Abstract
We characterized and evaluated the functional attributes of three yeast high-confidence protein-protein interaction data sets derived from affinity purification/mass spectrometry, protein-fragment complementation assay, and yeast two-hybrid experiments. The interacting proteins retrieved from these data sets formed distinct, partially overlapping sets with different protein-protein interaction characteristics. These differences were primarily a function of the deployed experimental technologies used to recover these interactions. This affected the total coverage of interactions and was especially evident in the recovery of interactions among different functional classes of proteins. We found that the interaction data obtained by the yeast two-hybrid method was the least biased toward any particular functional characterization. In contrast, interacting proteins in the affinity purification/mass spectrometry and protein-fragment complementation assay data sets were over- and under-represented among distinct and different functional categories. We delineated how these differences affected protein complex organization in the network of interactions, in particular for strongly interacting complexes (e.g. RNA and protein synthesis) versus weak and transient interacting complexes (e.g. protein transport). We quantified methodological differences in detecting protein interactions from larger protein complexes, in the correlation of protein abundance among interacting proteins, and in their connectivity of essential proteins. In the latter case, we showed that minimizing inherent methodology biases removed many of the ambiguous conclusions about protein essentiality and protein connectivity. We used these findings to rationalize how biological insights obtained by analyzing data sets originating from different sources sometimes do not agree or may even contradict each other. An important corollary of this work was that discrepancies in biological insights did not necessarily imply that one detection methodology was better or worse, but rather that, to a large extent, the insights reflected the methodological biases themselves. Consequently, interpreting the protein interaction data within their experimental or cellular context provided the best avenue for overcoming biases and inferring biological knowledge.
The collection of proteins and protein assemblies in a cell constitutes a vital and integral part of the machinery required to sustain all cellular functions and processes (1). Given that most proteins are part of one or more protein complexes, protein-protein interactions are essential in understanding the nature of protein-mediated biological processes. Therefore, because of the large number of potential protein interactions, high-throughput technologies are essential for generating whole-cell maps of these interactions (2, 3). Several large-scale protein interaction data sets of the yeast Saccharomyces cerevisiae have been determined using different high-throughput technologies, namely the following: (1) affinity purification followed by mass spectroscopy (AP/MS)1 (4–7), (2) protein-fragment complementation assay (PCA) (8), and (3) yeast two-hybrid (Y2H) (9–11). Each approach detects and reports interactions in a distinct manner. The Y2H and PCA techniques detect binary interactions, whereas the AP/MS techniques purify and identify protein complexes. All three methods at some point rely on modified protein constructs to identify protein interactions. For example, although the AP/MS uses tagged bait proteins to bind to prey proteins in the native cellular environment, followed by affinity purification and mass spectrometry detection of proteins, both Y2H and PCA rely on separate protein complementation schemes to ultimately report on whether a protein pair is interacting. In addition, the AP/MS and PCA methods identify interactions at approximate physiological cellular protein concentrations, the concentrations of the interacting partners in the Y2H screens are not necessarily comparable to that found in the native environment. Furthermore, the Y2H method requires that interacting partners are present in the nucleus in order for their interaction to be detectable.
The reliability of each technique has been extensively reviewed in the literature and comprehensive analyses have often resulted in contrasting conclusions (8, 11–17). For example, the overlap of Y2H screens by different laboratories is often small (18), suggesting high false-negative rates, whereas AP/MS screens infer a substantial fraction of indirect interactions (11), suggesting high false positive rates. However, it is generally accepted that any measure of reliability is not absolute and largely dependent on the nature of the selected gold standard reference set (11, 19). Several studies have attempted to identify subsets of high-confidence interactions in the raw AP/MS (6, 7, 14, 20, 21) and the Y2H data (11). To date, only one comprehensive PCA data set exists for yeast (8), limiting assessments of interlaboratory variability and reproducibility of this method (22). Recently, Yu and colleagues consolidated three Y2H data sets into a single high-confidence set and showed that this set is more enriched with interactions found in the manually curated binary gold standard (BGS) data set than the combined set from two AP/MS studies (11). More recently, our group developed a novel approach to score pair-wise protein associations derived from AP/MS data sets (14). This procedure, termed interaction detection based on shuffling (IDBOS), computes a co-occurrence significance score for two proteins by comparing the number of times they are experimentally observed to co-purify with those obtained from commensurate randomized simulations. This data set identifies binary interactions as well as, or better than, the high confidence consolidated Y2H set and previous high-confidence data sets based on AP/MS purifications. These results are of particular importance because, unlike previous studies (7, 20), the IDBOS procedure, which generates the high-confidence AP/MS set is a purely numerical approach that requires no training set or machine learning, resulting in data sets that are less likely to be biased by previous knowledge. Topological analyses reveal stark differences with respect to the modularity of the networks between different data sets, in particular, our high-confidence AP/MS network exhibits densely connected regions of proteins (14) indicative of functional modules (23–25). Here, the IDBOS-derived high-confidence data set is compared and contrasted with the consolidated Y2H and the PCA high-confidence data sets.
The apparent dependence of the protein interaction data or network on the detection methodology raises two fundamental questions: what are the different methods actually detecting and how does this influence the downstream analyses and interpretation of the data as protein interaction data? The core question of whether two proteins bind together is a thermodynamic question that involves the binding free energy associated with the bound protein complex as opposed to two infinitely separated, noninteracting, proteins (26–29). This quantity relates to the ability of two proteins to bind. Even if this information were available, the chemical and biological state of a cell will dynamically determine if binding actually takes place (30). The uncertainty increases if we were to probe this interaction via an experimental technique that would affect the chemical and biological state of the cell as well as the proteins' microenvironment. Therefore, given that the results are not independent of the experimental techniques and that each experimental technique yields a completely different set of interactions, how can one interpret the data in any meaningful fashion? The most general approach is to keep each data source separate and identify biological insights captured by all methods. However, this approach is hampered by a lack of sufficient overlap between the existing data sets and, hence, could still produce contradictory conclusions about the biology associated with or derived from the underlying interaction data.
Herein, we have analyzed and compared three categories of high-confidence high-throughput protein interaction data sets to highlight the apparent differences in biological content associated with these data sets. The high-confidence nature of these data sets ensures that the experimental uncertainty associated with large genomic-scale interaction screens is minimized and allows us to focus on the inherent methodological differences. We found that there were marked differences in terms of which proteins were retrieved from these screens, how their interactions were distributed among different functional classes of proteins, and, in particular, that there were strong methodological biases for the retrieval of proteins belonging to small versus large interaction complexes. We mapped known Munich Information Center for Protein Sequences (MIPS) (31, 32) protein complexes onto each high-confidence protein-protein interaction network to highlight the distinct higher-order organization between functional components in the data sets. These differences were partly reflective of the experimental technology and were germane to the downstream analyses and interpretations of each high-confidence network in terms of correlation of protein abundance among interacting proteins pairs and the location of essential proteins in the composite protein interaction network. In the latter case, we derived a consensus analysis that minimized the experimental biases and showed that the essentiality-connectivity correlation was present in these data sets.
In summary, we quantified the differences between three high-confidence protein-protein interaction networks in yeast and showed how the different methodologies affect the biological interpretation of the data. Specific interactions and conclusions derived from selected protein interactions are, at the current state of knowledge and experimental capacity, strongly tied to the underlying experimental platforms and, hence, comparisons of biological insights derived from data sets with different biological characteristics may be contradictory without accounting for the underlying experimental biases themselves.
RESULTS AND DISCUSSION
We investigated three high-confidence high-throughput protein-protein interaction data sets (AP/MS, PCA, and Y2H), an unfiltered, raw interaction data set (raw-AP/MS), and the manually curated BGS set which focuses on binary protein interactions. The Methods Section describes the selection and salient features of these data sets, whereas columns 1–3 of Table I summarize the number of proteins and interactions contained in these data sets. Herein, we first discuss the classification of detected proteins and their interactions based on the detection methodology, and then we highlight the impact of the observed differences in the biological properties of the interacting proteins.
