Abstract
Protein post-translational modifications (PTMs) at the lysine residue, such as lysine methylation, acetylation, and ubiquitination, are diverse, abundant, and dynamic. They play a key role in the regulation of diverse cellular physiology. Here we report discovery of a new type of lysine PTM, lysine malonylation (Kmal). Kmal was initially detected by mass spectrometry and protein sequence-database searching. The modification was comprehensively validated by Western blot, tandem MS, and high-performance liquid chromatography of synthetic peptides, isotopic labeling, and identification of multiple Kmal substrate proteins. Kmal is a dynamic and evolutionarily conserved PTM observed in mammalian cells and bacterial cells. In addition, we demonstrate that Sirt5, a member of the class III lysine deacetylases, can catalyze lysine demalonylation and lysine desuccinylation reactions both in vitro and in vivo. This result suggests the possibility of nondeacetylation activity of other class III lysine deacetylases, especially those without obvious acetylation protein substrates. Our results therefore reveal a new type of PTM pathway and identify the first enzyme that can regulate lysine malonylation and lysine succinylation status.
Cellular function and physiology are largely determined by the inventory of all proteins in a cell, its proteome. The collection and characterization of the proteome is critical to understanding cellular mechanisms and diseases. Proteomes in eukaryotic cells consist of over a million molecular species of proteins, easily orders of magnitude more complex than the corresponding genomes (1, 2). There are two major mechanisms for expanding the coding capacity of the human genome: mRNA splicing and protein post-translational modifications (PTMs)1.
PTMs (more than 300 types) are complex and fundamental mechanisms of cellular regulation, and have been associated with almost all known cellular pathways and disease processes (1, 2). As an example, protein phosphorylation, the most well-studied PTM, is present in more than one third of human proteins, the phosphorylation status of which can potentially be regulated by ∼500 human protein kinases and ∼150 phosphatases (3, 4). The modification mainly occurs at several amino acid residues: serine, threonine, tyrosine, and histidine. Protein phosphorylation makes its substrate residues more acidic, hydrophilic, and induces a charge change from +1 charge to –1 (at physiological pH), which in turn modulates the structure and functions of substrate proteins.
The high complexity of PTMs is also reflected by diverse modifications at ε-amine group of lysine residue, including methylation, acetylation, and ubiquitination. These lysine PTMs have been shown to play an important role in cellular regulations (5, 6). Recently, we identified a new type of PTM at lysine residues, lysine succinylation (7). Like phosporylation, lysine succinylation also induces a change of two negative charges in lysine residue, from a +1 charge to a –1 charge at physiological pH. This PTM is dynamic, abundant, and evolutionarily conserved from bacteria to mammalian cells. We anticipate that there will be more than a thousand lysine succinylated substrates in mammalian cells.
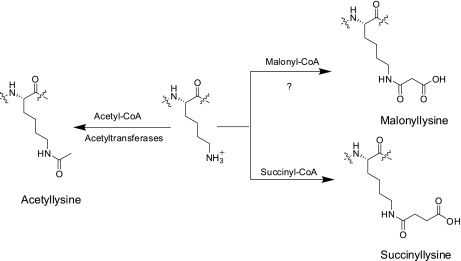
Here we report the identification and verification of a new type of PTM, lysine malonylation (Fig. 1). We extensively confirmed this new modification using a variety of chemical and biochemical methods. We showed that Sirt5 can catalyze lysine demalonylation and desuccinylation reactions, in peptide and protein substrates, both in vivo and in vitro. Taken together, we conclusively establish lysine malonylation as a new type of evolutionarily conserved PTM pathway and Sirt5 is a regulatory enzyme for lysine malonylation and lysine succinylation.
Fig. 1.
Chemical structures of lysine, acetyllysine, succinyllisine, malonyllysine, and the candidate CoA for lysine malonylation.
EXPERIMENTAL PROCEDURES
Materials
Peptides were synthesized through custom synthesis using racemic or enantiomeric protected amino acid residues. The synthesis of the protected amino acid residues was described in detail in supplementary Methods, including Fmoc-Lys(mono-tert-butyl malonate)-OH, Fmoc-Lys(succinyl)-OH. Ni-NTA agarose beads were purchased from Qiagen (Valencia, CA); modified sequencing-grade trypsin from Promega (Madison, WI); C18 ZipTips from Millipore Corporation (Billerica, MA); water and acetonitrile from Burdick and Jackson (Meskegon, MI); protein A beads from GE Healthcare; isotopic [1, 2, 3-13C] sodium malonate was purchased from Cambridge Isotope Laboratories (Andover, MA). The recombinant HDACs except Sirt5 were purchased from BPS Bioscience (San Diego, CA). Wt Sirt5 and its inactive mutant proteins were expressed and purified in house. Experimental procedure for expression and purification of Sirt5 were included in the supplementary Methods. Expression plasmids for Sirt5 wt and its inactive mutant (H158Y) were constructed by Wei Gu (Columbia University) and Eric Verdin (UCSF), respectively. Other chemicals were obtained from the following suppliers: Sigma-Aldrich: trifluoroacetic acid (99%), formic acid (>98%), NH4HCO3 (>99%), trichloroacetic acid (6.1 N), iodoacetamide, dithothreitol, bovine serum albumin (BSA), sodium dodecyl sulfate, Tris-Cl, MgCl2 (ACS grade), NaCl (ACS grade), KCl (ACS grade), β-nicotinamide adenine dinucleotide hydrate (NAD+), nicotinamide (ACS grade), trichostatin A (TSA), sirtinol (ACS grade); Fisher: NaHCO3 (ACS grade), NaOH (ACS grade), CH3CN (HPLC grade), HCl solution (37.3%), glycine (ACS grade), MeOH (ACS grade), acetone (ACS grade), DMEM medium and hydrogen peroxide (ACS grade).
