Romanini M. E., Cadoni G., Benedetti-Panici P., Paludetti G., Scambia G., Mancuso S. (1996) Prognostic significance of epidermal growth factor receptor in laryngeal squamous cell carcinoma. Br. J. Cancer 74, 1253–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weinstein I. B. (2002) Cancer: Addiction to oncogenes: The Achilles heal of cancer. Science 297, 63–64 [DOI] [PubMed] [Google Scholar]
- 60. Bischoff J. R., Plowman G. D. (1999) The Aurora/Ipl1p kinase family: Regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 9, 454–459 [DOI] [PubMed] [Google Scholar]
- 61. Inohara N., del Peso L., Koseki T., Chen S., Núñez G. (1998) RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J. Biol. Chem. 273, 12296–12300 [DOI] [PubMed] [Google Scholar]
- 62. Wykosky J., Gibo D. M., Stanton C., Debinski W. (2005) EphA2 as a novel molecular marker and target in glioblastoma multiforme. Mol. Cancer Res. 3, 541–551 [DOI] [PubMed] [Google Scholar]
- 63. Landen C. N., Kinch M. S., Sood A. K. (2005) EphA2 as a target for ovarian cancer therapy. Expert Opin. Ther. Targets 9, 1179–1187 [DOI] [PubMed] [Google Scholar]
- 64. Duxbury M. S., Ito H., Zinner M. J., Ashley S. W., Whang E. E. (2004) EphA2: A determinant of malignant cellular behavior and a potential therapeutic target in pancreatic adenocarcinoma. Oncogene 23, 1448–1456 [DOI] [PubMed] [Google Scholar]
- 65. Brannan J. M., Dong W., Prudkin L., Behrens C., Lotan R., Bekele B. N., Wistuba I., Johnson F. M. (2009) Expression of the receptor tyrosine kinase EphA2 is increased in smokers and predicts poor survival in non-small cell lung cancer. Clin. Cancer Res. 15, 4423–4430 [DOI] [PubMed] [Google Scholar]
- 66. Rivera R. S., Gunduz M., Nagatsuka H., Gunduz E., Cengiz B., Fukushima K., Beder L. B., Pehlivan D., Yamanaka N., Shimizu K., Nagai N. (2008) Involvement of EphA2 in head and neck squamous cell carcinoma: mRNA expression, loss of heterozygosity and immunohistochemical studies. Oncol. Rep. 19, 1079–1084 [PubMed] [Google Scholar]
- 67. Shao Z., Zhang W. F., Chen X. M., Shang Z. J. (2008) Expression of EphA2 and VEGF in squamous cell carcinoma of the tongue: Correlation with the angiogenesis and clinical outcome. Oral Oncol. 44, 1110–1117 [DOI] [PubMed] [Google Scholar]
- 68. Wykosky J., Debinski W. (2008) The EphA2 receptor and ephrinA1 ligand in solid tumors: Function and therapeutic targeting. Mol. Cancer Res. 6, 1795–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gene Expression Atlas: ArrayExpress database: http://www.ebi.ac.uk/gxa under accession number E-MTAB-62
- 70. Pardanani A., Lasho T., Smith G., Burns C. J., Fantino E., Tefferi A. (2009) CYT387, a selective JAK1/JAK2 inhibitor: In vitro assessment of kinase selectivity and preclinical studies using cell lines and primary cells from polycythemia vera patients. Leukemia 23, 1441–1445 [DOI] [PubMed] [Google Scholar]
- 71. Goldenberg-Furmanov M., Stein I., Pikarsky E., Rubin H., Kasem S., Wygoda M., Weinstein I., Reuveni H., Ben-Sasson S. A. (2004) Lyn is a target gene for prostate cancer: Sequence-based inhibition induces regression of human tumor xenografts. Cancer Res. 64, 1058–1066 [DOI] [PubMed] [Google Scholar]
- 72. Tourret J., McKeon F. (1996) Tyrosine kinases wee1 and mik1 as effectors of DNA replication checkpoint control. Prog. Cell Cycle Res. 2, 91–97 [DOI] [PubMed] [Google Scholar]
- 73. PosthumaDeBoer J., Wrdinger T., Graat H. C., van Beusechem V. W., Helder M. N., van Royen B. J., Kaspers G. J. (2011) WEE1 inhibition sensitizes osteosarcoma to radiotherapy. BMC Cancer 11, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schwartz D. L., Dong L. (2011) Adaptive radiation therapy for head and neck cancer-can an old goal evolve into a new standard? J. Oncol. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kim L. C., Song L., Haura E. B. (2009) Src kinases as therapeutic targets for cancer. Nat. Rev. Clin. Oncol. 6, 587–595 [DOI] [PubMed] [Google Scholar]
- 76. Song L., Morris M., Bagui T., Lee F. Y., Jove R., Haura E. B. (2006) Dasatinib (BMS-354825) selectively induces apoptosis in lung cancer cells dependent on epidermal growth factor receptor signaling for survival. Cancer Res. 66, 5542–5548 [DOI] [PubMed] [Google Scholar]
- 77. Christensen J. G., Schreck R., Burrows J., Kuruganti P., Chan E., Le P., Chen J., Wang X., Ruslim L., Blake R., Lipson K. E., Ramphal J., Do S., Cui J. J., Cherrington J. M., Mendel D. B. (2003) A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 63, 7345–7355 [PubMed] [Google Scholar]
- 78. Kononen J., Bubendorf L., Kallioniemi A., Barlund M., Schraml P., Leighton S., Torhorst J., Mihatsch M. J., Sauter G., Kallioniemi O. P. (1998) Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 4, 844–847 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The protein identification data from this study are available from the following ftp site: ftp://ftp.lrz.de/transfer/proteomics/.