Table I. Annotation coherence among interacting proteins for selected protein interaction data sets. For each top-level annotation class of “Function,” “Location,” and “Complex” in the Munich Information Center for Protein Sequences, we classified an interaction as intra-annotation if both proteins were annotated and shared at least one common annotation item, or interannotation if both proteins were annotated but no common annotation was shared. The sum of the intra- and inter-annotation can add up to less than the total number of interactions due to cases where at least one protein lacks any annotation. The number in parenthesis gives the percentage of the total number of interactions in Column 3. The Methods Section provides descriptions and characteristics of the separate data sets. Abbreviations: AP/MS, affinity purification/mass spectrometry; PCA, protein-fragment complementation assay; Y2H, yeast two-hybrid.
| Network | Number of Proteins | Number of Interactions | Function |
Location |
Complex |
|||
|---|---|---|---|---|---|---|---|---|
| Intra-annotation | Inter-annotation | Intra-annotation | Inter-annotation | Intra-annotation | Inter-annotation | |||
| Manually curated binary interaction | ||||||||
| BGS | 1061 | 1239 | 1176 (95%) | 41 (3%) | 1092 (88%) | 83 (7%) | 521 (42%) | 102 (8%) |
| High-confidence high-throughput | ||||||||
| AP/MS | 1274 | 7879 | 6722 (85%) | 1007 (13%) | 6914 (88%) | 626 (8%) | 2462 (31%) | 819 (10%) |
| PCA | 1076 | 2530 | 1315 (52%) | 732 (29%) | 1802 (71%) | 530 (21%) | 161 (6%) | 109 (4%) |
| Y2H | 1962 | 2703 | 1161 (43%) | 834 (31%) | 1691 (63%) | 580 (21%) | 148 (6%) | 113 (4%) |
| Raw high-throughput | ||||||||
| AP/MS | 2551 | 18,043 | 9537 (53%) | 7315 (40%) | 12,486 (69%) | 5048 (28%) | 1393 (8%) | 2731 (15%) |
Classification of Detected Proteins and Their Interactions
Functional Diversity in Protein Interaction Data Sets
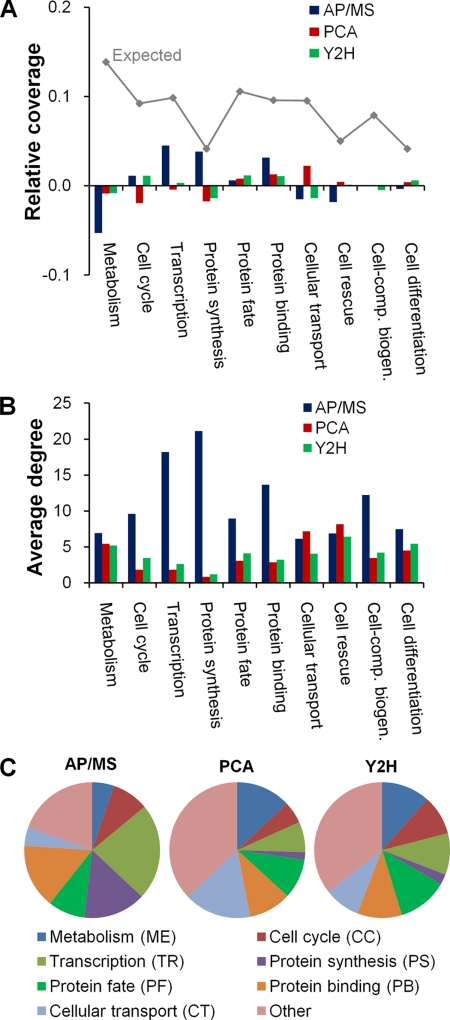
Although genomic-scale protein-protein interaction detection campaigns are by design intended to probe as many interactions as possible, it is well known that the retrieved interactions do not actually overlap. Even for the high-confidence data sets analyzed herein, the overlap between interactions in the AP/MS:Y2H, AP/MS:PCA, and Y2H:PCA sets in Table I were 175, 182, and 66, respectively. Out of all 13,112 high-confidence interactions in Table I, only 18 interactions (0.1%) were common among all three sets. This was despite a relatively larger partial overlap between the constituent proteins among the three data sets, with overlaps between the AP/MS:Y2H, AP/MS:PCA, and Y2H:PCA data sets in Table I of 545, 357, and 440, respectively, with 182 proteins (4.2%) common among all three sets. Although the overlap was larger than for the interactions themselves, each protein set was quite distinct and a functional classification of these protein sets allowed us to generalize the different characteristics between the data sets. Fig. 1A shows the relative distribution of functional classes for all S. cerevisiae proteins for ten selected MIPS functional classes (labeled “Expected”) and the difference from these values for the interacting proteins from the three high-confidence data sets, AP/MS, PCA, and Y2H. Across all 17 MIPS functional classes, the Y2H and PCA data sets showed the smallest deviation between the measured and “Expected” values with root mean squared differences of 0.03 and 0.04, respectively, compared with 0.09 for the AP/MS data. We found the largest deviations in the AP/MS data set associated with under-representation of metabolic proteins and cell rescue, and over-representations in the categories of transcription, protein synthesis, and protein binding. For the PCA data set, the largest under-representation was of cell-cycle proteins and protein synthesis proteins, and the largest over-representation was of cellular transport proteins. If the selection of tested protein is unbiased, these differences should reflect the unavoidable experimental biases associated with each detection method. For example, the under-representation of metabolic proteins in the AP/MS data can be rationalized by the fact that metabolic proteins, in general, do not function in large complexes or bind strongly to other proteins (33); hence, it is expected that the affinity purification step will most likely result in loss of these proteins. Similarly, proteins that function in larger, tightly organized assemblies involved in transcription or protein synthesis would be preferentially included in the AP/MS data set, whereas the restriction imposed on the PCA and Y2H reporter systems would not favor these protein classes.
Fig. 1.
Functional diversity among proteins and interactions present in the high-confidence data sets. A, The relative distribution of all proteins from Saccharomyces cerevisiae, as annotated according to the Munich Information Center for Protein Sequences functional categories, is shown by the gray line labeled “Expected.” It denotes the relative fraction (coverage) of all yeast proteins that belong to a given category. The deviations of the AP/MS, PCA, and Y2H high-confidence data sets from this distribution are shown by the different colored bars, e.g. proteins labeled “Metabolism” are relatively under-represented in the AP/MS data set. B, The average degree of the proteins in a given functional category for each high-confidence data set. C, The relative distribution of protein-protein interactions in the different functional categories for each high-confidence data set.
The number of detected interactions each protein has with other proteins was strongly dependent on the functional class membership and detection methodology. Fig. 1B shows the average number of interactions (or degree) distributed among the same functional categories as in Fig. 1A for the three data sets. There are large and clear differences between the AP/MS data set on the one hand and the PCA and Y2H data sets on the other hand, particularly for proteins involved in cell cycle, transcription, protein synthesis, protein fate, protein binding, and cell-component biogenesis. Fig. 1C shows the relative distribution of these interactions among all detected interactions for each experimental technique. These relative distributions indicate that the data sets contained clearly differentiated sets of distinct sets of interactions distributed among varying functional classes. Thus, the AP/MS interaction data were skewed toward proteins in transcription (23%) and protein synthesis (15%), the PCA data were skewed toward proteins in cellular transport (16%) and metabolism (13%), and the two largest groups in the Y2H data were proteins in metabolism (12%) and protein fate (12%). This confirms that the different high-confidence interaction data sets were associated with proteins in varying functional classes whose proteins had a distinct number of interactions, resulting in a unique distribution of interactions among specific protein sets.
Fig. 2 shows the projection of the three high-confidence data sets on the protein-protein interaction matrix encompassing all possible binary interactions that can be formed based on the yeast proteome. The proteins are ordered according to their MIPS functional categorization and follow the same order as in Fig. 1, i.e. metabolism, cell cycle, etc. It is well known that the sparsity of the matrix is due to the relatively low estimates of the yeast interactome (∼105 unique protein-protein interactions) compared with the total number of possible pair-wise interactions (∼107) (34, 35). The interactions mapped out in Fig. 2 show that the different data sets covered distinct parts of the interaction space, with some functional categories relatively well covered whereas others were disproportionately sparse. This observation is due partly to biological reasons and partly to the methodological differences in detecting the interactions. Thus, we expected that direct interactions among cell cycle proteins (CC) and proteins involved in protein synthesis (PS) would be absent. We confirmed this expectation for all three experimental platforms. Similarly, the intersection between PS and protein fate (PF) should be, and was, relatively free from interactions in all three data sets. The methodological biases noted in Fig. 1 were also evident in the interaction mapping in Fig. 2; the functional categories of PS and protein binding (PB) were well represented by the AP/MS data set, whereas interactions among metabolic proteins (ME) were relatively sparse among all data sets. Fig. 2 also shows the high proportion of interactions within the cellular transport (CT) category associated with the PCA data set.
Fig. 2.
High-confidence data set coverage of the protein-protein interaction matrix. We have mapped each interaction in the AP/MS, PCA, and Y2H data sets to the yeast proteome protein-protein interaction matrix defined by all possible binary interactions. The proteins were ordered according to their Munich Information Center for Protein Sequences functional categories. We have indicated the location of proteins belonging to the categories of metabolism (ME), cell cycle (CC), transcription (TR), protein synthesis (PS), protein fate (PF), protein binding (PB), and cellular transport (CT). We have enlarged the symbols of each interaction to make the differences among the data sets and functional categories more visible. The different methodologies retrieved different interaction sets influenced by the underlying experimental platform, e.g. AP/MS recovered tightly bound protein complexes associated with transcription, protein synthesis, and proteins binding, whereas PCA recovered many more weakly bound interactions, e.g. those involved in cellular transport.
The protein-protein interaction matrix can also be used to gauge the level of influence biological functions have on the detected interactions. A randomly generated selection of interactions, on the same order of magnitude as the number of interactions among the high-confidence data sets (shown in Table I), will not show any preferential clustering among or between functional categories in the matrix (shown in Fig. 2). We characterized this property by calculating the distribution of distances between each two points within a data set in the matrix and comparing the distributions of the data sets (supplementary Fig. S1). This analysis showed that the interactions in the Y2H data set were distributed most similarly to those in the randomly selected interaction data compared with the AP/MS and PCA data sets. This does not indicate that the Y2H data were random; rather, it demonstrated that the interactions detected in this data set were not biased toward any particular functional set or classification (11). This implied that the experimental detection of these interactions was not influenced by the biological function of the tested proteins and, therefore, provided a markedly unbiased test of the proteins' capability to interact. This leads to increased confidence in the ability of this data set to provide a foundation for a stricter thermodynamic evaluation of the proteins' ability to interact under the given experimental conditions.