Chemical Synthesis, Peptide Synthesis, and Anti-Kmal Antibody
Chemical synthesis of Fmoc-Lys (mono-tert-butyl malonate)-OH and fluorescent compounds for assaying lysine demalonylation and lysine desuccinylation activities were included in the supplementary Methods. The peptides were synthesized by commercial custom synthesis using Fmoc-Lys (mono-tert-butyl malonate)-OH. Generation of anti-Kmal antibody was described in supplementary Methods.
Western Blot and Dot Blot Analysis
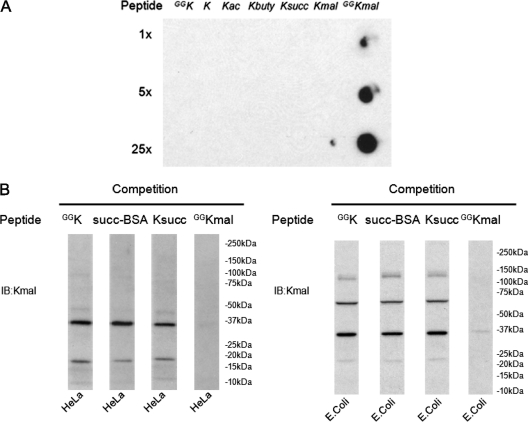
The specificity of the sequence-specific antibody was demonstrated by dot-blot assay (Fig. 2A). To carry out Western blot, 40 μg of each cell lysate was resolved in gradient SDS-PAGE gel, transferred to polyvinylidene difluoride (PVDF) membrane; to carry out dot-blot assay, a gradient amount of peptide was spotted to the nitrocellulose (NC) membrane. Then the membrane was blocked in TBST buffer (1x TBS containing 0.05% Tween 20) with 5% BSA and probed with appropriate primary antibodies and secondary antibodies coupled to horseradish peroxidase. The competition experiments were performed by incubating the primary antibodies with a synthetic peptide bearing either an unmodified lysine or a modified lysine residue. Signals were revealed by ECL Western blot detection system.
Fig. 2.
Detection of lysine malonylation by Western blot. A, Specificity of the anti-Kmal antibody. Dot-blot assay was carried out using anti-Kmal antibody by incubation with peptide libraries bearing a fixed unmodified K, Kac, Kbuty, Ksucc, Kmal, two glycine-rich peptides (CEGGGGKmalGGSG) bearing a unmodified K (GGK), and a malonylated K (GGKmal), respectively. Each peptide library contains 13 residues CXXXXXKXXXXXX, where X is a mixture of 19 amino acids (excluding cysteine), C is cysteine, and the seventh residue is a fixed lysine residue: unmodified lysine (K), acetyllysine (Kac), butyryllysine (Kbuty), succinyllysine (Ksucc), malonyllysine (Kmal). GGK indicates a glycine-rich peptide, CEGGGGKGGSG, whereas GGKmal is a malonylated glycine-rich peptide, CEGGGGKmalGGSG, in which the lysine residue is malonylated. B, Western blot of the whole cell lysates from HeLa cell and E. coli. Western blot was performed using the whole cell lysates from HeLa cell and E. coli with competition of modified CEGGGGKmalGGSG, unmodified CEGGGGKGGSG, succinylated BSA, or peptide library bearing a succinylated lysine(Ksucc).
Isotopic Labeling of Kmal Using [1, 2, 3-13C] malonate
HeLa cells were treated with isotopic sodium [1, 2, 3-13C] malonate (20 mm in DMEM medium, pH 7.5) for 24 h. The cells were harvested and washed twice with ice-cold phosphate-buffered saline (PBS) and then lysed with SDS sample buffer. The protein lysate was used either for Western blot analysis or for identifying Kmal peptides by immunoprecipitation using anti-Kmal antibody and mass spectrometry.
