Abstract
Dengue virus (DENV), an emerging mosquito-transmitted pathogen capable of causing severe disease in humans, interacts with host cell factors to create a more favorable environment for replication. However, few interactions between DENV and human proteins have been reported to date. To identify DENV-human protein interactions, we used high-throughput yeast two-hybrid assays to screen the 10 DENV proteins against a human liver activation domain library. From 45 DNA-binding domain clones containing either full-length viral genes or partially overlapping gene fragments, we identified 139 interactions between DENV and human proteins, the vast majority of which are novel. These interactions involved 105 human proteins, including six previously implicated in DENV infection and 45 linked to the replication of other viruses. Human proteins with functions related to the complement and coagulation cascade, the centrosome, and the cytoskeleton were enriched among the DENV interaction partners. To determine if the cellular proteins were required for DENV infection, we used small interfering RNAs to inhibit their expression. Six of 12 proteins targeted (CALR, DDX3X, ERC1, GOLGA2, TRIP11, and UBE2I) caused a significant decrease in the replication of a DENV replicon. We further showed that calreticulin colocalized with viral dsRNA and with the viral NS3 and NS5 proteins in DENV-infected cells, consistent with a direct role for calreticulin in DENV replication. Human proteins that interacted with DENV had significantly higher average degree and betweenness than expected by chance, which provides additional support for the hypothesis that viruses preferentially target cellular proteins that occupy central position in the human protein interaction network. This study provides a valuable starting point for additional investigations into the roles of human proteins in DENV infection.
Viruses have limited genetic capacity and must rely on cellular factors to complete their life cycle. Thus, viruses interact with cellular proteins to acquire activities not encoded in the viral genome, to thwart host immune defenses, and to manipulate cellular pathways in order to create a more favorable environment for replication (for example, (1–4)). Conversely, host cells counter viral infection by expressing proteins that bind to and degrade or inhibit viral proteins (for example, (5)). However, for most viruses, little is known about the interactions between viral and cellular proteins that engender these effects. A continued effort to define the interactions between virus and host cell will provide a better understanding of how viruses reproduce and cause disease, enable comparisons of the strategies different viruses use to manipulate host cells, and may reveal novel targets for therapeutic intervention.
Recent technical advances have greatly increased the pace at which cellular cofactors of virus infection have been identified. The discovery of RNA interference (RNAi) and the development of genome-wide RNAi screening approaches enabled cellular genes to be systematically assayed for their effect on virus replication (6–20). High-throughput yeast two-hybrid assays and co-affinity purification plus mass spectrometry allowed protein-protein interactions to be identified on a large-scale. These high throughput technologies enabled the identification of virus-host cell interactions and the development of human protein interaction networks that provide a larger context to understand virus-host cell interactions (21–28). The two approaches to identify cellular cofactors of virus infection are complementary: RNAi screens provide a list of host factors that can affect virus replication either directly or indirectly, and virus-host protein interaction networks elucidate the direct interface between the virus and the host cell. In theory, when the two data sets are complete, the requirement of proteins identified in RNAi screens should be discernable through their associations with proteins that interact directly with viral proteins or nucleic acids.
In this study we constructed a network of interactions between dengue virus (DENV)1 and human proteins. DENV is an enveloped, positive-stranded RNA virus that belongs to a family of important human pathogens (the Flaviviridae) that includes hepatitis C virus (HCV), yellow fever virus, Japanese encephalitis virus, and West Nile virus. DENV is transmitted by Aedes aegypti, a mosquito that thrives in urban settings, which likely contributes to the rapid increase in both the number of human infections and the geographical distribution of DENV over the past two decades (29). Currently more than 50 million DENV infections occur each year and more than 2 billion people live in areas where dengue is transmitted (30). Dengue causes diverse symptoms that range from a mild flu-like illness (dengue fever) to severe disease characterized by hemorrhage and shock (dengue hemorrhagic fever and dengue shock syndrome) (31). There are currently no vaccines to prevent dengue infection and no specific antiviral drugs to treat infected patients.
DENV infection begins with the binding of the virion to the host cell via direct interactions between the DENV E protein and cell surface ligands including DC-SIGN, heparin sulfate, and the macrophage receptor or indirect interactions between DENV-antibody complexes and the Fc or complement receptors (32–36). Following entry by receptor-mediated endocytosis and uncoating, the 10.2 kb DENV RNA genome is translated as a single polyprotein that is subsequently processed by viral and cellular proteases to yield 10 smaller proteins. The first three comprise the structural proteins that form the DENV virion, whereas the other seven play nonstructural roles. After the initial period of translation, the RNA genome switches from translation to replication in response to an unknown trigger. Negative- and positive-strand RNA synthesis occurs within virus-induced replication complexes derived from ER membranes (37). The newly formed positive-strand RNA products then serve as templates for translation and RNA replication, and are incorporated into immature virions. Virions acquire their lipid envelope by budding through the ER membrane and complete their maturation as they pass through the Golgi network, ultimately resulting in the release of infectious progeny (38, 39).
Because of the small size of the DENV genome, numerous host cell factors are presumed to participate at multiple points in the DENV life cycle. Although recent large-scale and pathway-focused RNAi screens have implicated ∼200 human genes in DENV infection (13, 16, 40–42), the number of host factors that have been demonstrated to interact with DENV proteins is small, and no genome-wide screen has been reported to date. Thus, to systematically identify cellular proteins that bind to DENV proteins, we used the yeast two-hybrid assay to screen DENV genes against a human liver cDNA library (43). We report the identification of more than 130 new interactions between DENV and human proteins and the validation of a subset of these interactions through split-luciferase, siRNA, and colocalization experiments. Furthermore, we show that DENV, like HIV, herpes viruses, and HCV, preferentially interacts with cellular proteins that are centrally located in the human protein interaction network.
EXPERIMENTAL PROCEDURES
Plasmids, Library and Yeast Strains
Yeast were maintained under standard laboratory conditions (44). Open reading frames and open reading frame fragments were amplified from DENV serotype 2 strain 16681 with gene-specific primers bearing 5′ 20-bp extensions (forward 5′-CCAAACCCAAAAAAAGAGATC-3′; reverse 5′- GTTTTTCAGTATCTACGATTCA-3′) homologous to the yeast two-hybrid DNA-binding domain plasmid pOBD2 (45). PCR primer sequences are listed in supplemental Table S1. PCR products were cloned into linearized pOBD2 by in vivo homologous recombination in the yeast strain R2HMet (MATa ura3–52 ade2–101 trp1–901 leu2–3,112 his3–200 met2Δ::hisG gal4Δ gal80Δ) (supplemental Fig. S1) (43, 46). All inserts were verified by PCR and sequencing. DENV DNA-binding domain constructs were screened against a human liver yeast two-hybrid library cloned into the activation domain plasmid pOAD.103 in yeast strain BK100 (MATa ura3–52 ade2–101 trp1–901 leu2–3,112 his3–200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ, a derivative of PJ69–4A) (43, 47, 48). The original library contained 9.6 × 105 independent clones with an average insert size of 840 bp.
Yeast Two-Hybrid Screens
The optimal concentration of 3-amino-1,2,4-triazole (3-AT) to suppress yeast growth in the absence of an interacting activation domain fusion (self-activation) was determined by growth on synthetic dropout (S.D.) medium lacking tryptophan, leucine, uracil, and histidine (S.D.-TLUH) and containing increasing amounts of 3-AT (1, 3, 5, 10, 20, and 50 mm). The lowest concentration of 3-AT that was able to suppress yeast growth - typically 1 or 3 mm - was used for the yeast two-hybrid assays. Library screens were performed by mating as described by Soellick and Uhrig (49). All DNA-binding domain clones were screened at least twice against the human liver activation domain library. Colonies expressing interacting proteins were selected on S.D.-TLUH medium containing the concentration of 3-AT as determined above. The human gene inserts from the activation domain plasmids were PCR-amplified, sequenced from the 5′ end, and identified by querying the human RefSeq database (downloaded 3/4/08) using Cross_Match (129). To retest the yeast two-hybrid interactions, PCR products of activation domain inserts from unique interactions were recloned into pOAD.103 by in vivo recombination in the yeast strain BK100. In addition, all unique interactions identified in a similar screen of HCV proteins against the same human liver library were recloned in the same manner (the full data set of HCV-human interactions will be presented elsewhere; manuscript in preparation). The resulting yeast strains were arrayed in quadruplicate in 384-spot format, mated with yeast expressing DNA binding domain fusions, and selected for growth on: (1) S.D.-TLUH containing the minimum concentration of 3-AT to suppress background growth; (2) S.D.-TLUH containing 3-AT at a concentration one step above that required to suppress background growth; and (3) S.D. medium lacking tryptophan, leucine, uracil, and adenine (S.D.-TLUA). Yeast growth was assessed on a scale of 0 to 4, with 0 being no growth above that observed for the negative control and 4 being robust growth. To standardize the results from different screens, each plate included a set of control yeast strains that displayed a range of growth rates on S.D.-TLUH+3-AT and S.D.-TLUA. Interactions were scored as positive if at least three of the four spots for each interaction were scored as one or higher on both media.
Split-luciferase Assays
The split-luciferase assay was performed as described (50). Full length NS3 and NS5 were cloned into plasmid p424-BYDV-NFLUC by in vivo homologous recombination. Similarly human gene fragments were PCR amplified from activation domain plasmids and cloned into plasmid p424-BYDV-CFLUC. Fusion proteins were expressed separately in wheat germ extracts, mixed in PBS supplemented with 1% BSA and protease inhibitors (Roche), incubated 18 h at 4 °C, and assayed for luciferase activity. Statistical significance was based on an unpaired student's t test.
Replication of a DENV Replicon in siRNA-Treated Cells
A DENV-luciferase replicon was generated in which the structural genes were replaced by the Renilla luciferase gene fused to the first 20 amino acids of the core protein (R. Perera and R. J. Khun, unpublished data). siRNA assays were performed as previously described with slight modifications (51). siRNA sequences are listed in supplemental Table S2. Briefly, 1 × 106 Huh-7.5 cells in 0.05 ml of PBS pH 7.4 were electroporated with 125 picomoles of siRNA and 300 ng of DENV reporter replicon RNA for five pulses of 770 volts for 99 microseconds with 1 s intervals on a BTX 830 electroporator with 96-well attachment. 4 × 104 cells were plated per well in a 96-well plate and grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Cells were harvested at 6 and 48 h post-electroporation and the level of DENV replication was determined via use of the Renilla Luciferase Assay System per the manufacturer's instruction (Promega, Madison, WI). The 48:6 h luciferase ratio represents DENV replication relative to input RNA. In parallel, cells were assayed for cellular ATP levels to assess viability using a CellTiter-Glo Luminescent Cell Viability Assay kit per the manufacturer's instructions (Promega). Statistical significance was based on an unpaired student's t test.