In contrast, native protein interactions are sensitive to local chemical environments, protein concentrations, regulatory and nonequilibrium processes, ATP-levels, phosphorylation status, post-translational modifications, etc., which define the “natural” environment. Although one advantage of the AP/MS and PCA procedures is their probing of interactions in this natural environment, an unbiased selection of protein interactions similar to the Y2H methods was not achieved. Although this may provide an advantage in detecting interactions that actually occur in the cellular environment under the given experimental conditions, the lack of direct overlap between the AP/MS and PCA data sets highlights the sensitivity of these methods to experimentally specific cellular conditions. In the material below, we explored the consequences of these biases in selecting protein interactions in the three data sets.
Annotation Consistency of High-Confidence Data Sets
In order to characterize the retrieved protein sets (12, 36, 37), we further stratified the interaction data according to three different high-level MIPS annotation classes as follows: “Function,” “Location,” and “Complex.” Table I summarizes these analyses for a reference set of manually curated binary interactions (BGS), the three high-confidence protein interaction data sets, as well as for a raw high-throughput data set. For each of the annotation classes we have further subdivided the interaction data, based on whether interacting proteins share their annotations, into inter- and intra-annotation groups. For example, if both members of a protein-protein interaction pair belong to the same functional category of “metabolism,” we classified the pair as an intra-annotation pair; otherwise, we classified the pair as an inter-annotation pair. For the annotation class “Function,” we noted that the manually curated BGS data set was heavily skewed toward protein annotations belonging to the same versus different functional categories (95% versus 3%). The same holds true for the high-confidence AP/MS set (85% versus 13%) in sharp contrast to the corresponding raw data (53% versus 40%). In this case, the extraction of the high-confidence subset from the raw data resulted in recovering more protein interactions belonging to the same functional category. The similar fraction of intra- and interannotation pairs in the BGS and the high confidence AP/MS data set was not caused by overlapping interactions between these sets, as only 340 of the these interactions coincided. This implies that the AP/MS technique, although designed to detect complexes, actually possesses a large binary interaction character (14). Neither the PCA nor the Y2H data sets showed the same sharp distinction between intra- and interannotations of protein interaction pairs. Of course, the general statement that interacting proteins tend to belong to the same functional category holds, regardless of the methodology or any biases in selection of tested proteins. Similarly, relative observations also hold true for the annotation class “Location.” In the class “Complex,” which contains proteins known to be associated with specific protein complexes, the lack of a comprehensive set of protein annotations sharply reduced the number of protein interactions that we could classify, especially for the PCA and Y2H data sets. However, it was clear that both the reference standard (BGS) and the high-confidence AP/MS data sets were still enriched in intra-annotation, compared with interannotation complex pairs.
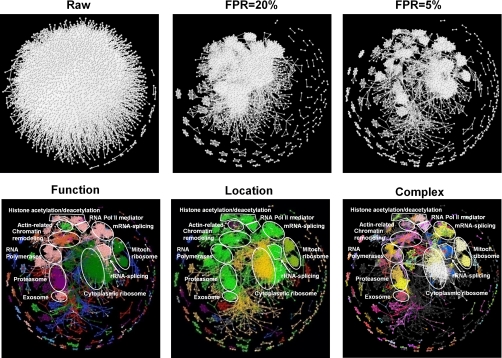
The observation that interacting proteins share a common function indicates that the interaction data itself could carry information about the organization of functional modules (23–25). Consistent with the high intra-annotation fraction in the AP/MS data set, Fig. 2 shows the densely populated diagonal of the protein interaction matrix. The AP/MS interactions data set was further characterized by distinct, densely connected off-diagonal regions, reflective of the high-confidence nature of this data set. The top row of Fig. 3 illustrates this point by showing the difference in connectivity between a low-confidence raw network and higher confidence networks (commensurate with 20 and 5% false positive rates (See Methods)) based on our IDBOS-analysis of the AP/MS data. The high intra-annotation fraction of this data set manifested itself as clusters of protein interactions among proteins with the same biological function, effectively defining, as well as identifying, clustered modules. The bottom row of Fig. 3 shows the highly clustered annotations for the “Function,” “Location,” and “Complex” annotation classes from Table I for the high-confidence AP/MS data set (commensurate with a 5% false positive rate). The modularity of the organization of this protein interaction network was evident by the concomitant clustering of functional properties (same color) to distinct sets of proteins. The interactions colored according to their “Complex” annotations are explored in more detail in the “MIPS Complex Annotation of Interaction Networks” Section.
Fig. 3.
Organization of interactions and biological processes in the AP/MS data set. The top row illustrates the effect of increasing the fidelity of the network representation by decreasing the false positive rate (FPR) associated with the interactions. The data set commensurate with a 5% false positive was designated as the high-confidence AP/MS data set in this work. The bottom row shows the projection of Munich Information Center for Protein Sequences (MIPS) high-level annotations color coded for different “Function,” “Location,” and “Complex” categories. We assigned each protein in the network only one of its MIPS function annotation item(s) to maximize the number of homogeneous interactions of the network using a Monte Carlo algorithm (See Methods). We also outlined selected major biological processes in these interaction maps. The complete color scheme and annotations for the “Complex” annotations are provided as an interactive and viewable map in the Supporting Material.
Protein Complex Size Dependence
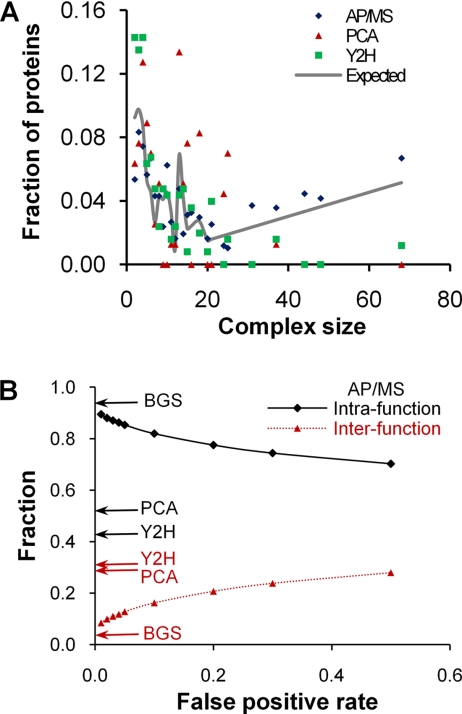
Proteins often assemble into larger functional multiprotein complexes that strongly determine how proteins interact and arrange themselves. Herein, we demonstrated the systematic biases associated with the detection techniques as manifested in the dependence on the number of interacting proteins retrieved from a given MIPS complex as a function of the size of the MIPS complex. Fig. 4A shows the relative fraction of proteins from intracomplex interactions detected for each high-confidence data set as a function of the size of the complex. Although there was some scatter in the data, it was clear that both the PCA and Y2H data were enriched with proteins from smaller-sized (less than 25) complexes compared with what would be expected from an analysis of all MIPS yeast proteins. Both the PCA and Y2H data lacked interaction data from larger-sized complexes. This is in contrast to the AP/MS data, which closely followed the Expected distribution present among all annotated yeast proteins, even for the larger size MIPS clusters. Although the general view is that AP/MS detects co-complexes and Y2H detects binary interactions, this is an over simplification. The high-confidence AP/MS data contain a wealth of known binary interactions and conversely, as shown here, the Y2H data capture many binary interactions among MIPS complexes with less than 25 members.
Fig. 4.
Complex size and intra- and interfunction detection. A, The relative distribution of sizes of Munich Information Center for Protein Sequences complexes in the Saccharomyces cerevisiae proteome is shown by the gray line labeled “Expected.” The y axis shows the fraction of proteins in a given data set that is associated with a specific complex size. We have indicated the corresponding distributions for proteins found in the complexes of each high-confidence data set by different symbols. Whereas both the PCA and Y2H data sets lacked representations among the large-sized clusters, the AP/MS data set roughly followed the “Expected” distribution derived from all yeast proteins. B, The influence of the false positive rate on the fraction of intra- and interfunction protein interactions detected in the AP/MS data set. For reference, we have indicated by arrows the PCA and Y2H values for intra- and interfunction annotation fractions from Table I on the y axis.