Protein Precipitation and In-Solution Tryptic Digestion
The protein lysate of interest was precipitated by conventional trichloroacetic acid method. The protein precipitate was digested by trypsin using a procedure we described previously (8). The protein precipitate was suspended in 100 mm NH4HCO3 (pH 8.0) and dissolved by sonication. Trypsin was added to the mixture at an enzyme-to-substrate ratio of 1:50 (w/w) and tryptic digestion was carried out at 37 °C for 16 h. The tryptic peptides were reduced by 5 mm dithothreitol and alkylated by 15 mm iodoacetamide. The mixture was digested again by adding an additional trypsin (1:100 w/w) and incubated at 37 °C for 3 h.
Affinity Enrichment and high-performance Liquid Chromatography (HPLC)/tandem MS (MS/MS) Analysis of Kmal Peptides
The Kmal peptides were enriched from proteolytic digests of protein cell lysate using a procedure described previously (8). Briefly, the tryptic peptides obtained above were resolubilized in NETN buffer (50 mm Tris-Cl, pH 8.0, 0.5% Nonidet P-40, 100 mm NaCl, 1 mm EDTA). Insoluble particles were removed by centrifugation at 50,000 × g for 10 min. Affinity purification was carried out by incubating the peptides with 10 μl of anti-Kmal antibody protein A immobilized agarose beads at room temperature for 4 h with gentle shaking. The beads were washed three times with NETN buffer and twice with ETN buffer (50 mm Tris-Cl, pH 8.0, 100 mm NaCl, 1 mm EDTA). The bound peptides were eluted from the beads by washing three times with 0.1% TFA. The isolated Kmal peptides were dried in SpeedVac. The resulting peptide sample was dissolved in HPLC buffer A (0.1% formic acid in water, v/v), desalted with C18 ZipTips and loaded onto an in-house packed reverse-phase C18 column (360 μm OD × 75 μm ID) connected to an Eksigent NanoLC-1D plus HPLC system (Eksigent Technologies). Peptide samples were analyzed with a 2-hr HPLC-gradient from 5 to 90% HPLC buffer B (0.1% formic acid in acetonitrile) at a flow rate of 200 nL/min. The eluted peptides were ionized and introduced into a LTQ Velos Orbitrap mass spectrometer using a nanospray source. Survey full-scan MS spectra (from m/z 350–2000) were acquired in the Orbitrap with resolution r = 30,000 at m/z 400. The twenty most intense ions were sequentially isolated in the linear ion trap and subjected to collision-activated dissociation with a normalized energy of 35%. The exclusion duration for the data-dependant scan was 36 s, the repeat count was 2, and the exclusion window was set at +2 Da and −1 Da. Charge state screening was enabled and singly charged ions are excluded for fragmentation. AGC settings were 1E6 for full scan Orbitrap analysis, 1E4 for MSn scan in ion trap and 6E4 for MSn scan in Orbitrap. Signal threshold for CID acquisition was set at 5000.
Data Interpretation
The acquired MS/MS spectra was analyzed by Mascot sequence alignment algorithm (version 2.1, Matrix Science, 9). Mass tolerances for precursor ions were set at 0.01 Da and for MS/MS were set at ±0.5 Da. Peak lists were generated by extract_msn.exe software (version 5.0, Thermo Scientific). All the data were searched against either International Protein Index human protein database (version 3.70, 87069 sequences) or NCBI RefSeq E. coli k12 protein database (4123 sequences). Cysteine alkylation by iodoacetamide, methionine oxidation, and lysine malonylation (lysine +86.00039 Da) were specified as variable modifications. Maximum of three missing cleavages were specified. All the results with Mascot score above 30 and E-value below 1 were manually verified to have high peptide-spectrum match quality and satisfy characteristic neutral-loss (–44Da) fragmentation pattern.
Verification of Kmal Peptides by HPLC/MS/MS
The in vivo peptide, the synthetic counterpart, and their mixture for each Kmal peptide were subjected to nano-HPLC/MS/MS analysis. Extensive washing was carried out to avoid sample carry over before each sample injection. Full-MS scans were acquired with resolution r = 30,000 at m/z 400, and targeted CID MS/MS spectra were acquired at a resolution of 7500 at m/z 400 in Orbitrap. The amounts of synthetic peptides were adjusted so that the intensities of the in vivo peptide were only within a few-fold difference from its synthetic counterpart.
In Vitro Demalonylation and Desuccinylation Fluorescence Assays
The reactions were performed in a final volume of 50 μl per well in a 96-well microplate. Briefly, one μl of Boc-Lys(Mal)-AMC or Boc-Lys(Succ)-AMC stocking solution (10 mm) and 200 ng HDACs (1∼11) were added to the reaction buffer (25 mm Tris-Cl, 130 mm NaCl, 3.0 mm KCl, 1 mm MgCl2, 0.1% PEG8000, pH = 8.0) and incubated at 37 °C for 2 h. For assaying activities of Sirtuins, 2 μg protein of interest and 1 μl of the NAD+ stocking solution (25 mm) were added to the reaction buffer. The reaction was then stopped by adding 25 μl of trypsin solution (25 mm Tris-Cl,130 mm NaCl, 3.0 mm KCl, 1 mm MgCl2, 30% isopropanol, 0.01 mg/ml trypsin, pH = 8.0). The resulting solvent was mixed well and incubated at 37 °C for another 2 h or appropriate time. The fluorescence was measured on a fluorescence plate reader (The Wallac 1420 Work station, IET Ltd, Vernon Hills, IL) with excitation set at 355 nm and emission measure at 460 nm.