Colocalization of Viral and Cellular Proteins
Mouse polyclonal antisera against DENV NS5 and NS2B-3 were generously provided by Dr. James Strauss. Huh-7 cells were plated on glass coverslips, infected with DENV-16681 (serotype 2) at a multiplicity of infection of 0.1–3, fixed with 3.7% paraformaldehyde in phosphate-buffered saline (PBS) (4 °C) and permeabilized with 0.25% Triton X-100 in PBS (formaldehyde-fixed cells only). After blocking with 10% goat serum or 3% bovine serum albumin in PBS/0.05% Triton X-100, cells were incubated with mouse antisera against DENV2 NS5, DENV2 NS2B-3, or dsRNA (English & Scientific Consulting Bt.) and rabbit antisera against human CALR (Sigma), γ-tubulin (Abcam, Cambridge, MA), or PCNT1 (Abcam). TRITC- (Thermo Scientific) or Alexa Fluor 568- (Invitrogen, Carlsbad, CA) labeled goat anti-mouse and FITC- (Thermo Scientific) or Alexa Fluor 488- (Invitrogen) labeled goat anti-rabbit antibodies were used to detect the primary antibodies. In some specimens, nuclei were visualized with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI). Cover slips were washed after each antibody treatment with PBS supplemented with 5% goat serum and 0.05% Triton X-100, mounted in Fluorsave (Calbiochem, San Diego, CA) mounting medium and imaged with a Bio-Rad MRC-1024 confocal laser scanning microscope with 60 × oil immersion objective or a Nikon C1+ upright confocal microscope with a 60 × (NA 1.4) oil immersion objective. Signal intensities of individual fluorophores were quantified using ImageJ software (http://rsbweb.nih.gov/ij/) (52). Colocalization of DENV proteins with CALR was analyzed in 12–25 cells from at least six fields using the JACoP plugin in ImageJ (53). For quantitation of NS5 and CALR colocalization, images were cropped to analyze only the colocalization in the cytoplasm; if present in the image, aggregates of CALR were also excluded. Colocalization of DENV protein or dsRNA with CALR is presented as the post-thresholding Mander's coefficient multiplied by 100, which corresponds to the percent overlap of the signal from the DENV protein with that of CALR.
Topological Analysis of Human Proteins Implicated in Virus Infections
We considered three versions of the human protein interaction network (HPIN), which we refer to as HPIN-1, HPIN-2, and HPIN-3. HPIN-1 was assembled by Dyer et al. (54) and consists of a nonredundant set of interactions obtained from BIND (55), DIP (56), HPRD (57), INTACT (58–60), MINT (61, 62), MIPS (63), and REACTOME (64, 65). HPIN-2 contains the set of binary interactions described in (21). HPIN-3 consists of human protein-protein interactions from (66) with a confidence score of 0.3 or higher. Each network displayed scale-free distribution of degree. The diameters of HPIN_1, HPIN_2, and HPIN_3 are 14, 17, and 15, respectively. Data sets of human cofactors of viral infection are listed in supplemental Table S3. Average degree and betweenness centrality were analyzed using the igraph package (http://igraph.sourceforge.net/) in R version 2.6.2 (http://www.r-project.org). Statistical significance was determined using the Wilcox-Mann-Whitney test (Rank order test) by randomly selecting the same number of nodes 1000 times from the HPIN. Fisher's exact test was employed to determine if hub proteins were over represented in each dataset. For this analysis we considered proteins with degrees in the 95th percentile and above as hub proteins (67).
Enrichment of Annotation Terms and Pathways
Enriched features were identified in the complete set of human proteins in this study and the subsets that bound to each DENV protein using the DAVID Bioinformatics Database (http://david.abcc.ncifcrf.gov/home.jsp) (68, 69), the Cytoscape plugin BINGO (70–72), and Ingenuity Pathway Analysis program (Ingenuity Systems, Redwood City, CA). Additional details are provided in supplemental methods.
RESULTS
Mapping the DENV-Human Protein Interaction Network
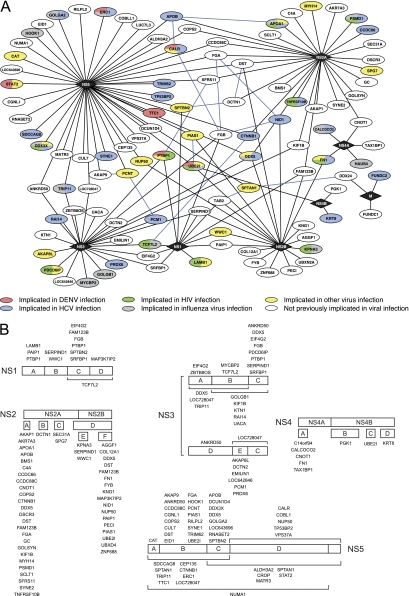
To identify human proteins that bound to DENV, we performed genome-wide yeast two-hybrid screens with DENV genes against a human liver cDNA library (supplemental Fig. S2 and supplemental Table 1). The human liver library was chosen because liver is a major target tissue during DENV infection and because DENV replicates well in human liver cell lines such as Huh-7 (31). Seven full-length DENV genes and 38 gene fragments were cloned into a yeast two-hybrid DNA-binding domain plasmid and screened at least twice against the human liver library (supplemental Fig. S3). From ∼1200 colonies picked for sequencing, 970 pairs of DENV-human interacting proteins representing 224 unique interactions were identified. To confirm the yeast two-hybrid interactions, the human gene fragments were recloned into the activation domain plasmid in fresh yeast cells and arrayed in 384-spot format; each unique fragment was represented by four independent spots in the array, thus allowing each interaction to be retested in quadruplicate (supplemental Fig. S2). To reduce the number of false-negatives and to better identify promiscuous activation domain fusions that can give rise to false positives, all 45 DENV DNA-binding domain fusions were tested by pair-wise mating against every human activation domain construct identified in the DENV yeast two-hybrid screens. Pairs of DENV and human proteins were tested for their ability to independently activate expression of the yeast two-hybrid reporter genes ADE2 and HIS3 by growth on media that lacked adenine or histidine, respectively. Only those pairs in which at least three spots grew on both selection media were classified as positives. Human gene fragments that stimulated yeast growth when paired with an empty DNA-binding domain plasmid or that yielded positives with more than 5% of the nonoverlapping bait constructs were classified as promiscuous activators and eliminated from the data set. To further increase the number of interactions identified, and thus to reduce the number of false-negatives, each DENV clone was also tested against 124 human genes identified in a similar screen with HCV (manuscript in preparation). In total, 188 interactions involving 34 DENV protein fragments and 105 human proteins were confirmed in the yeast two-hybrid retests (Fig. 1A and supplemental Table S4). These represent 139 unique interactions involving eight of the ten DENV proteins. Thirty-three interactions (24%) were identified by two or more DENV gene fragments, lending further confidence to these interactions (supplemental Table 4). The use of multiple fragments from the same DENV gene also enabled the protein interaction domains to be more narrowly defined, in some cases to regions as small as 100 amino acids (Fig. 1B and supplemental Tables S1 and S4).
Fig. 1.
DENV-human protein interaction network. A, the complete set of DENV-human protein interactions identified in this study are shown. Diamonds indicate viral proteins; ovals, human proteins; black lines, DENV-human protein interactions identified in this study; blue lines, previously identified interactions between human proteins from HPIN-1. Shading identifies proteins implicated in the replication of other viruses. Red, human proteins previously implicated in DENV replication; blue, human proteins implicated in HCV replication; green, human proteins implicated in HIV replication; gray, human proteins implicated in INFV replication; yellow, proteins implicated in the replication of other viruses; and white, proteins not previously implicated in virus replication. Additional details can be found in supplemental Table S5. B, DENV-human protein interactions by fragment. Human proteins identified in the DENV-human yeast two-hybrid screens are shown above or below the smallest viral protein fragment they interacted with in the yeast two-hybrid retests. Brackets indicate fragments composed of two or more smaller fragments (for example, NS1 fragment CD). Complete results from the yeast two-hybrid screens listing all viral fragments that interacted with a particular human protein can be found in supplemental Table S4.
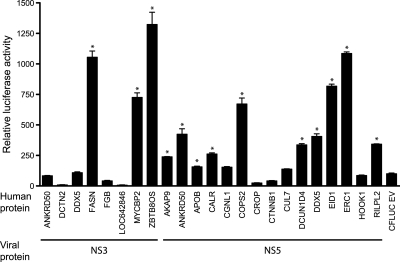
We employed the split-luciferase assay to validate the interactions through an independent approach (Fig. 2) (73). In this system, putative interacting proteins are expressed as fusions to N- and C-terminal fragments of luciferase. If the fused proteins bind to each other, the luciferase fragments reassociate to produce a functional enzyme. For these experiments we used the N- and C-terminal firefly luciferase fragments optimized for their ability to confer high signal to noise ratios and robust enzymatic activity (74). We previously adapted this system for use with in vitro translated proteins produced in wheat germ extracts (50). DENV proteins fused to the N-terminal luciferase fragment (NFLUC) and human proteins fused to the C-terminal luciferase fragment (CFLUC) were expressed in wheat germ extracts, mixed, and tested for luciferase activity after overnight incubation. With the exception of DDX5, all viral and human proteins were expressed at similar levels relative to each other and at equal or lower levels compared with NFLUC and CFLUC containing no insert (supplemental Fig. S4). As a negative control, we tested each human protein from Fig. 2 against the HCV core protein. HCV core has been reported to bind to more than 45 human proteins (22), but did not interact with any of the human proteins from Fig. 2 in our yeast two-hybrid screens. The NFLUC-HCV core construct used in this experiment was functional in the split-luciferase assay, yielding a 104-fold increase in luciferase activity when incubated with CFLUC-DDX3X; this fragment of DDX3X encompassed the previously mapped HCV core binding site on DDX3X (78), but was distinct from the DDX3X fragment that interacted with DENV NS5. None of the human proteins produced a significant increase in luciferase activity when paired with NFLUC-HCV core. In contrast, 17 of the 23 DENV-human protein pairs yielded significantly higher levels of luciferase activity relative to the empty N-terminal luciferase vector control and 16 had a significantly higher level of luciferase activity relative to the empty C-terminal luciferase vector control (Fig. 2). Thirteen pairs (57%) yielded a significant increase in luciferase activity relative to both empty vector controls (p value < 0.02); we consider this set to represent true positives in the split-luciferase assay. This rate of confirmation is similar to those observed for other high quality large-scale yeast two-hybrid screens, which range from 20 to 85% (22, 75–77).