Apart from experimental constraints in the PCA methodology, which limits interaction detection to within an 8-nm resolution, protein crowding affects interaction detection in both PCA and Y2H. Both of these detection methods explicitly rely on the reconstitution of the biological activity of a particular protein (transcription factor GAL4 for Y2H and the DFR1 enzyme for PCA). Even if two investigated proteins, as components of a large native cellular complex, can interact and constitute a functional unit, other components may bind to this unit disturbing the activity reconstitution and consequently preventing detection (8, 11). The probability of such disturbance is roughly proportional to the number of components in the complex, i.e. the complex size, effectively preventing the detection of interactions within large complexes using Y2H and PCA methods. The probability of this happening would be larger at the complex's native cellular location because of higher abundances of other complex components, e.g. this may partially explain why we did not observe any of the interactions between RNA polymerase II components in any of the Y2H set (11). Methodological biases in detecting interactions among specific RNA synthesis complexes are characterized in more detail in the “MIPS Complex Annotation of Interaction Networks” Section. The AP/MS data sets strictly do not report an interaction per se, but rather a colocalization of proteins in bait-prey affinity purifications. Although these interactions should be less sensitive to the location of the tagged baits, the influence of tagging cannot be ignored because it has been previously demonstrated that intraspecies overlap between AP/MS data sets from different laboratories remains low (38). Therefore, whereas the high-confidence subsets of the PCA and Y2H data sets can evaluate whether a protein pair interacts, the AP/MS experimental data can also characterize nonbinary protein interactions. This analysis confirms that AP/MS data may be more suitable for understanding interactions and biological relationships related to structural complexes and larger molecular machines, whereas the Y2H and PCA data sets capture many binary interactions of smaller-size complexes.
False Positive Rate of AP/MS Interactions
The question whether one data set or experimental technique is more accurate than another cannot be directly addressed by the analyses presented herein. However, one advantage of our derivation of high-confidence interactions from the raw AP/MS data is that we can select data at a given false positive rate (See Methods), e.g. as shown in Fig. 3. As a further analysis of the data in Table I, we calculated the fraction of interacting protein pairs that contain only inter- or only intra-functional annotations for the AP/MS data set as a function of the false positive rate. Fig. 4B shows the decrease (increase) of the intra (inter)-function fraction with an increasing false positive rate, i.e. at a lower accuracy. The limiting behavior at the highest accuracy (lowest false positive rate) approaches the fractions found in the manually curated data set of high-confidence interactions (BGS). Intrafunction annotation fractions for the AP/MS data set remained consistently higher than that for either the PCA or the Y2H data set, whereas the inter-function annotation fractions leveled out at ∼30% at higher false positive rates, similar to the PCA and Y2H data sets. However, one should note that the fractions of intra- and interfunction interactions are not indicative of the false positive rates in the PCA and Y2H data sets themselves. Although the construction of the high-confidence AP/MS data set did not discriminate between inter- or intrafunction protein-protein interactions, there was a systematic enrichment of intrafunction interactions at the low false positive rates. As previously reported (14), the binary data contained in our high-confidence AP/MS data comprise interactions of the same quality as the manually curated data set (BGS). Increasing confidence (lowering the false positive rate) in the AP/MS data increased the “binary” flavor of the interactions. This was reflected both in the closer concordance with the intra- and interfunction fractions (Fig. 4B) and in the fraction of proteins retained for complexes with less than 25 members (data not shown).
Biological Properties Associated with Interacting Proteins
We observed that the high-confidence high-throughput interaction data sets contained strong remnants and signatures reflecting their experimental creation with respect to functional characteristics of the proteins retrieved and in the size of the underlying protein complex from which the interactions were detected. We now turn our attention to describing how the different interaction data sets reconstitute important functional components of the cell and their interactions, how the different methods capture the cellular abundance between interacting proteins, and how methodological biases influence the connectivity of essential proteins in the reconstructed interaction networks.
MIPS Complex Annotation of Interaction Networks
The objective of detecting and cataloging a genome-wide range of protein-protein interactions present in a cell is to understand how these interactions mediate biological processes. The premise of creating interaction networks is that biological events can be partially deconstructed and understood by sets of direct protein interactions. Because cellular processes are often performed and mediated by protein complexes and assemblies, it is expected that investigating the overall organization of protein interaction networks reconstituted from interaction data should both recover known associations and provide additional insights into these processes. Consequently, we mapped known MIPS protein complexes (39) onto each reconstituted high-confidence protein interaction network to explore the biological implication of the network organization. An interactive map of the annotated high-confidence networks for use with the chemical structure viewer Jmol (http://www.jmol.org/) is provided to facilitate further explorations (see Live Cellular Machinery Map in the Supplementary Information). The networks exhibited different degrees of patterns of connections between sets of proteins, providing a wealth of information on interactions between complexes and possible protein regulation of their biological function.
Verification of known associations between functional modules serves to validate the inherent biological content of high-confidence high-throughput networks (4–11). Given the high degree of MIPS annotated intrafunction interactions in the AP/MS data set, this reconstructed network provided well-separated and distinct modules in comparison to the PCA and Y2H data sets. Fig. 5 shows that the known protein complexes from MIPS (with high-throughput complexes excluded, see Methods) were recovered well in the AP/MS data set. The preponderance of annotated intra- versus inter-complex interactions, accentuated the modular nature of the network, with many complexes of known biological connections linked to each other via direct protein interactions. For example, the two 20S and 19/22S regulator units of the proteasome were directly linked to each other. The central and dominant part of the connected network consisted mainly of protein complexes responsible for RNA synthesis and other RNA-related processes, protein synthesis, protein degradation, and DNA replication and repair. As transport proteins were under-sampled in the AP/MS data set (Fig. 1), many of the transport systems (e.g. Transport Protein Particle, Golgi Transport, GIM complexes, t-SNAREs, v-SNAREs, AP-2, AP-3, etc.) were not connected to the central network and were represented as isolated units on the periphery of this map.
Fig. 5.
The “Complex” annotated protein interaction map. Known complexes from the Munich Information Center for Protein Sequences (MIPS) database and their biological relations were apparent in the reconstructed protein-protein interaction network derived from the high-confidence AP/MS data set. We colored components in a same MIPS complex the same, with gray nodes representing unannotated proteins. This reconstructed network captured a global organization of protein complexes, in particular for protein assemblies related to RNA synthesis, chromatin remodeling, DNA replication and repair, protein synthesis, and protein and RNA degradation (proteasome and exosome). Proteins in transport complexes were connected among themselves, but the proteins that they transport were not captured in this mapping. Some labels have been removed for clarity, and the complete annotations are provided in the Supplementary Material.
A global comparison between individual interactions and functional complex interactions at the protein level between the three different high-confidence protein interaction data sets is not productive because of the almost negligible overlap of interactions. Instead, we have focused on a comparison of interactions between select functional complexes associated with “RNA synthesis related complexes” and a separate comparison of the “Protein transport machineries” to highlight the inherent methodological biases of the three high-throughput techniques.
RNA Synthesis Related Complexes
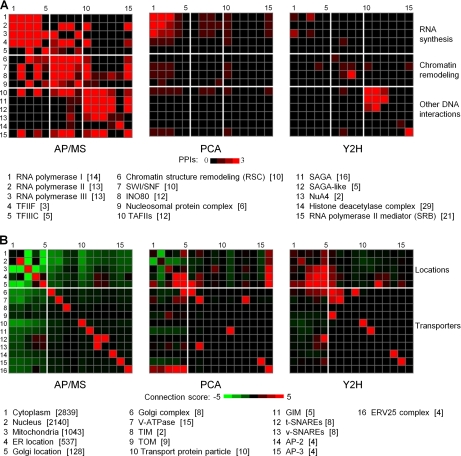
RNA synthesis is closely connected to chromatin remodeling and other biological processes that control DNA. Fig. 5 shows that RNA synthesis complexes formed a highly interconnected cluster, including RNA polymerases I, II, and III, Transcription factor complexes II F (TFIIF) and III C (TFIIIC), which were connected via direct protein-protein interactions with many other functional complexes. Fig. 6A shows a comparison of the number of detected intra- and intercomplex protein-protein interactions among and between selected MIPS annotated complexes. In particular, Fig. 6A highlights the intra- and interconnectivity among RNA synthesis, chromatin remodeling, and other DNA interacting protein complexes for the three high-confidence interaction data sets. Consistent with the higher intra-annotation fraction of the AP/MS data (Table I and Fig. 2), the diagonal elements for this data set were the most populated. Therefore, the AP/MS data show that these three biological processes are highly intraconnected via protein-protein interactions; the PCA data capture part of these interactions among RNA synthesis complexes and the Y2H data capture protein interactions mainly among the DNA-associated protein complexes, such as TATA-binding protein associated factors (TAFIIs) and SAGA complexes. This is also consistent with the observation that Y2H methods preferentially detect protein interactions that contains specific DNA-binding motifs (40).
Fig. 6.
Connectivity among and between Munich Information Center for Protein Sequences complexes. A, The number of interactions between proteins among the constituent functional complexes associated with RNA synthesis (rows and columns 1–5), Chromatin remodeling (rows and columns 6–9), and Other DNA interactions (rows and columns 10–15) are color-coded from none (dark) to three or more (bright red). The number of proteins in each of the 15 complexes is given in square brackets below the graph. Abbreviations: TFIIF, Transcription factor complexes II F; TFIIIC, Transcription factor complexes III C; SWI/SNF, SWItch/Sucrose NonFermentable; TAFIIs, TATA-binding protein associated factors; SAGA, Spt/Ada/GCN5/acetyltransferase; NuA4, nucleosome acetyltransferase of H4. B, The link between the cellular locations of proteins and the different protein transport assemblies that interact with these proteins. The figure shows the connection Z-score (See Methods) associated with co-occurring protein labels of interacting proteins as a function of their locations (rows and columns 1–5) and association with different transporter complexes (rows and columns 6–16). Abbreviations: ER, Endoplasmic reticulum; TIM, the inner mitochondrial membrane protein translocase; TOM, transport across the outer membrane; GIM, prefoldin protein complex; t-SNARE, target SNAP (Soluble NSF Attachment Protein) Receptor; v-SNARE, vesicle SNAP (Soluble NSF Attachment Protein) Receptor; AP-2, Adaptor protein complex-2; AP-3, Adaptor protein complex-3; ERV25, ER Vesicle 25.