RESULTS
We recently identified lysine succinylation as a new type of PTM. Ksucc is an abundant, evolutionarily conserved, and dynamic PTM (7). When we carried out proteomic screening of Ksucc substrates by affinity enrichment using an anti-Ksucc antibody, HPLC/MS/MS, and protein sequence-alignment, we detected the possible presence of a malonylated peptide from a low-quality MS/MS spectrum. To directly test for Kmal peptides, we generated an anti-Kmal antibody, which was shown to have high specificity against lysine malonylation. Anti-Kmal antibody detected its antigen peptide (CEGGGGKmalGGSG) and peptide library bearing a fixed malonylated lysine(Kmal) but not peptide libraries bearing a fixed unmodified K, acetylated lysine, butyrylated lysine, succinylated lysine (Ksucc), or a glycine-rich peptide bearing an unmodified K (Fig. 2A).
Detection of Kmal in E. coli and HeLa Cells
To test whether Kmal exists in cells, we performed Western blot analysis using anti-Kmal antibody on the whole cell lysates from E. coli and HeLa cells. Multiple bands were detected that could be efficiently competed away by its antigen, a peptide with a Kmal residue, but not the corresponding unmodified peptide, succinylated BSA, or peptide library bearing a fixed succinylated lysine (Fig. 2B, supplemental Fig. S1). When exposed longer, multiple weaker bands can be detected with the antibody (data not shown). These results suggest that the Kmal PTM is present in both mammalian and bacterial cells.
Identification of Kmal Substrates by HPLC/MS/MS
To identify potential Kmal substrates and malonylation sites in proteins, we used a method previously described for the proteomics of lysine acetylation (8). In this experiment, we used anti-Kmal antibody to enrich Kmal peptides from a tryptic digest of HeLa whole-cell lysate and E. coli whole-cell lysate. The resulting Kmal peptides were analyzed by HPLC/MS/MS. The generated MS/MS spectra were analyzed by protein sequence alignment to identify the possible Kmal peptides. A careful manual verification was used to ensure correct peptide identification using a procedure we previously described (10). All the MS/MS spectra are included in supplementary Information (supplementary Data Set S1, S2). This study led to the identification of 25 peptides from 17 proteins (Table I) of HeLa cell and three peptides from three proteins of E. coli (supplemental Table S1) that each contains a mass shift of 86.00 Da. And up to 30% of the identified peptides are malonylated in HeLa cells.
Table I. List of all the Kmal peptides identified by MS from HeLa cell.
| Protein | Mascot scorea | Peptide sequence | m/z | z |
|---|---|---|---|---|
| GAPDH | 47 | TVDGPSGKLWR | 651.3270 | 2+ |
| 46 | GALQNIIPASTGAAKAVGK | 927.0113 | 2+ | |
| SLC25A5 | 53 | YKQIFLGGVDK | 677.3573 | 2+ |
| 58 | YFPTQALNFAFKDK | 888.4359 | 2+ | |
| 57 | DFLAGGVAAAISKTAVAPIER | 1072.0756 | 2+ | |
| 36 | LAADVGKAGAER | 622.3176 | 2+ | |
| NSUN2 | 59 | VINTGIKVWCR | 716.3767 | 2+ |
| ACTA1 | 39 | DSYVGDEAQSKR | 720.8150 | 2+ |
| PGK1 | 38 | FHVEEEGKGK | 623.2904 | 2+ |
| ENO1 | 51 | IEEELGSKAK | 595.2995 | 2+ |
| 41 | TAIGKAGYTDK | 605.8017 | 2+ | |
| MDH2 | 37 | NLGIGKVSSFEEK | 747.3761 | 2+ |
| DBI | 47 | AKWDAWNELK | 673.8235 | 2+ |
| GPI | 51 | ELQAAGKSPEDLER | 814.8927 | 2+ |
| HSPA9 | 83 | QATKDAGQISGLNVLR | 878.9620 | 2+ |
| ALDOA | 56 | ALQASALKAWGGK | 698.8729 | 2+ |
| 51 | VDKGVVPLAGTNGETTTQGLDGLSER | 900.7822 | 3+ | |
| HSPD1 | 45 | ISKGANPVEIR | 635.3432 | 2+ |
| TUT1 | 35 | AFNQGKIFK | 569.7979 | 2+ |
| HSPE1 | 71 | GKGGEIQPVSVK | 642.8438 | 2+ |
| 51 | SQGKVLQATVVAVGSGSK | 901.4864 | 2+ | |
| 49 | VLQATVVAVGSGSKGK | 793.9456 | 2+ | |
| HSP90AB1 | 46 | EMLQQSKILK | 652.3508 | 2+ |
| EPRS | 52 | AYVDDTPAEQMKAER | 905.4015 | 2+ |
| KRT8 | 43 | TLNNKFASFIDK | 742.3757 | 2+ |
a All the results were manually verified; Mascot cut-off score = 30.