Fig. 2.
Validation of DENV-human protein interactions in the split-luciferase assay. DENV and human proteins were expressed in wheat germ extracts as fusions to the N- and C-terminal fragments of firefly luciferase, respectively. In vitro translated DENV and human proteins were mixed in PBS supplemented with 1% BSA and protease inhibitors (Roche) and incubated overnight at 4 °C. As negative controls, each viral protein was tested against the empty C-terminal luciferase vector and each human protein was tested against the empty N-terminal luciferase vector. Binding reactions were set up in duplicate and luciferase activity was measured twice for each reaction. Graph shows the average of the four luciferase readings normalized to the luciferase activity of the DENV protein with the empty C-terminal luciferase negative control, which was set at 100 relative light units (RLU). Error bars indicate the S.E. Asterisks indicate pairs of interacting proteins that yielded luciferase activity that was significantly greater than both negative controls (student's unpaired t test, p value <0.02).
Comparison to Existing Sets of Human Proteins Implicated in Virus Infection
Although only a few DENV-human protein-protein interactions have been reported in small scale studies ((79) and references therein; (80–83)), several recent reports have begun to identify human cofactors of DENV infection on a more systematic basis. Using the bacterial two-hybrid system, 31 interactions involving the DENV structural proteins C, M, and E were identified (84). More than 4000 interactions involving 2321 human proteins have been predicted by computational approaches (79). In addition, two large-scale and three smaller, focused RNAi screens identified a total of 219 human proteins that affected DENV replication (13, 16, 40–42). As is the case for most large-scale screens (for example, (8, 22)), the overlap between the sets of human proteins identified in previous screens for DENV cofactors and with the yeast two-hybrid screen described here is small (Fig. 1A, supplemental Table S5). Of the 20 DENV-human protein interactions identified in the literature, only the interaction between NS5 and STAT2 was also detected in this study (85, 86). Other proteins in our data set (UBE2I, CALR, and PTBP1) were reported to interact with DENV proteins, though the interacting viral proteins differ from those identified here (87–91); these host factors may bind to multiple DENV proteins, as has previously been observed for other viruses (21), or may represent false-positives. PTBP1, which has been reported to bind to NS4A, also colocalized with NS1 and NS3 in DENV-infected cells (92), consistent with our finding that PTBP1 interacted with NS1 and NS3 in the yeast two-hybrid assay. None of the interactions from the bacterial two-hybrid were present in our yeast two-hybrid data set, though this is not surprising because only DENV structural proteins were screened in the bacterial two-hybrid study and relatively few of our interactions involved these proteins. Seven of the 4000 computationally predicted interactions were also detected by our yeast two-hybrid screens; an additional 35 human proteins from the computational study are also present in our data set, but interacted with different viral proteins than predicted (79). Finally, three proteins found to be required for DENV replication in siRNA screens interacted with DENV proteins in this study (13, 16). One additional protein shared between our yeast two-hybrid screen and an siRNA screen (TTC1) was required for DENV replication in Drosophila but not human cells (16).
Comparisons to sets of cellular cofactors of viral infection reported in other large-scale screens or in the literature revealed more extensive overlaps, with a total of 45 proteins from our DENV-human interactome being reported to either bind to viral proteins or to be required for replication of at least one virus (Fig. 1A and supplemental Table S5). The largest overlap was observed with human proteins implicated in HCV infection. Six human proteins identified by large-scale RNAi screens and 17 proteins that interacted with HCV were present in the DENV yeast two-hybrid data set. Extensive overlap with human genes implicated in HIV replication was also noted (four genes from RNAi screens and 16 HIV interacting proteins). Multiple proteins, including APOAI, DDX3X, FN1, PDCD6IP, PIAS1, PSMD1, STAT2, and UBE2I, have been implicated in the life cycles of at least three other viruses, though in the case of APOA1, FN1, and PSMD1, very little is known about their functional roles. Together these results indicate that our yeast two-hybrid screens have identified human proteins relevant to virus infection and suggest that novel proteins identified here are also likely to contribute to DENV replication.
Enriched Features of the Human Proteins Targeted by DENV
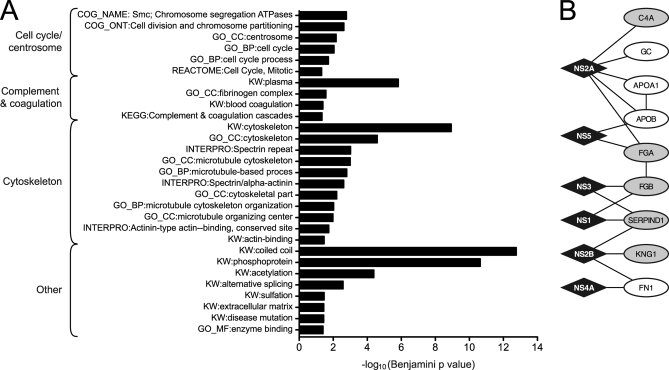
The set of human proteins that interacted with DENV proteins in this study and subsets that interacted with each individual protein were submitted for analysis with the DAVID Bioinformatics Database to identify annotation terms that were over-represented (68, 69) (Fig. 3A, supplemental Tables S6 and S7). Similar enrichment analyses were performed using BINGO (70) and Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA), and yielded similar results (data not shown). Plasma proteins involved in complement activation and the coagulation cascade were among the enriched annotation terms of the human proteins that interacted with DENV (Fig. 3B). These pathways were also enriched among the partners of DENV structural proteins (84) and have been associated with severe dengue disease (31). Recent evidence suggests that the complement system, in addition to acting as a link between the adaptive and innate immune system, may have a significant role as a viral antagonist and as a major target in viral immune evasion (reviewed in (93)). DENV proteins inhibit complement activation (94). In addition, the levels of complement proteins, including C4, were altered in patients with severe DENV disease (95).
Fig. 3.
Enriched annotation terms and pathways among the human proteins that interacted with DENV. A, Enriched terms identified by the DAVID Bioinformatics Database in the complete set of human proteins that interacted with DENV proteins in this study (68, 69). Graph shows the –log10-transformed Benjamini-corrected p values for each term. Terms were grouped according to their functional similarity. Enriched features of human proteins that interacted with individual viral proteins are listed in supplemental Table S6. B, Subnetwork of human plasma proteins that interacted with DENV. Diamonds indicate DENV proteins. Ovals represent human proteins. Shading indicates human proteins annotated with the term “Complement and coagulation cascade.”
Other enriched features in this study included proteins linked to the cytoskeleton, centrosomes, and chromosome segregation (supplemental Fig. S5). Though many viruses target the cytoskeleton (96–99), when we performed the same enrichment analysis described above on human proteins identified in large-scale interactome projects for Epstein-Barr virus (EBV), HCV, HIV, and influenza virus (INFV), only HCV showed a significant enrichment of terms related to the cytoskeleton. Similarly, centrosomes are known to be exploited by retroviruses and large DNA viruses (reviewed by (100)), but were only enriched among the cellular partners of DENV and HCV. Based on this informatic analysis, we performed immunofluoresence assays on DENV-infected Huh7 cells with antibodies against NS5 and two centrosomal proteins, γ-tubulin and PCNT1. γ-tubulin did not interact with any DENV protein in our yeast two-hybrid screens, whereas PCNT1 interacted with NS5. Consistent with the enrichment analysis, NS5 accumulated in dividing cells in the region immediately surrounding γ-tubulin and PCNT1 (supplemental Fig. S6). Additional discussion of enriched features in the DENV-human interactome is included in supplemental materials and supplemental Fig. S7.
To determine if siRNA screens for DENV cofactors identified proteins from the same pathways, we performed the same enrichment analyses on the set of human genes identified by (13, 16). Although no significantly enriched terms were found in both data sets, combining the human proteins identified in RNAi screens with those from the yeast two-hybrid data sets revealed six additional enriched annotations relating to protein localization and vesicle transport (supplemental Table S8).
Inhibiting Expression of Human Proteins that Interacted with DENV Reduced DENV Replication
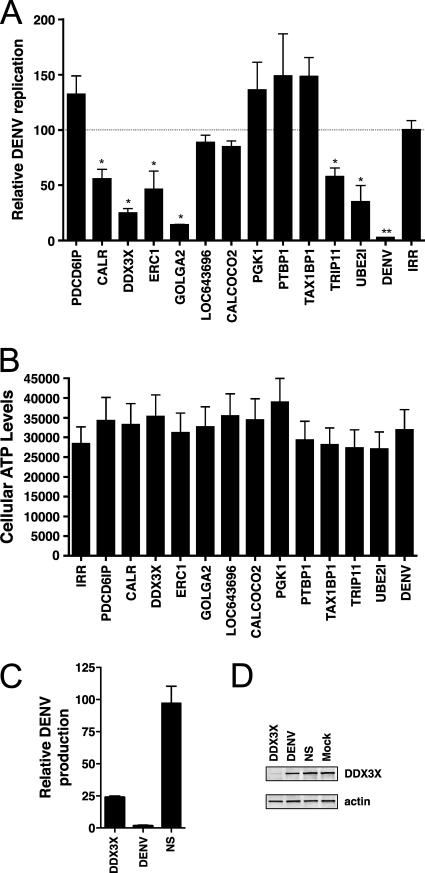
To further examine the contribution of the human proteins identified in this study to DENV replication, we inhibited the expression of 12 cellular genes by RNAi. siRNAs were electroporated into Huh7.5 cells along with RNA from a DENV replicon in which the nonstructural genes were replaced by Renilla luciferase. Because luciferase activity is proportional to virus replication and no progeny virus are produced, this construct can be used to evaluate the effect of virus mutations and cellular perturbations on DENV RNA replication in the absence of confounding effects from reinfection. Inhibiting expression of six out of the 12 genes (CALR, DDX3X, ERC1, GOLGA2, TRIP11, and UBE2I) caused a statistically significant reduction in the level of luciferase activity compared with cells electroporated with an irrelevant siRNA (Fig. 4A). The reduction in viral replication was not because of cellular toxicity of the siRNA (Fig. 4B). We further examined the effect of DDX3X in the context of virally infected cells. In agreement with our results from the replicon experiment, inhibiting DDX3X in Huh7 cells caused a ∼75% reduction in the amount of progeny virus produced compared with cells treated with an irrelevant siRNA control (Fig. 4C). Western blotting confirmed that DDX3X was depleted in DDX3X siRNA treated cells, but not in cells treated with DENV or irrelevant siRNAs (Fig. 4D). Because the replicon RNA is transfected into cells and no virus particles are formed, these data indicate that CALR, DDX3X, ERC1, GOLGA2, TRIP11, and UBE2I the cellular proteins affects a step related to translation of the viral RNA or viral RNA synthesis. However, because the assay does not assess viral entry or virion assembly and release, cellular proteins affecting these steps will be missed. Indeed, PDCD6IP (ALIX), which has been implicated in the acquisition of the membrane envelope by multiple viruses (101), had no effect on luciferase levels in the replicon assay.