The off-diagonal elements of Fig. 6A address protein interaction links between different functional complexes and draw attention to the different types of interactions retrieved by the three high-throughput methods. Chromatin remodeling is required to initiate and conduct RNA synthesis and DNA replication and repair. Fig. 5 shows the overall location of chromatin remodeling and RNA polymerase proteins of the AP/MS data set, whereas Fig. 6A shows the detailed complex-complex interaction mapping for the three high-confidence networks. The interaction data support the RNA polymerases II and III connection with chromatin remodeling modules, via RNA polymerases II interacting with the nucleosome remodeling complexes SWI/SNF and INO80 and via RNA polymerases III interacting with the chromatin structure remodeling (RSC) complex and nucleosomal protein complexes (histones) (41, 42). DNA interacting assemblies, such as TAFIIs and RNA polymerase II mediator complex (SRB), were also directly linked via protein-protein interactions to RNA synthesis complexes, whereas others, such as, histone acetyltransferase complexes [SAGA (43) and NuA4 (44)] and histone deacetylase complex were indirectly linked (43–46). As shown in Fig. 1, the AP/MS data set was relatively enriched in proteins and protein interactions associate with cell cycle processes compared with the PCA or Y2H data. Hence, the organization of the AP/MS interaction network captured the biological association of the functional components of the cell cycle via the assembly of direct protein-protein interactions to a higher degree than the PCA and Y2H networks.
Protein Transport Machineries
Fig. 5 shows the relative isolation of transport complexes in the AP/MS network reconstruction. Fig. 6B shows an overall comparison between detected protein interactions within transport complexes in the three high-confidence data sets as well as the interaction tendency between the transporters and other proteins in different cellular compartments. Herein, we reported the co-occurrences of MIPS labels (“Locations” and “Transporters”) between interacting proteins based on Z-score calculations (See Methods), where the green color indicates avoidance and the red color indicates preferential co-occurrence. This score measures the likelihood that proteins in different annotation categories are associated and interact with each other. For the transporters themselves, the AP/MS data recovered the intracomplex interactions to a higher degree than in the corresponding PCA and Y2H data sets. Conversely, interactions between these transport complexes and proteins located in the cytoplasm, nucleus, mitochondria, endoplasmic reticulum (ER), and Golgi were strongly avoided in the AP/MS data set. The PCA and Y2H data sets did not show this avoidance; instead, many more interactions between transporter complexes and other cellular proteins were present in these data sets. As a specific example, members of the p24 family are engaged in protein transport between the ER and the Golgi apparatus. Protein transport is executed via COPII-coated vesicles, where four of the p24 member proteins (ERV25, EMP24, ERP1, and ERP2) are known to form the ERV25 complex that line the vesicles and interact with cytoplasmic coat proteins (47). Annotated as a cellular transport protein, ERV25 was associated with 43 interactions in the PCA data set, but with only five interactions in the AP/MS data set. Fig. 6B also captured the gross features of this interaction imbalance with a virtual absence of interactions in the AP/MS data between the ERV25-complex and proteins in the five locations shown, compared with an abundance of interactions in the PCA data. Whereas none of these interactions were present in the Y2H data set, only one protein (ERP1) among these 43 PCA proteins coincided with the AP/MS set, suggesting that a large portion of the detected PCA interactions were weak or transient physical associations, such as those between the transporters and the protein objects that they transport.
The raw-AP/MS data set contains similar weak associations between cellular machinery components and the objects they operate upon. For example, as a sub-unit of the peripheral membrane domain of the vacuolar H+-ATPase (V-ATPase), VMA2 was associated with 512 and six interactions in the raw and high-confidence AP/MS data sets, respectively. The high number of interactions in the raw data set was most likely a true reflection of the cellular function of the protein, i.e. V-ATPase is an enzyme with remarkably diverse functions in eukaryotic organisms and it acidifies a wide array of intracellular organelles by pumping protons across plasma membranes. Although other V-ATPase-interactions were present, none of the VMA2 specific interactions was present in the high-confidence PCA or the Y2H data sets (Fig. 6B). In general, although PCA can capture some of the transient interactions very accurately, the IDBOS-analysis of the AP/MS purification data cannot distinguish transient or true weak interactions from low-confidence promiscuous associations.
Although the overall coverage of interaction in the high-confidence AP/MS data set remained small compared with the complete interactome, we could still recover a rough outline of many of the components of the cellular machinery and connections among and between these components. The clusters of proteins mapped-out in Fig. 5 defined biologically distinct components and assemblies that constituted the bulk of the cellular machinery. These biological units were, in turn, connected with other units and delineated a global organization of the working components of the cell. Thus, the high-confidence AP/MS data set yielded different, complementary insights into protein properties, protein assemblies, and how they are cross-connected in the cell compared with the PCA and Y2H high-confidence data sets.
Protein Abundance of Interacting Protein Pairs
Previous research has shown that proteins in stable complexes tend to have similar mRNA expression profiles (48, 49). It is natural to assume that two interacting proteins are more likely to be of similar abundance than two noninteracting proteins. This assumption may now be directly tested with the advent of protein abundance measurements such as the recent measurement of yeast cells in rich media (50). For all the proteins in our study we retrieved the corresponding proteins' cellular abundance data from the study of Newman et al. (50). Fig. 7A shows the calculated Spearman correlation coefficients between abundances of interacting proteins for the five data sets in Table I. In order to verify that these correlations were not spuriously related to the observed interactions, we calculated the correlation coefficients using 1000 randomly rewired versions of each protein interaction data set and detected no significant correlations. Using the same cellular abundance data, we also “colored” proteins in the high-confidence AP/MS derived network from Fig. 3 according to their abundance. Fig. 7B shows the corresponding abundance map, which demonstrates that interactions were much more likely to occur between proteins of similar abundance.
Fig. 7.
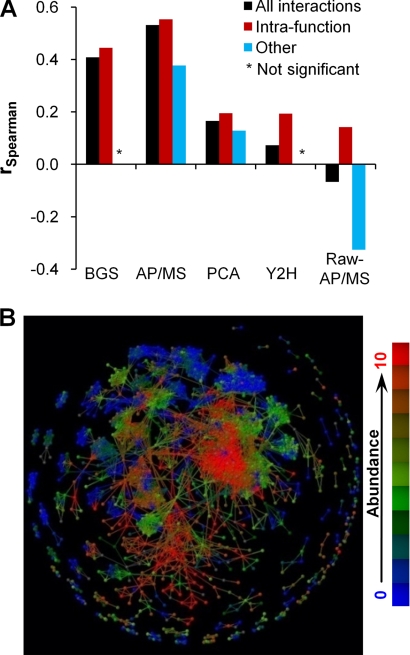
Protein abundance correlation between interacting protein pairs. A, Spearman rank-correlation coefficients rSpearman between abundance ranks of interacting proteins. The histograms are shown for all interactions (All interactions), the subset that encompasses interactions that share the same intrafunction annotations (Intrafunction), and all remaining ones that do not (Other). We indicated correlations that were not statistically significant by a star (*). B, Abundance map of the AP/MS data set. Abundance values (50) were divided into 11 classes from 0 (smallest) to 10 (largest), and each class was represented by a color. Class 0 is the collection of proteins whose abundances were too small to be detected, and classes 1–10 were equally divided among proteins whose abundance could be detected. Interacting proteins in each visible cluster tended to have the same abundance values.
The curated binary interaction data set (BGS) showed a clear positive correlation of the protein abundance of the interacting pairs, indicating that the assumption that interacting proteins having similar concentrations tend to interact with each other was a reasonable. All high-confidence data sets exhibited statistically significant positive correlations to varying degrees. However, the magnitude of the positive correlations for the high-confidence AP/MS, PCA, and Y2H data sets, 0.53 (p value < 10−700), 0.17 (p value < 10−20), and 0.07 (p value < 10−5), respectively, varied substantially. Using the intra- and interannotation schemes shown in Table I, we calculated the corresponding intra-function correlation values. Fig. 7A shows that the high-confidence AP/MS data retained the higher abundance correlation for this subset of interactions, whereas the PCA and Y2H intrafunction correlation values increased to roughly 0.2 (p values < 10−30). The relatively lower correlation values of the intrafunction Y2H and PCA data sets with respect to the AP/MS ultimately depended on the functional characteristics of the proteins sets retrieved by the different methodologies. For example, the enrichment of interactions in the AP/MS data set of highly coordinated functions, such as cell cycle, transcriptions, and protein synthesis, resulted in an enhanced abundance correlation. Likewise, the enrichment of interactions in the PCA data set of transport functions, which do not require the expression of proteins at the same level, resulted in a decreased abundance correlation.