Taken together, the results of Western blot analysis, competition assay, and proteomics screening using HPLC/MS/MS suggest the existence of Kmal in both mammalian and bacterial cells.
Verification of Kmal Using Synthetic Peptides
Identical peptides will exhibit the same MS/MS patterns and same HPLC retention times in HPLC/MS/MS analysis. Accordingly, the analysis of corresponding synthetic peptides built with the PTM of interest is the best method to validate that a natural peptide bears a new PTM.
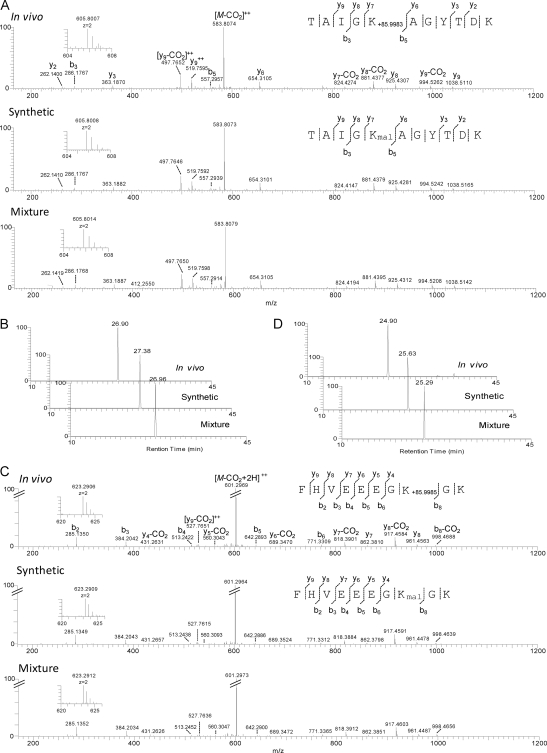
To verify the Kmal peptides identified in Table I, we randomly selected five Kmal peptides for chemical synthesis. The resulting peptides were pair-wise analyzed using HPLC/MS/MS between the in vivo peptide and its synthetic counterpart. As an example, the MS/MS spectrum of a peptide isolated from HeLa cell enolase (TAIGK+85.9988DaAGYTDK) has the same MS/MS spectrum as that of a synthetic peptide with a malonyl group (TAIGKmalAGYTDK), and a mixture of the two peptides (Fig. 3A). In addition, the in vivo peptide co-elutes with the synthetic peptide in HPLC (Fig. 3B). These two lines of evidence confirmed that the mass shift of 85.9988 Da in the peptide, TAIGK+85.9988DaAGYTDK, is caused by lysine malonylation.
Fig. 3.
Verification of Kmal peptides by HPLC/MS/MS of synthetic peptides. A, High-resolution CID MS/MS spectra of an α-enolase tryptic peptide (TAIGK+85.9983Da AGYTDK) with a mass shift of + 85.9983 Da (top), the synthetic TAIGKmalAGYTDK (middle), and their mixture (bottom). Inset shows the mass-to-charge ratios (m/z) of the precursor ions. A neutral loss of CO2 (44 Da) indicated decarboxylation. B, Extracted ion chromatograms (XICs) of the in vivo-derived peptide (top), its synthetic counterpart (middle), and their mixture (bottom). C, High-resolution CID MS/MS spectra of a PGK1 tryptic peptide (FHVEEEGK+85.9985DaGK) with a mass shift of + 85.9985 Da (top), the synthetic FHVEEEGKmalGK (middle), and their mixture (bottom). Inset shows m/z of precursor ions. D, XICs of the in vivo-derived peptide (top), its synthetic counterpart (middle) and their mixture (bottom).
Likewise, we also confirmed lysine malonylation in other four peptides by HPLC and MS/MS analysis using their corresponding synthetic peptides (Figs. 3C, 3D, and supplemental Figs. S2–S4).
Validation of Kmal Using Isotopic Labeling
Acetate and succinate can be converted into acetyl CoA and succinyl CoA, which can be used in turn by cells for lysine acetylation and succinylation reactions, respectively (7, 11). We argue that malonate may also be used by cells in a similar fashion as malonyl-CoA synthetase was reported previously (12, 13).