Fig. 4.
Inhibition of DENV replication in cells with reduced expression of human genes. A, Luciferase production from a DENV replicon in siRNA treated cells. siRNAs targeting the indicated human genes, DENV NS5, or an irrelevant sequence were electroporated into Huh-7.5 cells along with DENV luciferase replicon RNA. Luciferase levels were measured in cell lysates prepared at 6 and 48 h after electroporation. DENV replication is represented as the ratio of the luciferase values at 48 and 6 h and normalized to the irrelevant siRNA control, which was defined as 100%. Values are averages of at least three replicates ± S.E. * p ≤ 0.05, ** p ≤ 0.001. B, Cell viability following siRNA treatment. siRNAs targeting the indicated genes or an irrelevant sequence (IRR) were electroporated into Huh-7.5 cells with DENV luciferase replicon RNAs. Cellular ATP levels were measured at 48 h using the CellTiter-Glo Luminescent Cell Viability Assay kit (Promega). Values are averages of at least three replicates ± S.E. C, DENV production in DDX3X siRNA-treated cells. Huh7 cells were transfected with the indicated siRNAs and infected with DENV at an MOI of 3. Supernatant was harvested at 24 h post infection and titered by plaque assay. Titers were normalized to nonspecific siRNA control (NS). D, Western blot analysis of DDX3X levels in siRNA-treated cells from part C. Bottom panel shows actin levels as a loading control.
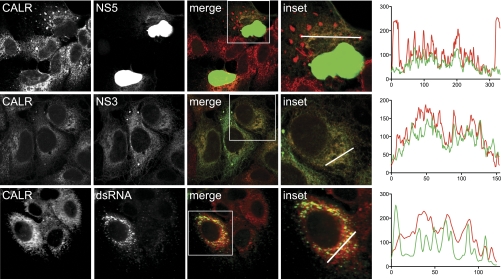
Colocalization of CALR with DENV Proteins During Viral Infection
CALR was recently reported to bind to and colocalize with the DENV envelope protein (E) in infected cells, and to be required for DENV replication (91). Based on these results, it was hypothesized that CALR functioned at a step late during infection related to virus assembly (91). Our results using a DENV replicon confirm a role for CALR in DENV replication, but indicate that CALR acts at an earlier step in the DENV life cycle. To begin to address the functional role of CALR in DENV replication, we performed immunofluorescence assays on endogenously expressed CALR, NS3, NS5, and dsRNA in infected cells (Fig. 5, supplemental Fig. S8). The antisera against NS5 and NS3 recognized a single band of the expected size in lysates from infected cells, and did not cross-react with any cellular proteins on Western blots or in immunofluorescence assays (supplemental Fig. S9). NS5, which interacted with CALR in both the yeast two-hybrid assay and the split-luciferase assay, colocalized with CALR in the cytoplasm of infected cells (71 ± 13% overlap). CALR did not relocalize to the nucleus where the vast majority of NS5 was found (102), suggesting that its functional role during infection is in the cytoplasm, where RNA synthesis occurs. DENV NS3 (70 ± 8% overlap) and dsRNA (an intermediate formed during viral RNA replication) also colocalized with CALR (89 ± 7% overlap). In some infected cells, CALR was concentrated in globular aggregates similar to features previously observed in DENV-infected cells (supplemental Fig. S8) (103–105). These structures became more abundant at later times after infection and were not present in mock-infected cells nor in uninfected cells on the same slide. NS3, but not NS5 or dsRNA, also accumulated in these structures. Combined with the observation that reducing CALR expression inhibited luciferase production from the DENV replicon, these results are consistent with a direct role for CALR in DENV RNA replication.
Fig. 5.
Colocalization of CALR with DENV proteins and dsRNA in infected cells. Huh7 cells were infected with DENV at an MOI of 1, fixed 36 h post infection, and probed with antibodies against CALR and DENV NS5 (top row), DENV NS3 (middle row), or dsRNA (bottom row). Pseudo-colored merged images are shown in column 3, with CALR in red and NS5, NS3, and dsRNA in green. Inset shows a higher magnification of the region in the white box of the merged images. The white line indicates the region sampled for quantification of the fluorescent intensity of CALR (red lines) and NS3 or NS5 (green lines), as shown in the graphs on the right. Nuclear-localized NS5, which accounts for ∼95% of the total NS5 present in the infected cell, is overexposed to enable visualization of cytoplasmic NS5. Images represent a single optical slice.
Network Analysis of DENV-Human Protein Interactions
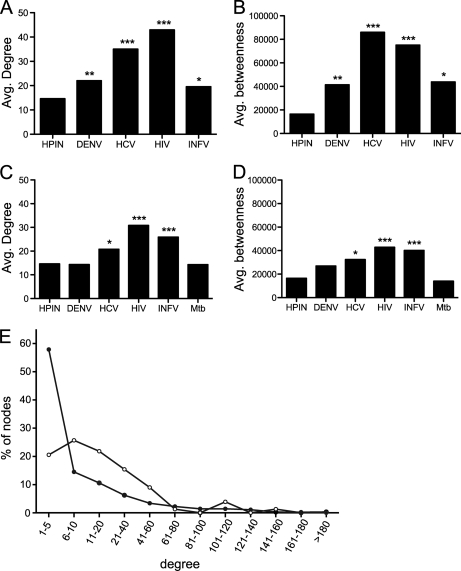
We next sought to determine if DENV preferentially targeted particular classes of human proteins in the human protein interaction network (HPIN). Previous reports have found that viral proteins tend to interact with cellular proteins centrally located in the HPIN. As a measure of local network properties, we compared the average degree of the human proteins in our DENV-human network with randomized sets from the HPIN containing the same number of proteins. Three independently derived sets of human protein-protein interactions, which we refer to as HPIN-1, −2, and −3 (21, 54, 66), were examined and yielded similar results (Fig. 6, supplemental Fig. S10, and supplemental Table S9). As with the EBV, HCV, and INFV interaction networks (17, 21, 22), the average degree of human proteins that bound to DENV proteins was higher than expected by chance (Fig. 6A). However, this was not due to interactions with hub proteins (defined here as human proteins with degrees in the 95th percentile or greater (67)). In contrast to the human proteins that interacted with HCV, HIV, and INFV, which showed a statistically significant enrichment in hub proteins in at least one HPIN, the number of hub proteins in the DENV-human interactome was not significantly higher than expected by chance in any network. Rather, DENV preferentially interacted with human proteins with an intermediate number of binding partners (between 6 and 60 interactions in HPIN-1, Fig. 6E). The average degree of the human proteins in the DENV-host data set was comparable to that of human proteins in the INFV interaction network and lower than the average degrees of human proteins that bound to either HCV or HIV. This difference may reflect the fact that the DENV- and INFV-host interaction networks have fewer human proteins and are therefore likely to be less complete than the HCV and HIV networks, or may be because of technical differences in how the interaction networks were generated.
Fig. 6.
Elevated degree and betweenness in cellular proteins implicated in virus infection. A, average degree (k) of human proteins that bound to viral proteins from DENV (this study), HCV, HIV, and INFV. B, Average betweenness centrality (b) of human proteins that bound to viral proteins from DENV (this study), HCV, HIV, and INFV. C, average degree (k) of human proteins that were identified in siRNA screens for cellular cofactors of DENV, HCV, HIV, INFV, and Mycobacterium tuberculosis. D, average betweenness centrality (b) of human proteins that were identified in siRNA screens for cellular cofactors of DENV, HCV, HIV, INFV, and Mycobacterium tuberculosis. Average degree and betweenness centrality were calculated for each data set and compared with the same values from a randomly chosen set of human proteins of the same size. Asterisks indicate statistical significance. * p ≤ 0.05, ** p ≤ 0.001, *** p ≤ 1 × 106. E, degree distribution of human proteins in the DENV-human protein interaction network (open circles) and the human-human protein interaction network (closed circles). Proteins were binned according to the number of binding partners as indicated.
As a measure of the global network properties of the human proteins that bound to DENV, we examined the average betweenness centrality of the proteins in our data set. Betweenness centrality, as its name implies, is a measure of how “central” a protein is in the network, i.e. the number of shortest paths that must pass through a particular node. Proteins with high betweenness are sometimes referred to as bottlenecks because they are links between different modules in the cell and are more likely to be essential (106). As was observed for HCV (22), the average betweenness centrality of human proteins in the DENV-human interaction network was higher than expected by chance when compared with randomized networks of the same size (p ≤ 0.0004) (Fig. 6B). Similar to the average degree of DENV-interacting proteins, the average betweenness centrality of the proteins in our dataset was comparable to the average betweenness centrality of the human proteins that bound to INFV and lower than that of human proteins that bound to HCV or HIV. Surprisingly, the human proteins with the highest degree and betweenness centrality in our data set also tended to bind to multiple viral proteins (supplemental Fig. S11).
We next compared these results with sets of human proteins implicated in virus replication by large-scale siRNA screens. Because these proteins were identified based solely on their impact on virus replication, many are likely to be only indirectly involved in the virus lifecycle and would therefore be expected to have average degree and betweenness centrality values similar to those of the entire HPIN. Indeed, this was the case for human proteins identified in siRNA screens for host factors that affect DENV replication (Figs. 6C and 6D, supplemental Fig. S10). However, the average degree, number of hub proteins, and betweenness centrality of human proteins identified in siRNA screens for co-factors of HIV and INFV, and to a lesser extent HCV, were significantly higher than the human network as a whole (Figs. 6C and 6D, supplemental Fig. S10). The effect may be specific to viruses, since the average degree and betweenness centrality of proteins identified in an siRNA screen for cellular cofactors of Mycobacterium tuberculosis were indistinguishable from those in the entire human protein interaction network (Figs. 6C and 6D, supplemental Fig. S10) (107).