In contrast to the high-confidence data sets, we observed a negative correlation in the raw AP/MS data set. This correlation is an artifact of the experimental methodology and it illustrates how technology can mislead a casual interpretation of the data. In the raw affinity purification data, there is a positive correlation between the number of interactions in which a protein is engaged and the cellular abundance of the proteins (49), i.e. proteins with many interaction partners (hubs) are typically high-abundance proteins and proteins with few interactions are typically low-abundance proteins. However, there are fewer hub proteins that result in hub-proteins in the raw data interacting with many more nonhub proteins. Thus, the raw AP/MS data showed an overall inverse correlation, i.e. interactions between high- and low-abundance proteins are more likely to occur than between similarly abundant proteins. Fig. 7A shows that neglecting protein pairs that we inferred to have positive abundance correlations in the raw-AP/MS data set, e.g. those occurring within the same functions (Intra-function), and evaluating the abundance correlation in the remaining set (Other, rSpearman = –0.34, p value < 10−300) enhanced this effect.
The application of an intrafunction constraint to select interacting proteins clearly increased the correlation of all these data sets, but it also highlighted the inherent differences in the interaction data present in the high-confidence data sets. We found, to a higher degree than for the PCA and Y2H data sets, that the high-confidence AP/MS data were enriched with interactions whose proteins were present at the same concentrations. To further confirm the assumption that interacting proteins should be present in roughly the same concentrations, we assessed whether interactions in the AP/MS data sets were sensitive to their cellular location. The expectation was that interactions present in the same cellular locations (intralocation) should have significantly higher correlation values between proteins than those that do not share the same cellular location (interlocation). We calculated the correlation values for interactions in the intra- and interlocation set for the AP/MS to be 0.55 (p value 10−729) and 0.10 (p value 0.004), respectively. This was consistent with complexes isolated from the AP/MS purifications retaining their natural composition, commensurate with their native location, and not contaminated with protein complexes from different cellular locations.
Essentiality of Proteins in the Different Interaction Data Sets
Relating the essentiality of a protein to its position in a protein interaction network has highlighted the uncertain nature of biological interpretations of data-driven reconstructions of protein interaction networks. Because the time of the earliest observation, based on one of the first Y2H sets (10), that hubs that have more interactors are more likely to be essential (51), other investigations have been carried out and confirmed a positive correlation between essentiality and degree, i.e. the essentiality-connectivity rule (52, 53). However, different explanations to this rule also emerged: (1) hubs play a vital role in maintaining the network connectivity (54); (2) essentialities of proteins come from their interactions, and hubs are more likely to be involved in essential interactions (55); (3) rejecting explanations 1 and 2 above, Zotenko and colleagues demonstrated instead that hubs tend to be essential because of their memberships in essential modules (56), a previously suggested concept (21). In line with explanation 3, another study has shown that large complexes tend to be essential (57). However, a recent investigation of the consolidated Y2H set and the union of two original AP/MS sets, invalidated the essentiality-connectivity rule (11), indicating the dependence of this rule on the underlying interaction networks. Herein, we quantified this dependence, highlighted the inherent differences among the high-confidence data sets, and provided a consensus analysis to finally infer a consistent conclusion from all data sets.
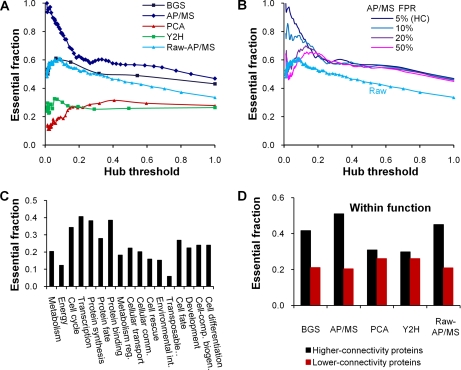
Fig. 8A shows the fraction of essential proteins in the set of proteins defined by a specific hub-threshold, where a hub-threshold of 0.1 means that we selected the highest top 10% connected proteins as hubs. The difference in essentiality-connectivity correlations among all the protein interaction data sets from Table I is quite evident. The PCA data exhibited an opposite behavior, in that, the more connected the proteins, the smaller was the fraction of essential proteins. In contrast, our high-confidence AP/MS data showed a strong correlation between essentiality and connectivity. In line with the investigation by Yu et al. (11), we confirmed that previously constructed high-confidence AP/MS sets (6, 7, 20) did not have such a strong correlation. Consistent with the apparent neutral sampling of interaction in the Y2H data set, the essential fraction was mainly independent of the hub-threshold. The manually curated binary data set and the raw AP/MS data exhibited similar modest correlations at lower hub-thresholds, i.e. among highly connected proteins.
Fig. 8.
Distribution of essential proteins. A, The fraction of essential proteins among hub proteins as a function of hub threshold, represented as the fraction of (hub) proteins with degrees larger than a given degree among all proteins of the studied protein interaction network. For example, a hub threshold of 0.1 means that we selected the highest top 10% connected proteins as hubs. B, The essential fraction of proteins in the AP/MS data are shown for several different false positive rates, indicating the sensitivity of the essentiality-connectivity correlation to the confidence level of the data. C, The fraction of all yeast proteins that were essential as a function of the Munich Information Center for Protein Sequences (MIPS) function categories. D, More connected proteins within the same MIPS function category tended to be essential for all five data sets. For each MIPS complex, we calculated the average degree and extracted proteins of above-average degrees (Higher-connectivity proteins) into a group and those with below-average degrees (Lower-connectivity proteins) into another group. After scanning all MIPS complexes, we calculated the fraction of essential proteins in each group. The consensus conclusion indicated that higher-connectivity proteins were indeed more essential than lower-connectivity ones.
Given the pervasive belief that protein interaction data sets are filled with “noise,” we investigated the consequences of imposing a “high-confidence” filter in the AP/MS data set. Fig. 8B shows the effect on the essentiality-connectivity correlation as a function of the false positive rate. Decreasing the confidence from the designated high-confidence level at a 5% false positive rate significantly weakened the essentiality-connectivity correlation. At false positive rates at or above 20%, the essentiality-connectivity correlation was similar to that of the raw data for hub-thresholds less than 0.1. To determine whether the high correlation for the high-confidence AP/MS data set stemmed from the essentiality of large-sized complexes, we used MIPS complexes to investigate the complex-size essentiality correlation (data not shown). Contradicting a previous study that used their self-defined complexes and which suggested a positive correlation (57), we found that the global complex-size essentiality correlation did not exist and that there was a positive correlation only for small complexes comprising less than 10 proteins.
Instead, we found that the largest influence on the essentiality-connectivity correlation in the data sets was the dependence of the functions of the retrieved proteins in the different data sets. Fig. 8C shows the essential fraction of all yeast proteins annotated by MIPS, which indicated that proteins involved in “Transcription,” “Protein synthesis,” and “Protein binding” tended to be more essential than other groups. Comparing this figure to the retrieved proteins in the high-confidence data sets and their interactions in Fig. 1C, makes it clear that conflicting essentiality-connectivity correlations of the high-confidence data sets were closely related with their opposite interaction frequency profiles in functional categories. For example, as mentioned above, AP/MS sampled a large fraction of interactions involved in transcription, whereas PCA sampled much fewer of these interactions. The negative essentiality-connectivity correlation in the PCA data set in Fig. 8A was a direct consequence of the relatively higher number of interactions sampled in the “Cellular transport” category (Fig. 1C) and these proteins' relatively low fraction of essential proteins (Fig. 8C). To exclude the effect of the interaction sampling difference, we investigated which proteins were more likely to be essential within a given functional category. Thus, for each data set, we separated proteins in each MIPS category with degrees above and below the category average into two groups, and found that the above-average group (“Higher-connectivity proteins” in Fig. 8D) was significantly more enriched with essential proteins. This consensus observation demonstrated that when removing sampling biases, more connected components in a functional category tended to be essential.
CONCLUSIONS
Recent advances in high-throughput experimental techniques have identified large numbers of possible protein-protein interactions. Although the protein sets tested for interactions overlap, the actual detected protein interactions among these data sets do not overlap to any significant degree. Consequently, extracting and interpreting biological information from networks reconstructed from such data depend on the underlying detection methodology. Using protein-protein interaction networks in systems biology approaches to study phenotypic behavior requires that the networks contain the relevant proteins responsible for the underlying biological processes (17, 58). Herein, our systematic classification of interactions according to their functional characteristics and each technology's ability to detect these interactions, shed insight into the underlying biases of the content of these data sets. The comparison between three different high-confidence high-throughput data sets derived from different methodologies provided a quantitative measure of the functional biases of the retrieved interactions and proteins. This confirmed the unbiased nature of the Y2H data, preferential retrieval of co-complex interactions from AP/MS, and the ability of the PCA method to detect many transient interactions. However, we also noted the large extent of binary interactions present in the high-confidence AP/MS, and the presence of many binary interactions from complexes that were retrieved in the Y2H and PCA data sets. The high-confidence protein interaction data sets were associated with biologically conflicting properties, such as protein abundance and essentiality, biased by the underlying detection methodology. These biases determined, to large extent, the biological insights derived from each data set and were more influential than the topological properties of the network themselves. Although these biases could be removed for certain analyses, e.g. consistently relating protein connectivity with gene essentiality, much work remains in defining the properties and the range of suitable applications of the experimentally determined, partially complete interactome in systems biology studies.