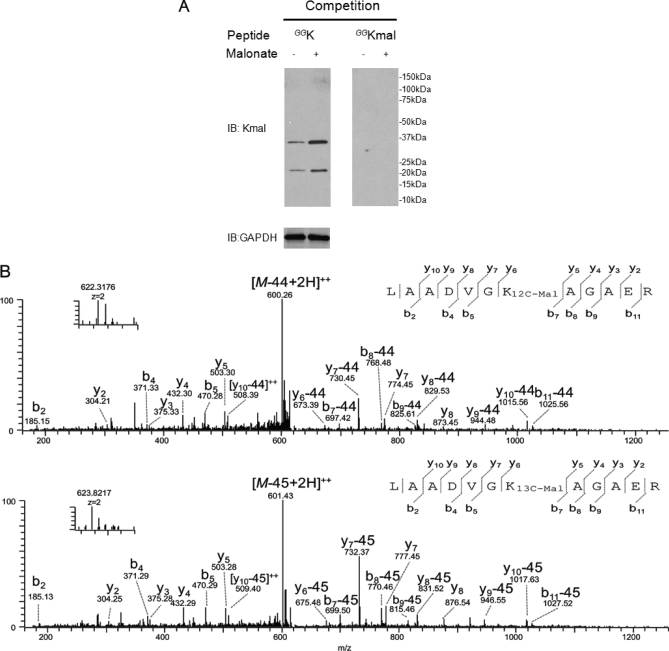
To test this possibility, we first fed HeLa cells with 20 mm malonate for 24 h. Lysine malonylation status was then evaluated in HeLa whole-cell lysate by Western blot analysis with our anti-Kmal antibody. Our result showed that lysine malonylation was enhanced by treatment with sodium malonate (Fig. 4A).
Fig. 4.
Isotopic labeling of Kmal peptides. A, Western blot of whole cell lysates from HeLa cells treated with (+) or without (-) malonate. Western blot was performed with competition of either modified CEGGGGKmalGGSG or unmodified CEGGGGKGGSG. B, CID MS/MS spectra of a 12C-Kmal peptide, LAADVGK12C-malAGAER (top). The neutral loss of the carbon dioxide (CO2) is designated as “-44”; MS/MS spectra (bottom) of a K13C-mal peptide, LAADVGK13C-malAGAER, from HeLa cells treated with [1, 2, 3-13C] malonate. The neutral loss of the carbon-13 dioxide (13CO2) is designated as “–45”.
To further examine if malonate can be converted to a high energy molecule such as malonyl CoA, that is in turn used by cells for lysine malonylation, we labeled HeLa cells with 20 mm isotopic sodium [1, 2, 3-13C]-malonate for 24 h. We extracted protein from whole-cell lysate that was then digested with trypsin. The resulting tryptic peptides were subjected to affinity enrichment using the anti-Kmal antibody, and enriched peptides were analyzed by HPLC/MS/MS and peptide sequence alignment for peptide identification. The isotopically labeled Kmal peptides can easily be identified by an additional 3.0 Da-mass shift over the lysine malonylation. This study led to the identification of Kmal-peptides that were labeled with [1, 2, 3-13C]-malonyl (Fig. 4B and supplementary Data Set S3). These results implied that malonate can be converted to an activated intermediate, which could be in turn used by cells as a malonyl donor for lysine malonylation reaction.
A Loss of CO2 is a Unique Feature in MS/MS Spectra of Kmal Peptides
Peptides bearing a PTM may have a unique mass signature in MS/MS spectra that is dependent on the PTM structure. For example, a phosphopeptide bearing a phosphorylation site in either a serine or threonine residue tends to lose the phosphate group, leading to a –98 Da satellite peak in MS/MS spectra (14, 15). This is mainly caused by fragmentation of the fragile phosphate bond in either the ionization source or mass analyzer during HPLC/MS/MS analysis. Likewise, the O-GlcNAc group is also very easily lost in a mass spectrometer, leading to a satellite peak with a neutral loss of 203 Da (16, 17).
When we analyzed MS/MS of Kmal peptides, we found that almost all the peptide fragment ions in MS/MS spectra of synthetic peptides bearing a Kmal residue have a satellite peak with a mass loss of 44 Da, which corresponds to the loss of CO2. We believe that this is caused by thermal decarboxylation, a process that readily occurs among derivatives of malonic acid (18). During this reaction, the malonylated lysine side chain first forms the enol intermediate through a cyclic transition state, which quickly tautomerizes to a carbonyl group (supplemental Fig. S5). This unique mass signature can be used for confirming the identification of peptides bearing a Kmal site.
Identification of Sirt5 as a Lysine Demalonylation and Desuccinylation Enzyme In Vitro
The status of cellular lysine acetylation is tightly regulated by two groups of enzymes with opposing enzymatic activities, lysine acetyltransferases (KATs) and lysine deacetylases (HDACs). Because not all of the HDACs have deacetylase activities, we hypothesize that some HDACs may be involved in lysine demalonylation.