DISCUSSION
We report here the first genome-wide analysis of DENV-human protein-protein interactions. This study more than tripled the number interactions between DENV and human proteins and identified 93 human proteins not previously linked to DENV replication, 60 of which have not been linked to any other virus. Eight of the ten DENV proteins yielded reproducible interactions with between three and forty-three human proteins. From a technical standpoint, this study differed from previous virus-host cell protein interaction projects in several ways that serve to minimize both false-positives and false-negatives. Each DENV gene was screened as a series of fragments, which enabled interactions to be identified that were missed when full-length genes were used, provided additional support for many interactions that were identified with multiple overlapping fragments, and allowed binding regions to be more narrowly defined. Each DENV DNA binding domain fusion was also retested against every human activation domain clone identified in the DENV screens on two yeast two-hybrid selection media. Though time consuming, this step identified additional interactions that were missed in library screens, increased confidence that the interactions were specific, and revealed activation domain fusions that behaved promiscuously even though they did not interact with the empty DNA binding domain vector. Finally, each DENV clone was screened against human activation domain clones identified in a screen with HCV genes. This approach is important because yeast two-hybrid library screens—like all currently available techniques to discover protein-protein interactions—tend to miss many interactions, which makes it difficult to draw meaningful conclusions when comparing cellular protein interactions identified with different viruses. By generating arrays of cellular proteins identified in yeast two-hybrid library screens with viral proteins and screening those arrays with proteins from different viruses, direct comparison of virus-host interactomes will be possible.
Over 40% of the human proteins reported in this study to interact with DENV proteins have been implicated in the life cycles of at least one other virus. The greatest overlap was with proteins linked to HCV infection. However, the overlap between DENV and HCV extended beyond shared proteins to shared annotation terms. Centrosome and cytoskeleton proteins were enriched among the partners of DENV and HCV, but not among the partners of the unrelated viruses EBV, HIV, or INFV. The similarity between the human proteins targeted by DENV and HCV is not unexpected given the evolutionary relatedness of these viruses and the similarity of their life cycles. However, DENV also interacted with 20 proteins that either bound to HIV proteins or that were required for HIV replication. The overlap between DENV and HIV cellular cofactors is consistent with the hypothesis that retroviruses and RNA viruses share similar replication strategies (108), but may also reflect the fact that many more host factors have been reported for HIV than for any other virus.
Half of the proteins targeted in our siRNA experiments were required for optimal replication of the DENV replicon. In a related siRNA screen, fatty acid synthase (FASN) was identified as a cellular co-factor of DENV replication (42). FASN interacted with NS3 in our yeast two-hybrid screen and was observed to relocalize to sites of DENV RNA replication in infected cells. We further demonstrated that the presence of NS3 increased FASN enzymatic activity in vitro, suggesting that DENV binds to FASN to exploit and enhance its role in lipid biosynthesis (42). In a parallel yeast two-hybrid study, we inhibited expression of a set ten human proteins that interacted with both DENV and HCV proteins and identified five additional proteins that reduced DENV replication (manuscript in preparation). Thus, 12 of the 23 human proteins we tested to date in siRNA experiments were required for DENV replication (11% of the human proteins identified in this screen). For comparison, 95 human proteins were identified in yeast two-hybrid screens with INFV and all were tested for their effects on INFV replication in similar siRNA experiments (17). Of these, three were required for INFV replication and eight exerted a negative effect, for a total of 11 (12%) proteins that affected INFV replication (17). Similarly, in large-scale siRNA screens for host factors affecting viral replication, the average hit rate was 1% (6, 7, 10–16, 18–20). Thus, the results from our initial siRNA experiments strongly suggest our DENV-human interactome is enriched in proteins relevant to DENV infection. Consistent with this conclusion, 57% of our interactions were confirmed in the split-luciferase assay, which is comparable to other high quality yeast two-hybrid studies (75–77).
A Role for CALR in DENV Replication
CALR (calreticulin) is a multifunctional protein with diverse roles in protein folding, calcium homeostasis and regulation of gene expression (109, 110). Though the best-characterized function of CALR is as a glycoprotein chaperone in the lumen of the endoplasmic reticulum, CALR also retrotranslocates from the ER to the cytoplasm, where it plays critical roles in regulating adhesion and gene expression (111–114). CALR has been implicated at multiple points in the life cycles of diverse viruses. Through its chaperone activity, CALR binds to and promotes the folding or maturation of viral glycoproteins (115–118). CALR also binds to the 3′ UTR of rubella virus positive strand genomic RNA, though this interaction is apparently not required for rubella virus replication (119, 120). During apoptosis, CALR translocates to the cell surface, where it acts as a signal to promote phagocytosis by surrounding cells (121). Viruses may disrupt this process in order to prevent destruction of the infected cell (122).
While this study was in progress, CALR was reported to bind to DENV envelope (E) protein and was shown to be required for DENV virus production (91). Based on this data, it was proposed that CALR's critical role in viral production was related to virus protein folding or virion assembly (91). Our results are consistent with a requirement for CALR in DENV replication and do not rule out a role for CALR at a late step during infection. However, our data indicates that CALR is also involved at a step prior to virus maturation, potentially in DENV RNA synthesis. We found that inhibiting expression of CALR reduced luciferase production from the DENV replicon. Because the E gene was deleted from the replicon and no viral particles were produced, CALR must be acting at an earlier step in the viral life cycle. CALR interacted with the DENV RNA-dependent RNA polymerase NS5 in both the yeast two-hybrid assay and the split-luciferase assay in this study, and was previously found to interact with the 3′ UTR of DENV negative strand RNA (123). Consistent with a role for CALR in DENV RNA synthesis, CALR colocalized with NS5 in the cytoplasm, where RNA synthesis occurs, but not in the nucleus where the majority of NS5 resides. CALR also colocalized with NS3, which binds to NS5, and with dsRNA, which is produced as an intermediate during viral RNA synthesis.
Golgi Apparatus Proteins Are Required for DENV Replication
Seven Golgi-associated proteins interacted with DENV proteins, of which three (ERC1/ELKS, GOLGA2/GMAP130, and TRIP11/GMAP210) were targeted with siRNAs and shown to affect DENV replication in our study (Fig. 2) (124–127). Another 13 Golgi proteins were previously identified as DENV cellular cofactors in RNAi screens (13, 16). Though the Golgi is known to be important for viral protein processing and DENV virion maturation (38, 39), the observation that ERC1 was required for replication of the DENV replicon, indicates that ERC1 must function at a step prior to virus assembly. ERC1 is recruited to the Golgi through its interaction with Rab6A' and appears to be involved in endosome to Golgi transport (124). However, ERC1 is a multifunctional protein that also interacts with IKK and regulates NF-κB mediated gene expression (130–132). Similarly, GOLGA2 and TRIP11 are also multifunctional proteins that are involved in processes unrelated to the Golgi. It is possible that one of these alternative functions is being exploited by DENV.
Network Properties of Human Proteins Implicated in Virus Infections
Preferential targeting of highly connected proteins and proteins with high betweenness may enable viruses to efficiently control the flux of infection-relevant information passing through the host interactome (54, 128). Topological analyses of the proteins targeted by DENV further supports the hypothesis that viruses have evolved strategies to target nodes of higher connectivity and centrality. As previously observed for human proteins that interacted with EBV, HCV, HIV, and INFV (17, 21, 22, 54, 128), human proteins that interacted with DENV proteins had higher average degree and betweenness than those in the HPIN as a whole. Similar patterns were found in a larger scale analysis of more than a hundred host-virus interactomes (128). However, DENV did not interact with the most highly connected human proteins, but rather those with an intermediate number of partners. This was true regardless of whether we considered human protein interaction networks derived exclusively from binary interaction data or from networks that included interactions from complex purification experiments.
We repeated the same network analyses on sets of human proteins identified as cofactors of viral infection in siRNA screens with the expectation that they would serve as negative controls. Because many of the proteins in the siRNA data sets are likely to be only indirectly involved in viral infection, we predicted that they would have similar degree and betweenness values as the entire network. Surprisingly, human proteins implicated in viral infection through siRNA screens also tended to have higher average degree and betweenness centrality. Taken at face value, these results suggest that human proteins with high centrality are preferentially required for virus infection. However, the increase in average degree and betweenness centrality may simply reflect the presence of human proteins that directly interact with viral proteins in the siRNA data sets. As discussed above, cellular proteins that interact with viral proteins tend to have higher degree and betweenness centrality, so their presence would serve to increase the average values of the data set as a whole. Because there is little overlap between proteins that interact with either HIV, HCV, or INFV and those that have been implicated in viral replication through siRNA screens, this explanation would suggest that many human proteins that interact with viral proteins have not yet been identified. A third possibility is that inhibiting expression of highly connected proteins is more likely to cause changes in the host cell that lead to a measurable effect on virus replication.
CONCLUSION
The set of DENV-human protein interactions is enriched in proteins that have been implicated in the replication of other viruses and implicates dozens of new proteins and pathways in DENV infection. These interactions provide a valuable starting point for additional investigations into the roles of host factors in DENV replication and for comparisons to other virus-host interaction networks.
Acknowledgments
We thank Charles Rice for providing Huh-7.5 cells, James Strauss for providing anti-DENV NS2B-3 and NS5 antisera, and the Purdue Bioscience Imaging Facility for assistance with microscopy. This project was initiated with funding provided by the Ralph W. and Grace M. Showalter Research Trust to D.J.L. and R.J.K.
Footnotes
* G.R. is supported by the NIH/NIAID Region V “Great Lakes” Regional Center of Excellence for Bio-defense and Emerging Infectious Diseases Research (1-U54-AI-057153). N.H. is funded by NIH training grant T32 AI065382-01. R.J.K. acknowledges support from the National Institute of Allergy and Infectious Disease (AI45976). D.J.L. acknowledges support from the National Institute of General Medical Sciences (GM092829).
 This article contains supplemental Figs. S1 to S11, Tables S1 to S9, Procedures and Discussion.
This article contains supplemental Figs. S1 to S11, Tables S1 to S9, Procedures and Discussion.