A proper and consistent interpretation of biological properties derived from high-throughput protein interaction data sets requires careful consideration of potential methodological biases incorporated in the data. A facile way to quantify this is to compare the functional characteristics of the constituent proteins by comparing the relative distribution of these proteins among functional categories. Although analyses of data with the same protein characteristics should yield consistent insights, apparent contradictory results derived from different data sets may tell us more about the underlying experimental biases themselves than the underlying biology.
MATERIALS AND METHODS
Protein Interaction Data Sets
We investigated the following protein interaction data sets, comprising three high-confidence high-throughput data sets derived from AP/MS, PCA, and Y2H experiments, an unfiltered, raw interaction data set (denoted as raw-AP/MS), and a manually curated set focusing on binary protein interactions (BGS). Columns 1–3 of Table I summarize the number of proteins and interactions contained in the following five data sets:
AP/MS
We have previously developed and applied the IDBOS method to extract and analyze high-confidence protein interaction networks from each of three individual AP/MS data sets (14). It was determined that the data of Gavin et al. (6) showed the highest specificity of protein associations and no abundance bias, i.e. high-abundance proteins tend not to have more interactions, and for these reasons we opted to study this data set as the best representation of a high-confidence AP/MS data. This data set is also included in the supplemental material as an annotated, downloadable file.
PCA
The recent PCA strategy detects in vivo protein interactions via fusions to enzyme fragments that, when reconstituted, restores catalytic activity and, consequently, cell growth (59). This methodology does not depend upon the expression of a reporter protein as required in Y2H screens. The PCA technique was applied on a genome-wide scale for yeast and yielded many new, previously undiscovered protein interactions (8). Although we are aware of only one large-scale interaction data set determined using the PCA technique, the reported 98% positive predictive value of these interactions defines them as a high-confidence data set.
Y2H
The Y2H method was the first high-throughput technology to assess protein interactions on a genomic scale, and Yu et al. have consolidated three large-scale Y2H data sets (10, 11, 60) into one high-quality representative S. cerevisiae data set (11). This data set was representative of high-confidence Y2H interactions.
Raw-AP/MS
To illustrate the importance of using a high-confidence network, we also analyzed the raw interaction data of Gavin et al. (6), where we used the spoke model to construct a corresponding protein interaction set. In this model, we retained the bait-prey pairs from the purification and did not include all possible prey-prey pairs.
BGS
The BGS interaction data set is a manually curated set of high-confidence physical binary interactions that represent direct protein associations, rather than indirect ones (11). This interaction set has been shown to have considerable overlaps with high-throughput Y2H data sets (11).
High-Confidence AP/MS Protein Interaction Network
Our IDBOS procedure for AP/MS data can be summarized as follows (14): For a given affinity purification data set in which individual purifications are specified, we counted, for each unique protein pair i and j, the total number of times they co-occurred in the same purification oij. This analysis corresponds to a matrix enumeration of possible interacting protein pairs within each purification. We then constructed 106 randomized, or shuffled, purification sets and computed average shuffled co-occurrences, ōij, and associated standard deviations, σij, over these sets. A shuffled purification set was constructed by shuffling, or exchanging, pairs of prey proteins in the data set. The co-occurrence score (CSij) for each protein pair was then determined as the Z-score of the observed co-occurrences given by:
 |
In order to gauge the significance of these scores, we also constructed the scores associated with randomly shuffled pairs themselves. First, we constructed an additional 105 shuffled sets in the same manner as that described above. Second, for each shuffled set, we determined the Z-scores for protein pairs having a shuffled co-occurrence of greater than one as:
 |
where cijn (> 1) denote the co-occurrence of proteins i and j in the nth shuffled set, and ōij and σij denote the mean co-occurrences and standard deviations, respectively, determined from the shuffled sets as in Equation 1. We can then determine the CS-score cutoff that yields a particular false positive rate by comparing the normalized experimental (PE) and random (PR) score distributions generated from the data. For a given score threshold ζ, we computed the fractions of protein pairs in the commensurate random (fR) and experimental (fE) distributions that have a higher score than ζ as:
 |
and
 |
We then approximated the false positive rate (FPR) as the ratio of these fractions at a given score threshold ζ, as:
 |
For example, for a false positive rate of 5% we computed the corresponding score cutoff ζ0.05 for the high-confidence AP/MS data set to be 5.95. We compiled high-confidence data sets at different false positive rates by including only interactions having higher CS scores than their respective cutoffs.
Protein Annotation and Essentiality Data
The Munich Information Center for Protein Sequences (MIPS) protein data were downloaded from the MIPS database (ftp://ftpmips.gsf.de/yeast/catalogues/) (31). We used the top-level protein function (funcat/) and location (subcellcat/) annotation labels to characterize proteins. The function labels ranged from metabolism to cell differentiation, whereas the location labels ranged from extracellular to lipid particles. We only considered protein complexes (complexcat/) identified in small-scale experiments, by excluding complexes listed under category 550, labeled as “complexes by systematic analysis.” Essentiality data were merged from both MIPS (gene_disruption/) and the Saccharomyces Genome Database (http://www.yeastgenome.org/) (47), where a protein was considered essential if it were labeled so in at least one of the data sets.
Assignment of a Single Property to Multiply Annotated Proteins in Network Visualization
It is often the case that MIPS annotations assign more than one function and/or location to a protein. These “moonlighting” proteins presented no problem in our analyses; however, for the network visualization, it was convenient to select a unique annotation item for each protein from each MIPS annotation category. These were chosen in order to maximize the number of homogeneous interactions, i.e. those between proteins having the same annotation item. By assuming that each homogeneous interaction had energy –1 and others had energy 0, we transformed the problem into a typical energy-minimization one. The total system energy was minimized using a Metropolis Monte Carlo annealing algorithm on a random initial annotation configuration in which each moonlighting protein was randomly assigned one of its MIPS annotation labels. For a protein interaction network, we uniformly annealed the system from a temperature of 50 to 0.2 (for simplicity sake, both energy and temperature have the same unit) in 10,000 steps. A step consisted of a loop of operations on every moonlighting protein. In one operation, the current annotation label for the protein was temporally replaced by another, which was randomly selected from its set of MIPS annotation labels. The operation was accepted if the new system energy was lower, or accepted with a probability of exp(–ΔE/T) if the new system energy was higher, where ΔE denotes the energy change caused by the operation and T denotes temperature. The annotation configuration was updated when the operation was accepted and remained unchanged when the operation was rejected. The lowest-energy configuration in the simulation was chosen for the visualization of this data set.
Connection Z-Scores of Labels of Interacting Proteins
To gauge the statistical connections between the functional categories assigned to the constituent proteins in an interaction, we compared the originally annotated interactions with shuffled annotations. During a shuffle, all annotations assigned to a protein were kept as a single package. In a shuffle simulation, each annotated protein was randomly chosen to switch their annotation packages with another protein. For each high-confidence interaction data set, we carried out 10,000 simulations. The connection significance between annotation categories i and j was represented as a Z-score, as follows:
 |
where Cij denotes the number of interactions between a protein annotated with category i and another annotated with category j in the data set, C̄ij represents the average Cij over the 10,000 simulations, and σij is the corresponding standard deviation.
Acknowledgments
The opinions and assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the U.S. Army or the U.S. Department of Defense. This paper has been approved for public release with unlimited distribution.
Footnotes
* This work was supported by the Military Operational Medicine Research Program of the U.S. Army Medical Research and Materiel Command, Ft. Detrick, Maryland, as part of the U.S. Army's Network Science Initiative.
 This article contains supplemental Fig. S1 and Data S1 and S2.
This article contains supplemental Fig. S1 and Data S1 and S2.
1 The abbreviations used are:
- AP/MS
- affinity purification followed by mass spectroscopy
- BGS
- binary gold standard
- IDBOS
- interactions detected based on shuffling
- MIPS
- Munich Information Center for Protein Sequences
- PCA
- protein-fragment complementation assay
- Y2H
- yeast two-hybrid.