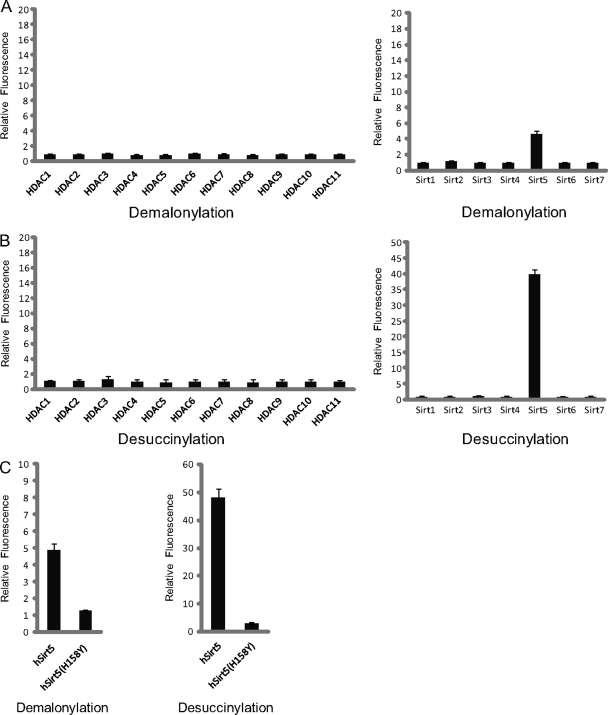
To test this hypothesis, we developed a fluorescence-based assay to measure the lysine demalonylation activity of all the HDACs. In this assay, a synthetic HDAC-mimic substrate was incubated with each of the 18 HDACs. A HDAC with lysine demalonylation activity would remove the malonyl group from the modified lysine residue, creating a proteolytic digestion site for trypsin. After a subsequent tryptic digestion, demalonylated substrates will generate an intense fluorescence signal. Using this assay, we tested the lysine demalonylation activity of all 18 HDACs (including HDAC 1–11 and Sirtuin 1–7). This assay showed that Sirt5 exhibits significant lysine demalonylation activity, and that its inactive mutant did not have significant demalonylation activity (Fig. 5A).
Fig. 5.
Sirt5 is an in vitro enzyme of lysine demalonylation and lysine desuccinylation. Fluorometric assay to detect in vitro lysine demalonylation activities (A) and lysine desuccinylation activities (B) among HDACs. C, Fluorometric assay to compare demalonylation and desuccinylation activities between wt Sirt5 and its inactive mutant.
Detection of lysine demalonylation activity of Sirt5 motivated us to evaluate HDAC family member's activities toward lysine succinylation, a PTM we previously discovered (7). Interestingly, Sirt5 is again the only HDAC that exhibits strong lysine desuccinylation activity in this assay (Fig. 5B). In contrast to lysine demalonylation and lysine desuccinylation activities, Sirt5 showed negligible activity toward lysine deacetylation in this assay, which is very different from high deacetylation activities of HDAC1∼3, 6, Sirt1∼3 (supplemental Fig. S7).
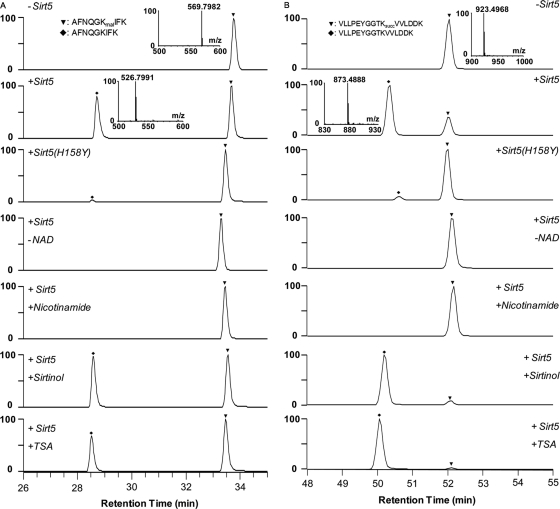
To further confirm the enzymatic activities of Sirt5 in vitro, we carried out a lysine demalonylation reaction using five synthetic peptides bearing Kmal residues (DSYVGDEAQSKmalR, IEEELGSKmalAK, TAIGKmalAGYTDK, AFNQGKmalIFK, AYVDDTPAEQMKmalAER). We used mass spectrometry to examine the demalonylation products of this reaction. As an example, the molecular weight of a synthetic peptide, AFNQGKmalIFK, from TUT1 is 905.4039Da (Fig. 6A). Upon incubation with Sirt5, an additional HPLC peak with a mass loss of ∼86 Da (with a loss of –43 m/z for a doubly charged ion) was detected in the presence of NAD+, corresponding to peptide that has lost a malonyl group. In contrast, the demalonylated peptide was not detected in a mock reaction and in a reaction using a Sirt5 inactive mutant or the same peptide bearing an acetylated lysine (supplemental Fig. S8A). The demalonylation reaction requires NAD+ and can be inhibited by nicotinamide (a class III HDAC inhibitor), but not trichostatin A (TSA, an inhibitor for class I/II HDACs) and sirtinol (an inhibitor of yeast Sir2 and human Sirt1) (Fig. 6A). This result indicates that Sirt5 can catalyze lysine demalonylation in a Kmal peptide in vitro and NAD+ is the cofactor of this reaction. Likewise, we also show that Sirt5 can catalyze a lysine demalonylation reaction in vitro for other four lysine malonylated substrates (supplemental Fig. S9).