1 The abbreviations used are:
- 3-AT
- 3-amino-1,2,4-triazole
- cDNA
- complementary DNA
- DAPI
- 4,6-diamidino-2-phenylindole dihydrochloride
- DENV
- dengue virus
- dsRNA
- double-stranded RNA
- EBV
- Epstein Barr virus
- ESCRT
- endosomal sorting complex required for transport
- HCV
- hepatitis C virus
- HIV
- human immunodeficiency virus
- HPIN
- human protein interaction network
- INFV
- influenza virus
- NS
- dengue virus nonstructural protein
- ORF
- open reading frame
- RNAi
- RNA interference
- SD–TLUA
- SD medium lacking tryptophan, leucine, uracil, and adenine
- SD–TLUH
- SD medium lacking lacking tryptophan, leucine, uracil, and histidine
- SD
- synthetic dropout medium
- siRNA
- small interfering RNA.
REFERENCES
- 1. Llano M., Saenz D. T., Meehan A., Wongthida P., Peretz M., Walker W. H., Teo W., Poeschla E. M. (2006) An essential role for LEDGF/p75 in HIV integration. Science 314, 461–464 [DOI] [PubMed] [Google Scholar]
- 2. Maertens G., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y. (2003) LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278, 33528–33539 [DOI] [PubMed] [Google Scholar]
- 3. Galluzzi L., Kepp O., Morselli E., Vitale I., Senovilla L., Pinti M., Zitvogel L., Kroemer G. (2010) Viral strategies for the evasion of immunogenic cell death. J. Intern Med. 267, 526–542 [DOI] [PubMed] [Google Scholar]
- 4. Chaurushiya M. S., Weitzman M. D. (2009) Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair 8, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Skaug B., Chen Z. J. (2010) Emerging role of ISG15 in antiviral immunity. Cell 143, 187–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brass A. L., Dykxhoorn D. M., Benita Y., Yan N., Engelman A., Xavier R. J., Lieberman J., Elledge S. J. (2008) Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926 [DOI] [PubMed] [Google Scholar]
- 7. Brass A. L., Huang I. C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., Ryan B. J., Weyer J. L., van der Weyden L., Fikrig E., Adams D. J., Xavier R. J., Farzan M., Elledge S. J. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bushman F. D., Malani N., Fernandes J., D'Orso I., Cagney G., Diamond T. L., Zhou H., Hazuda D. J., Espeseth A. S., König R., Bandyopadhyay S., Ideker T., Goff S. P., Krogan N. J., Frankel A. D., Young J. A., Chanda S. K. (2009) Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog 5, e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hao L., Sakurai A., Watanabe T., Sorensen E., Nidom C. A., Newton M. A., Ahlquist P., Kawaoka Y. (2008) Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454, 890–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karlas A., Machuy N., Shin Y., Pleissner K. P., Artarini A., Heuer D., Becker D., Khalil H., Ogilvie L. A., Hess S., Mäurer A. P., Muller E., Wolff T., Rudel T., Meyer T. F. (2010) Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463, 818–822 [DOI] [PubMed] [Google Scholar]
- 11. König R., Stertz S., Zhou Y., Inoue A., Hoffmann H. H., Bhattacharyya S., Alamares J. G., Tscherne D. M., Ortigoza M. B., Liang Y., Gao Q., Andrews S. E., Bandyopadhyay S., De Jesus P., Tu B. P., Pache L., Shih C., Orth A., Bonamy G., Miraglia L., Ideker T., Garcia-Sastre A., Young J. A., Palese P., Shaw M. L., Chanda S. K. (2010) Human host factors required for influenza virus replication. Nature 463, 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Känig R., Zhou Y., Elleder D., Diamond T. L., Bonamy G. M., Irelan J. T., Chiang C. Y., Tu B. P., De Jesus P. D., Lilley C. E., Seidel S., Opaluch A. M., Caldwell J. S., Weitzman M. D., Kuhen K. L., Bandyopadhyay S., Ideker T., Orth A. P., Miraglia L. J., Bushman F. D., Young J. A., Chanda S. K. (2008) Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krishnan M. N., Ng A., Sukumaran B., Gilfoy F. D., Uchil P. D., Sultana H., Brass A. L., Adametz R., Tsui M., Qian F., Montgomery R. R., Lev S., Mason P. W., Koski R. A., Elledge S. J., Xavier R. J., Agaisse H., Fikrig E. (2008) RNA interference screen for human genes associated with West Nile virus infection. Nature 455, 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Q., Brass A. L., Ng A., Hu Z., Xavier R. J., Liang T. J., Elledge S. J. (2009) A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc. Natl. Acad. Sci. U.S.A. 106, 16410–16415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ng T. I., Mo H., Pilot-Matias T., He Y., Koev G., Krishnan P., Mondal R., Pithawalla R., He W., Dekhtyar T., Packer J., Schurdak M., Molla A. (2007) Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology 45, 1413–1421 [DOI] [PubMed] [Google Scholar]
- 16. Sessions O. M., Barrows N. J., Souza-Neto J. A., Robinson T. J., Hershey C. L., Rodgers M. A., Ramirez J. L., Dimopoulos G., Yang P. L., Pearson J. L., Garcia-Blanco M. A. (2009) Discovery of insect and human dengue virus host factors. Nature 458, 1047–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shapira S. D., Gat-Viks I., Shum B. O., Dricot A., de Grace M. M., Wu L., Gupta P. B., Hao T., Silver S. J., Root D. E., Hill D. E., Regev A., Hacohen N. (2009) A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139, 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tai A. W., Benita Y., Peng L. F., Kim S. S., Sakamoto N., Xavier R. J., Chung R. T. (2009) A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vaillancourt F. H., Pilote L., Cartier M., Lippens J., Liuzzi M., Bethell R. C., Cordingley M. G., Kukolj G. (2009) Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology 387, 5–10 [DOI] [PubMed] [Google Scholar]
- 20. Zhou H., Xu M., Huang Q., Gates A. T., Zhang X. D., Castle J. C., Stec E., Ferrer M., Strulovici B., Hazuda D. J., Espeseth A. S. (2008) Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4, 495–504 [DOI] [PubMed] [Google Scholar]
- 21. Calderwood M. A., Venkatesan K., Xing L., Chase M. R., Vazquez A., Holthaus A. M., Ewence A. E., Li N., Hirozane-Kishikawa T., Hill D. E., Vidal M., Kieff E., Johannsen E. (2007) Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U.S.A. 104, 7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Chassey B., Navratil V., Tafforeau L., Hiet M. S., Aublin-Gex A., Agaugué S., Meiffren G., Pradezynski F., Faria B. F., Chantier T., Le Breton M., Pellet J., Davoust N., Mangeot P. E., Chaboud A., Penin F., Jacob Y., Vidalain P. O., Vidal M., André P., Rabourdin-Combe C., Lotteau V. (2008) Hepatitis C virus infection protein network. Mol. Syst. Biol. 4, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart I. I., Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 25. Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F. H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksöz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E. E. (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 [DOI] [PubMed] [Google Scholar]
- 26. Uetz P., Dong Y. A., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S. V., Roupelieva M., Rose D., Fossum E., Haas J. (2006) Herpesviral protein networks and their interaction with the human proteome. Science 311, 239–242 [DOI] [PubMed] [Google Scholar]
- 27. Behrends C., Sowa M. E., Gygi S. P., Harper J. W. (2010) Network organization of the human autophagy system. Nature 466, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mackenzie J. S., Gubler D. J., Petersen L. R. (2004) Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10, S98–109 [DOI] [PubMed] [Google Scholar]
- 30. Guha-Sapir D., Schimmer B. (2005) Dengue fever: new paradigms for a changing epidemiology. Emerg Themes Epidemiol 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martina B. E., Koraka P., Osterhaus A. D. (2009) Dengue virus pathogenesis: an integrated view. Clin. Microbiol Rev. 22, 564–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen Y., Maguire T., Hileman R. E., Fromm J. R., Esko J. D., Linhardt R. J., Marks R. M. (1997) Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 3, 866–871 [DOI] [PubMed] [Google Scholar]
- 33. Miller J. L., de Wet B. J. M., Martinez-Pomares L., Radcliffe C. M., Dwek R. A., Rudd P. M., Gordon S. (2008) The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog 4, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Navarro-Sanchez E., Altmeyer R., Amara A., Schwartz O., Fieschi F., Virelizier J. L., Arenzana-Seisdedos F., Despres P. (2003) Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep 4, 723–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Porterfield J. S. (1986) Antibody-dependent enhancement of viral infectivity. Adv. Virus Res. 31, 335–355 [DOI] [PubMed] [Google Scholar]
- 36. Tassaneetrithep B., Burgess T. H., Granelli-Piperno A., Trumpfheller C., Finke J., Sun W., Eller M. A., Pattanapanyasat K., Sarasombath S., Birx D. L., Steinman R. M., Schlesinger S., Marovich M. A. (2003) DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 197, 823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C. K., Walther P., Fuller S. D., Antony C., Krijnse-Locker J., Bartenschlager R. (2009) Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5, 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu I. M., Zhang W., Holdaway H. A., Li L., Kostyuchenko V. A., Chipman P. R., Kuhn R. J., Rossmann M. G., Chen J. (2008) Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319, 1834–1837 [DOI] [PubMed] [Google Scholar]
- 39. Li L., Lok S. M., Yu I. M., Zhang Y., Kuhn R. J., Chen J., Rossmann M. G. (2008) The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319, 1830–1834 [DOI] [PubMed] [Google Scholar]
- 40. Rothwell C., Lebreton A., Young Ng C., Lim J. Y., Liu W., Vasudevan S., Labow M., Gu F., Gaither L. A. (2009) Cholesterol biosynthesis modulation regulates dengue viral replication. Virology 389, 8–19 [DOI] [PubMed] [Google Scholar]
- 41. Ang F., Wong A. P., Ng M. M., Chu J. J. (2010) Small interference RNA profiling reveals the essential role of human membrane trafficking genes in mediating the infectious entry of dengue virus. Virol J. 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heaton N. S., Perera R., Berger K. L., Khadka S., Lacount D. J., Kuhn R. J., Randall G. (2010) Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 17345–17350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. LaCount D. J., Vignali M., Chettier R., Phansalkar A., Bell R., Hesselberth J. R., Schoenfeld L. W., Ota I., Sahasrabudhe S., Kurschner C., Fields S., Hughes R. E. (2005) A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 438, 103–107 [DOI] [PubMed] [Google Scholar]