REFERENCES
- 1. Alberts B. (1998) The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92, 291–294 [DOI] [PubMed] [Google Scholar]
- 2. Gentleman R., Huber W. (2007) Making the most of high-throughput protein-interaction data. Genome Biol. 8, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hakes L., Pinney J. W., Robertson D. L., Lovell S. C. (2008) Protein-protein interaction networks and biology–what's the connection? Nat. Biotechnol. 26, 69–72 [DOI] [PubMed] [Google Scholar]
- 4. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 5. Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sørensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 6. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 7. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrin-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 8. Tarassov K., Messier V., Landry C. R., Radinovic S., Serna Molina M. M., Shames I., Malitskaya Y., Vogel J., Bussey H., Michnick S. W. (2008) An in vivo map of the yeast protein interactome. Science 320, 1465–1470 [DOI] [PubMed] [Google Scholar]
- 9. Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U.S.A. 98, 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Qureshi-Emili A., Li Y., Godwin B., Conover D., Kalbfleisch T., Vijayadamodar G., Yang M., Johnston M., Fields S., Rothberg J. M. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 [DOI] [PubMed] [Google Scholar]
- 11. Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J. F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A. S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabasi A. L., Tavernier J., Hill D. E., Vidal M. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. von Mering C., Krause R., Snel B., Cornell M., Oliver S. G., Fields S., Bork P. (2002) Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417, 399–403 [DOI] [PubMed] [Google Scholar]
- 13. Bader J. S., Chaudhuri A., Rothberg J. M., Chant J. (2004) Gaining confidence in high-throughput protein interaction networks. Nat. Biotechnol. 22, 78–85 [DOI] [PubMed] [Google Scholar]
- 14. Yu X., Ivanic J., Wallqvist A., Reifman J. (2009) A novel scoring approach for protein co-purification data reveals high interaction specificity. PLoS Comput. Biol. 5, e1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ivanic J., Wallqvist A., Reifman J. (2008) Probing the extent of randomness in protein interaction networks. PLoS Comput. Biol. 4, e1000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fields S. (2005) High-throughput two-hybrid analysis. The promise and the peril. FEBS J. 272, 5391–5399 [DOI] [PubMed] [Google Scholar]
- 17. Brückner A., Polge C., Lentze N., Auerbach D., Schlattner U. (2009) Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci. 10, 2763–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rajagopala S. V., Hughes K. T., Uetz P. (2009) Benchmarking yeast two-hybrid systems using the interactions of bacterial motility proteins. Proteomics 9, 5296–5302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wodak S. J., Pu S., Vlasblom J., Séraphin B. (2009) Challenges and rewards of interaction proteomics. Mol. Cell. Proteomics 8, 3–18 [DOI] [PubMed] [Google Scholar]
- 20. Collins S. R., Kemmeren P., Zhao X. C., Greenblatt J. F., Spencer F., Holstege F. C., Weissman J. S., Krogan N. J. (2007) Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell. Proteomics 6, 439–450 [DOI] [PubMed] [Google Scholar]
- 21. Hart G. T., Lee I., Marcotte E. R. (2007) A high-accuracy consensus map of yeast protein complexes reveals modular nature of gene essentiality. BMC Bioinformatics 8, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petschnigg J., Snider J., Stagljar I. (2011) Interactive proteomics research technologies: recent applications and advances. Curr. Opin. Biotechnol. 22, 50–58 [DOI] [PubMed] [Google Scholar]
- 23. Sen T. Z., Kloczkowski A., Jernigan R. L. (2006) Functional clustering of yeast proteins from the protein-protein interaction network. BMC Bioinformatics 7, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lubovac Z., Gamalielsson J., Olsson B. (2006) Combining functional and topological properties to identify core modules in protein interaction networks. Proteins 64, 948–959 [DOI] [PubMed] [Google Scholar]
- 25. Ulitsky I., Shamir R. (2007) Identification of functional modules using network topology and high-throughput data. BMC Syst. Biol. 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carbonell P., Nussinov R., del Sol A. (2009) Energetic determinants of protein binding specificity: insights into protein interaction networks. Proteomics 9, 1744–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson M. E., Hummer G. (2010) Nonspecific binding limits the number of proteins in a cell and shapes their interaction networks. Proc. Natl. Acad. Sci. U.S.A. 108, 603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shi Y. Y., Miller G. A., Qian H., Bomsztyk K. (2006) Free-energy distribution of binary protein-protein binding suggests cross-species interactome differences. Proc. Natl. Acad. Sci. U.S.A. 103, 11527–11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katebi A. R., Kloczkowski A., Jernigan R. L. (2010) Structural interpretation of protein-protein interaction network. BMC Struct. Biol. 10, Suppl 1, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Przytycka T. M., Singh M., Slonim D. K. (2010) Toward the dynamic interactome: it's about time. Brief Bioinform. 11, 15–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mewes H. W., Amid C., Arnold R., Frishman D., Göldener U., Mannhaupt G., Münsterkötter M., Pagel P., Strack N., Stümpflen V., Warfsmann J., Ruepp A. (2004) MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 32, D41–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mewes H. W., Ruepp A., Theis F., Rattei T., Walter M., Frishman D., Suhre K., Spannagl M., Mayer K. F., Stümpflen V., Antonov A. (2011) MIPS: curated databases and comprehensive secondary data resources in 2010. Nucleic Acids Res. 39, D220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamada T., Bork P. (2009) Evolution of biomolecular networks: lessons from metabolic and protein interactions. Nat. Rev. Mol. Cell Biol. 10, 791–803 [DOI] [PubMed] [Google Scholar]
- 34. Hart G. T., Ramani A. K., Marcotte E. M. (2006) How complete are current yeast and human protein-interaction networks? Genome Biol. 7, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sambourg L., Thierry-Mieg N. (2010) New insights into protein-protein interaction data lead to increased estimates of the S. cerevisiae interactome size. BMC Bioinformatics 11, 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Przulj N., Wigle D. A., Jurisica I. (2004) Functional topology in a network of protein interactions. Bioinformatics 20, 340–348 [DOI] [PubMed] [Google Scholar]
- 37. Güldener U., Münsterkötter M., Oesterheld M., Pagel P., Ruepp A., Mewes H. W., Stümpflen V. (2006) MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 34, D436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gupta S., Wallqvist A., Bondugula R., Ivanic J., Reifman J. (2010) Unraveling the conundrum of seemingly discordant protein-protein interaction datasets. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2010, 783–786 [DOI] [PubMed] [Google Scholar]
- 39. Mewes H. W., Amid C., Arnold R., Frishman D., Güldener U., Mannhaupt G., Münsterkötter M., Pagel P., Strack N., Stumpflen V., Warfsmann J., Ruepp A. (2004) MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 32, D41–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saha S., Kaur P., Ewing R. M. (2010) The bait compatibility index: computational bait selection for interaction proteomics experiments. J. Proteome Res 9, 4972–4981 [DOI] [PubMed] [Google Scholar]
- 41. Studitsky V. M., Walter W., Kireeva M., Kashlev M., Felsenfeld G. (2004) Chromatin remodeling by RNA polymerases. Trends Biochem. Sci 29, 127–135 [DOI] [PubMed] [Google Scholar]
- 42. Klopf E., Paskova L., Solé C., Mas G., Petryshyn A., Posas F., Wintersberger U., Ammerer G., Schüller C. (2009) Cooperation between the INO80 complex and histone chaperones determines adaptation of stress gene transcription in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 29, 4994–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee S. K., Fletcher A. G., Zhang L., Chen X., Fischbeck J. A., Stargell L. A. (2010) Activation of a poised RNAPII-dependent promoter requires both SAGA and mediator. Genetics 184, 659–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Doyon Y., Selleck W., Lane W. S., Tan S., Côté J. (2004) Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24, 1884–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brand M., Leurent C., Mallouh V., Tora L., Schultz P. (1999) Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286, 2151–2153 [DOI] [PubMed] [Google Scholar]
- 46. Qiu H., Hu C., Zhang F., Hwang G. J., Swanson M. J., Boonchird C., Hinnebusch A. G. (2005) Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 25, 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cherry J. M., Adler C., Ball C., Chervitz S. A., Dwight S. S., Hester E. T., Jia Y., Juvik G., Roe T., Schroeder M., Weng S., Botstein D. (1998) SGD: Saccharomyces Genome Database. Nucleic Acids Res. 26, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jansen R., Greenbaum D., Gerstein M. (2002) Relating whole-genome expression data with protein-protein interactions. Genome Res. 12, 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ivanic J., Yu X., Wallqvist A., Reifman J. (2009) Influence of protein abundance on high-throughput protein-protein interaction detection. PLoS One 4, e5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Newman J. R., Ghaemmaghami S., Ihmels J., Breslow D. K., Noble M., DeRisi J. L., Weissman J. S. (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441, 840–846 [DOI] [PubMed] [Google Scholar]
- 51. Jeong H., Mason S. P., Barabási A. L., Oltvai Z. N. (2001) Lethality and centrality in protein networks. Nature 411, 41–42 [DOI] [PubMed] [Google Scholar]
- 52. Batada N. N., Hurst L. D., Tyers M. (2006) Evolutionary and physiological importance of hub proteins. PLoS Comput. Biol. 2, e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hahn M. W., Kern A. D. (2005) Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol. Biol. Evol. 22, 803–806 [DOI] [PubMed] [Google Scholar]
- 54. Albert R., Jeong H., Barabasi A. L. (2000) Error and attack tolerance of complex networks. Nature 406, 378–382 [DOI] [PubMed] [Google Scholar]
- 55. He X., Zhang J. (2006) Why do hubs tend to be essential in protein networks? PLoS Genet. 2, e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zotenko E., Mestre J., O'Leary D. P., Przytycka T. M. (2008) Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput. Biol. 4, e1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang H., Kakaradov B., Collins S. R., Karotki L., Fiedler D., Shales M., Shokat K. M., Walther T. C., Krogan N. J., Koller D. (2009) A complex-based reconstruction of the Saccharomyces cerevisiae interactome. Mol. Cell. Proteomics 8, 1361–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stelzl U., Wanker E. E. (2006) The value of high quality protein-protein interaction networks for systems biology. Curr. Opin. Chem. Biol. 10, 551–558 [DOI] [PubMed] [Google Scholar]
- 59. Michnick S. W., Ear P. H., Manderson E. N., Remy I., Stefan E. (2007) Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat. Rev. Drug Discov. 6, 569–582 [DOI] [PubMed] [Google Scholar]
- 60. Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U.S.A. 98, 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]