- 44. Guthrie C., Fink G. R. (1981) Guide to yeast genetics and molecular biology, Academic Press, San Diego, CA [Google Scholar]
- 45. Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Qureshi-Emili A., Li Y., Godwin B., Conover D., Kalbfleisch T., Vijayadamodar G., Yang M., Johnston M., Fields S., Rothberg J. M. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 [DOI] [PubMed] [Google Scholar]
- 46. Ma H., Kunes S., Schatz P. J., Botstein D. (1987) Plasmid construction by homologous recombination in yeast. Gene 58, 201–216 [DOI] [PubMed] [Google Scholar]
- 47. James P., Halladay J., Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vignali M., McKinlay A., LaCount D. J., Chettier R., Bell R., Sahasrabudhe S., Hughes R. E., Fields S. (2008) Interaction of an atypical Plasmodium falciparum ETRAMP with human apolipoproteins. Malar J. 7, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Soellick T. R., Uhrig J. F. (2001) Development of an optimized interaction-mating protocol for large-scale yeast two-hybrid analyses. Genome Biol. 2, RESEARCH0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brown H. F., Wang L., Khadka S., Fields S., LaCount D. J. (2011) A densely overlapping gene fragmentation approach improves yeast two-hybrid screens for Plasmodium falciparum proteins. Mol. Biochem. Parasitol 178, 56–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Randall G., Panis M., Cooper J. D., Tellinghuisen T. L., Sukhodolets K. E., Pfeffer S., Landthaler M., Landgraf P., Kan S., Lindenbach B. D., Chien M., Weir D. B., Russo J. J., Ju J., Brownstein M. J., Sheridan R., Sander C., Zavolan M., Tuschl T., Rice C. M. (2007) Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U.S.A. 104, 12884–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Image Processing with ImageJ. Biophotonics International 11, 36–42 [Google Scholar]
- 53. Bolte S., Cordelieres F. P. (2006) A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc 224, 213–232 [DOI] [PubMed] [Google Scholar]
- 54. Dyer M. D., Murali T. M., Sobral B. W. (2008) The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog 4, e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bader G. D., Betel D., Hogue C. W. (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 31, 248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xenarios I., Salwinski L., Duan X. J., Higney P., Kim S. M., Eisenberg D. (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 30, 303–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peri S., Navarro J. D., Amanchy R., Kristiansen T. Z., Jonnalagadda C. K., Surendranath V., Niranjan V., Muthusamy B., Gandhi T. K., Gronborg M., Ibarrola N., Deshpande N., Shanker K., Shivashankar H. N., Rashmi B. P., Ramya M. A., Zhao Z., Chandrika K. N., Padma N., Harsha H. C., Yatish A. J., Kavitha M. P., Menezes M., Choudhury D. R., Suresh S., Ghosh N., Saravana R., Chandran S., Krishna S., Joy M., Anand S. K., Madavan V., Joseph A., Wong G. W., Schiemann W. P., Constantinescu S. N., Huang L., Khosravi-Far R., Steen H., Tewari M., Ghaffari S., Blobe G. C., Dang C. V., Garcia J. G., Pevsner J., Jensen O. N., Roepstorff P., Deshpande K. S., Chinnaiyan A. M., Hamosh A., Chakravarti A., Pandey A. (2003) Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 13, 2363–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Aranda B., Achuthan P., Alam-Faruque Y., Armean I., Bridge A., Derow C., Feuermann M., Ghanbarian A. T., Kerrien S., Khadake J., Kerssemakers J., Leroy C., Menden M., Michaut M., Montecchi-Palazzi L., Neuhauser S. N., Orchard S., Perreau V., Roechert B., van Eijk K., Hermjakob H. (2010) The IntAct molecular interaction database in 2010. Nucleic Acids Res. 38, D525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S., Vingron M., Roechert B., Roepstorff P., Valencia A., Margalit H., Armstrong J., Bairoch A., Cesareni G., Sherman D., Apweiler R. (2004) IntAct: an open source molecular interaction database. Nucleic Acids Res. 32, D452–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kerrien S., Alam-Faruque Y., Aranda B., Bancarz I., Bridge A., Derow C., Dimmer E., Feuermann M., Friedrichsen A., Huntley R., Kohler C., Khadake J., Leroy C., Liban A., Lieftink C., Montecchi-Palazzi L., Orchard S., Risse J., Robbe K., Roechert B., Thorneycroft D., Zhang Y., Apweiler R., Hermjakob H. (2007) IntAct–open source resource for molecular interaction data. Nucleic Acids Res. 35, D561–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chatr-aryamontri A., Ceol A., Palazzi L. M., Nardelli G., Schneider M. V., Castagnoli L., Cesareni G. (2007) MINT: the Molecular INTeraction database. Nucleic Acids Res. 35, D572–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. (2002) MINT: a Molecular INTeraction database. FEBS Lett. 513, 135–140 [DOI] [PubMed] [Google Scholar]
- 63. Mewes H. W., Amid C., Arnold R., Frishman D., Güldener U., Mannhaupt G., Munsterkotter M., Pagel P., Strack N., Stumpflen V., Warfsmann J., Ruepp A. (2004) MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 32, D41–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Matthews L., Gopinath G., Gillespie M., Caudy M., Croft D., de Bono B., Garapati P., Hemish J., Hermjakob H., Jassal B., Kanapin A., Lewis S., Mahajan S., May B., Schmidt E., Vastrik I., Wu G., Birney E., Stein L., D'Eustachio P. (2009) Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 37, D619–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vastrik I., D'Eustachio P., Schmidt E., Gopinath G., Croft D., de Bono B., Gillespie M., Jassal B., Lewis S., Matthews L., Wu G., Birney E., Stein L. (2007) Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 8, R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yu J., Finley R. L., Jr. (2009) Combining multiple positive training sets to generate confidence scores for protein-protein interactions. Bioinformatics 25, 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Batada N. N., Reguly T., Breitkreutz A., Boucher L., Breitkreutz B.-J., Hurst L. D., Tyers M. (2006) Stratus Not Altocumulus: A New View of the Yeast Protein Interaction Network. PLoS Biology 4, e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 69. Huang, da W., Sherman B. T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M. W., Lane H. C., Lempicki R. A. (2007) DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35, W169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Maere S., Heymans K., Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 71. Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A. R., Vailaya A., Wang P. L., Adler A., Conklin B. R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G. J., Ideker T., Bader G. D. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ozawa T., Kaihara A., Sato M., Tachihara K., Umezawa Y. (2001) Split luciferase as an optical probe for detecting protein-protein interactions in mammalian cells based on protein splicing. Anal Chem. 73, 2516–2521 [DOI] [PubMed] [Google Scholar]
- 74. Paulmurugan R., Gambhir S. S. (2007) Combinatorial library screening for developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein-protein interactions. Anal Chem. 79, 2346–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pilot-Storck F., Chopin E., Rual J. F., Baudot A., Dobrokhotov P., Robinson-Rechavi M., Brun C., Cusick M. E., Hill D. E., Schaeffer L., Vidal M., Goillot E. (2010) Interactome mapping of the phosphatidylinositol 3-kinase-mammalian target of rapamycin pathway identifies deformed epidermal autoregulatory factor-1 as a new glycogen synthase kinase-3 interactor. Mol. Cell Proteomics 9, 1578–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Boxem M., Maliga Z., Klitgord N., Li N., Lemmens I., Mana M., de Lichtervelde L., Mul J. D., van de Peut D., Devos M., Simonis N., Yildirim M. A., Cokol M., Kao H. L., de Smet A. S., Wang H., Schlaitz A. L., Hao T., Milstein S., Fan C., Tipsword M., Drew K., Galli M., Rhrissorrakrai K., Drechsel D., Koller D., Roth F. P., Iakoucheva L. M., Dunker A. K., Bonneau R., Gunsalus K. C., Hill D. E., Piano F., Tavernier J., van den Heuvel S., Hyman A. A., Vidal M. (2008) A protein domain-based interactome network for C. elegans early embryogenesis. Cell 134, 534–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Braun P., Tasan M., Dreze M., Barrios-Rodiles M., Lemmens I., Yu H., Sahalie J. M., Murray R. R., Roncari L., de Smet A. S., Venkatesan K., Rual J. F., Vandenhaute J., Cusick M. E., Pawson T., Hill D. E., Tavernier J., Wrana J. L., Roth F. P., Vidal M. (2009) An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods 6, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Owsianka A. M., Patel A. H. (1999) Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257, 330–340 [DOI] [PubMed] [Google Scholar]
- 79. Doolittle J. M., Gomez S. M. (2011) Mapping protein interactions between Dengue virus and its human and insect hosts. PLoS Negl Trop Dis 5, e954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Duan X., Lu X., Li J., Liu Y. (2008) Novel binding between pre-membrane protein and vacuolar ATPase is required for efficient dengue virus secretion. Biochem. Biophys. Res. Commun. 373, 319–324 [DOI] [PubMed] [Google Scholar]
- 81. Johansson M., Brooks A. J., Jans D. A., Vasudevan S. G. (2001) A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-beta and the viral helicase, NS3. J. Gen. Virol. 82, 735–745 [DOI] [PubMed] [Google Scholar]
- 82. Bhuvanakantham R., Li J., Tan T. T., Ng M. L. (2010) Human Sec3 protein is a novel transcriptional and translational repressor of flavivirus. Cell Microbiol 12, 453–472 [DOI] [PubMed] [Google Scholar]
- 83. Rawlinson S. M., Pryor M. J., Wright P. J., Jans D. A. (2009) CRM1-mediated Nuclear Export of Dengue Virus RNA Polymerase NS5 Modulates Interleukin-8 Induction and Virus Production. J. Biol. Chem. 284, 15589–15597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Folly B. B., Weffort-Santos A. M., Fathman C. G., Soares L. R. (2011) Dengue-2 structural proteins associate with human proteins to produce a coagulation and innate immune response biased interactome. BMC Infect Dis 11, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ashour J., Laurent-Rolle M., Shi P. Y., Garcia-Sastre A. (2009) NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83, 5408–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mazzon M., Jones M., Davidson A., Chain B., Jacobs M. (2009) Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J. Infect Dis 200, 1261–1270 [DOI] [PubMed] [Google Scholar]
- 87. Anwar A., Leong K. M., Ng M. L., Chu J. J., Garcia-Blanco M. A. (2009) The polypyrimidine tract-binding protein is required for efficient dengue virus propagation and associates with the viral replication machinery. J. Biol. Chem. 284, 17021–17029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chiu M. W., Shih H. M., Yang T. H., Yang Y. L. (2007) The type 2 dengue virus envelope protein interacts with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9). J. Biomed Sci. 14, 429–444 [DOI] [PubMed] [Google Scholar]
- 89. De Nova-Ocampo M., Villegas-Sepúlveda N., del Angel R. M. (2002) Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology 295, 337–347 [DOI] [PubMed] [Google Scholar]
- 90. Jiang L., Yao H., Duan X., Lu X., Liu Y. (2009) Polypyrimidine tract-binding protein influences negative strand RNA synthesis of dengue virus. Biochem. Biophys. Res. Commun. 385, 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Limjindaporn T., Wongwiwat W., Noisakran S., Srisawat C., Netsawang J., Puttikhunt C., Kasinrerk W., Avirutnan P., Thiemmeca S., Sriburi R., Sittisombut N., Malasit P., Yenchitsomanus P. T. (2009) Interaction of dengue virus envelope protein with endoplasmic reticulum-resident chaperones facilitates dengue virus production. Biochem. Biophys. Res. Commun. 379, 196–200 [DOI] [PubMed] [Google Scholar]
- 92. Agis-Juárez R. A., Galvan I., Medina F., Daikoku T., Padmanabhan R., Ludert J. E., del Angel R. M. (2009) Polypyrimidine tract-binding protein is relocated to the cytoplasm and is required during dengue virus infection in Vero cells. J. Gen. Virol. 90, 2893–2901 [DOI] [PubMed] [Google Scholar]
- 93. Stoermer K. A., Morrison T. E. (2011) Complement and viral pathogenesis. Virology 411, 362–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Avirutnan P., Punyadee N., Noisakran S., Komoltri C., Thiemmeca S., Auethavornanan K., Jairungsri A., Kanlaya R., Tangthawornchaikul N., Puttikhunt C., Pattanakitsakul S. N., Yenchitsomanus P. T., Mongkolsapaya J., Kasinrerk W., Sittisombut N., Husmann M., Blettner M., Vasanawathana S., Bhakdi S., Malasit P. (2006) Vascular leakage in severe dengue virus infections: a potential role for the nonstructural viral protein NS1 and complement. J. Infect Dis 193, 1078–1088 [DOI] [PubMed] [Google Scholar]
- 95. Nascimento E. J., Silva A. M., Cordeiro M. T., Brito C. A., Gil L. H., Braga-Neto U., Marques E. T. (2009) Alternative complement pathway deregulation is correlated with dengue severity. PLoS One 4, e6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Harries P. A., Schoelz J. E., Nelson R. S. (2010) Intracellular transport of viruses and their components: utilizing the cytoskeleton and membrane highways. Mol. Plant Microbe Interact 23, 1381–1393 [DOI] [PubMed] [Google Scholar]
- 97. Hsieh M. J., White P. J., Pouton C. W. (2010) Interaction of viruses with host cell molecular motors. Curr. Opin. Biotechnol 21, 633–639 [DOI] [PubMed] [Google Scholar]
- 98. Lyman M. G., Enquist L. W. (2009) Herpesvirus interactions with the host cytoskeleton. J. Virol. 83, 2058–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ward B. M. (2011) The taking of the cytoskeleton one two three: how viruses utilize the cytoskeleton during egress. Virology 411, 244–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wileman T. (2007) Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu Rev. Microbiol. 61, 149–167 [DOI] [PubMed] [Google Scholar]
- 101. Martin-Serrano J., Marsh M. (2007) ALIX catches HIV. Cell Host Microbe 1, 5–7 [DOI] [PubMed] [Google Scholar]
- 102. Kapoor M., Zhang L., Ramachandra M., Kusukawa J., Ebner K. E., Padmanabhan R. (1995) Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 270, 19100–19106 [DOI] [PubMed] [Google Scholar]
- 103. Chua J. J., Ng M. M., Chow V. T. (2004) The non-structural 3 (NS3) protein of dengue virus type 2 interacts with human nuclear receptor binding protein and is associated with alterations in membrane structure. Virus Res. 102, 151–163 [DOI] [PubMed] [Google Scholar]
- 104. Miller S., Kastner S., Krijnse-Locker J., Bühler S., Bartenschlager R. (2007) The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282, 8873–8882 [DOI] [PubMed] [Google Scholar]
- 105. Miller S., Sparacio S., Bartenschlager R. (2006) Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J. Biol. Chem. 281, 8854–8863 [DOI] [PubMed] [Google Scholar]
- 106. Yu H., Kim P. M., Sprecher E., Trifonov V., Gerstein M. (2007) The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 3, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kumar D., Nath L., Kamal M. A., Varshney A., Jain A., Singh S., Rao K. V. (2010) Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell 140, 731–743 [DOI] [PubMed] [Google Scholar]
- 108. Ahlquist P. (2006) Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol 4, 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gold L. I., Eggleton P., Sweetwyne M. T., Van Duyn L. B., Greives M. R., Naylor S. M., Michalak M., Murphy-Ullrich J. E. (2010) Calreticulin: non-endoplasmic reticulum functions in physiology and disease. FASEB J. 24, 665–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Michalak M., Corbett E. F., Mesaeli N., Nakamura K., Opas M. (1999) Calreticulin: one protein, one gene, many functions. Biochem. J. 344 Pt 2, 281–292 [PMC free article] [PubMed] [Google Scholar]
- 111. Afshar N., Black B. E., Paschal B. M. (2005) Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol. Cell Biol. 25, 8844–8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Burns K., Duggan B., Atkinson E. A., Famulski K. S., Nemer M., Bleackley R. C., Michalak M. (1994) Modulation of gene expression by calreticulin binding to the glucocorticoid receptor. Nature 367, 476–480 [DOI] [PubMed] [Google Scholar]
- 113. Coppolino M., Leung-Hagesteijn C., Dedhar S., Wilkins J. (1995) Inducible interaction of integrin alpha 2 beta 1 with calreticulin. Dependence on the activation state of the integrin. J. Biol. Chem. 270, 23132–23138 [DOI] [PubMed] [Google Scholar]
- 114. Coppolino M. G., Woodside M. J., Demaurex N., Grinstein S., St-Arnaud R., Dedhar S. (1997) Calreticulin is essential for integrin-mediated calcium signalling and cell adhesion. Nature 386, 843–847 [DOI] [PubMed] [Google Scholar]
- 115. Alefantis T., Flaig K. E., Wigdahl B., Jain P. (2007) Interaction of HTLV-1 Tax protein with calreticulin: implications for Tax nuclear export and secretion. Biomed Pharmacother 61, 194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Braakman I., van Anken E. (2000) Folding of viral envelope glycoproteins in the endoplasmic reticulum. Traffic 1, 533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Nakhasi H. L., Ramanujam M., Atreya C. D., Hobman T. C., Lee N., Esmaili A., Duncan R. C. (2001) Rubella virus glycoprotein interaction with the endoplasmic reticulum calreticulin and calnexin. Arch Virol. 146, 1–14 [DOI] [PubMed] [Google Scholar]
- 118. Otteken A., Moss B. (1996) Calreticulin interacts with newly synthesized human immunodeficiency virus type 1 envelope glycoprotein, suggesting a chaperone function similar to that of calnexin. J. Biol. Chem. 271, 97–103 [DOI] [PubMed] [Google Scholar]
- 119. Chen M. H., Frey T. K. (1999) Mutagenic analysis of the 3′ cis-acting elements of the rubella virus genome. J. Virol. 73, 3386–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Singh N. K., Atreya C. D., Nakhasi H. L. (1994) Identification of calreticulin as a rubella virus RNA binding protein. Proc. Natl. Acad. Sci. U.S.A. 91, 12770–12774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Obeid M., Tesniere A., Ghiringhelli F., Fimia G. M., Apetoh L., Perfettini J. L., Castedo M., Mignot G., Panaretakis T., Casares N., Métivier D., Larochette N., van Endert P., Ciccosanti F., Piacentini M., Zitvogel L., Kroemer G. (2007) Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat. Med. 13, 54–61 [DOI] [PubMed] [Google Scholar]
- 122. Kepp O., Senovilla L., Galluzzi L., Panaretakis T., Tesniere A., Schlemmer F., Madeo F., Zitvogel L., Kroemer G. (2009) Viral subversion of immunogenic cell death. Cell Cycle 8, 860–869 [DOI] [PubMed] [Google Scholar]
- 123. Yocupicio-Monroy M., Padmanabhan R., Medina F., del Angel R. M. (2007) Mosquito La protein binds to the 3′ untranslated region of the positive and negative polarity dengue virus RNAs and relocates to the cytoplasm of infected cells. Virology 357, 29–40 [DOI] [PubMed] [Google Scholar]
- 124. Monier S., Jollivet F., Janoueix-Lerosey I., Johannes L., Goud B. (2002) Characterization of novel Rab6-interacting proteins involved in endosome-to-TGN transport. Traffic 3, 289–297 [DOI] [PubMed] [Google Scholar]
- 125. Infante C., Ramos-Morales F., Fedriani C., Bornens M., Rios R. M. (1999) GMAP-210, A cis-Golgi network-associated protein, is a minus end microtubule-binding protein. J. Cell Biol. 145, 83–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Pernet-Gallay K., Antony C., Johannes L., Bornens M., Goud B., Rios R. M. (2002) The overexpression of GMAP-210 blocks anterograde and retrograde transport between the ER and the Golgi apparatus. Traffic 3, 822–832 [DOI] [PubMed] [Google Scholar]
- 127. Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., Warren G. (1995) Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131, 1715–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Navratil V., de Chassey B., Combe C. R., Lotteau V. (2011) When the human viral infectome and diseasome networks collide: towards a systems biology platform for the aetiology of human diseases. BMC Syst Biol. 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Green P. Cross_Match program (part of Phrap package from Phil Green's documentation, www.phrap.org)
- 130. Ducut Sigala J. L., Bottero V., Young D. B., Shevchenko A., Mercurio F., Verma I. M. (2004) Activation of transcription factor NF-kappaB requires ELKS, an IkappaB kinase regulatory subunit. Science 304, 1963–1967 [DOI] [PubMed] [Google Scholar]
- 131. Wu Z. H., Shi Y., Tibbetts R. S., Miyamoto S. (2006) Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science 311, 1141–1146 [DOI] [PubMed] [Google Scholar]
- 132. Wu Z. H., Wong E. T., Shi Y., Niu J., Chen Z., Miyamoto S., Tergaonkar V. (2010) ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol Cell 40, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]