Fig. 6.
Lysine demalonylation and desuccinylation activities of Sirt5 in peptide substrates. A, LC/MS detection of in vitro demalonylated peptide. A synthetic peptide, AFNQGKmalIFK (HRMS, m/z, 569.7982), from TUT1 was used as the substrate. B, LC/MS detection of an in vitro desuccinylated peptide. A synthetic peptide, VLLPEYGGTKsuccVVLDDK (HRMS, m/z 923.4968), from HSPE1 was used as substrate. The assays were carried out without Sirt5, with wt Sirt5, with Sirt5 inactive mutant (H158Y), without NAD+, with Sirt5 in the presence of nicotinamide (40 mm), sirtinol (200 μm), or TSA (2 μm). A triangle and a diamond indicate modified and unmodified peptides, respectively.
To further examine the desuccinylation activity of Sirt5 in vitro, we also carried out lysine desuccinylation reactions using three synthetic peptides bearing Ksucc residues (VLLPEYGGTKsuccVVLDDK, SQGKsuccVLQATVV, ETGVDLTKsuccDNMALQR). As an example, the molecular weight of a synthetic peptide, VLLPEYGGTKsuccVVLDDK, from HSPE1 is 1845.9879 Da (Fig. 6B). Upon incubation with Sirt5, an additional HPLC peak with a mass loss of ∼100 Da (with a loss of –50 m/z for a doubly charged ion) was detected in the presence of NAD+, corresponding to the peptide that has lost a succinyl group. In contrast, the desuccinylated peptide was not detected in a mock reaction and in a reaction using a Sirt5 enzyme-dead mutant. Like lysine demalonylation reaction, desuccinylation activity of Sirt5 requires NAD+ and can be inhibited by nicotinamide, but not TSA or sirtinol. Likewise, we also showed that Sirt5 can catalyze lysine desuccinylation reactions in vitro for other two lysine succinylated substrates (supplemental Fig. S10), but cannot for a lysine deacetylation reaction for the corresponding acetyled peptides (supplemental Fig. S11).
Sirt5 Can Catalyze Protein Lysine Demalonylation and Desuccinylation Both In Vitro and In Vivo
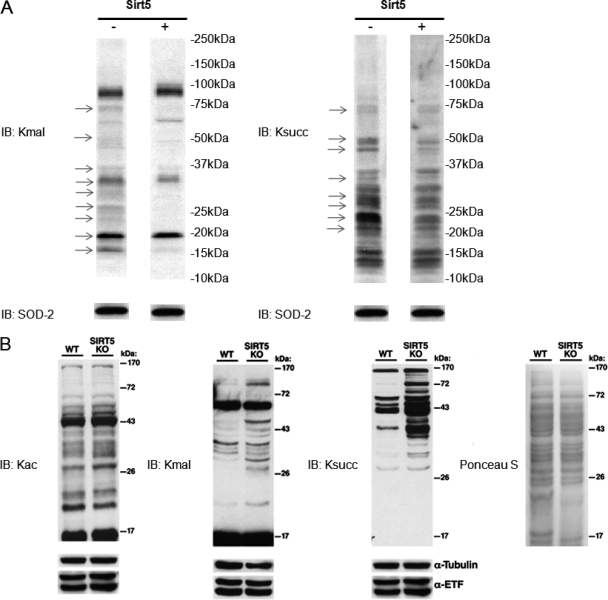
To examine the lysine demalonylation and lysine desuccinylation activities of Sirt5 in proteins, we resolved mitochondrial protein lysate in SDS-PAGE gel and transferred the proteins from the gel to the PVDF membrane. We then incubated the recombinant Sirt5 with the PVDF membrane (Fig. 7A). Western blot experiment showed that lysine malonylation and lysine succinylation levels are decreased in multiple protein bands. This result indicated that Sirt5 can catalyze the lysine demalonylation and lysine succinylation in vitro.
Fig. 7.
Sirt5 can catalyze protein lysine demalonylation and desuccinylation reactions both in vitro and in vivo. A, Sirt5 can catalyze in vitro demalonylation and desuccinylation reactions in mitochondrial proteins. Thirty micrograms of HeLa mitochondrial proteins were resolved by SDS-PAGE gel and transferred to the PVDF membranes. The membranes were incubated with or without 80 μg of Sirt5 in 2 ml of SDAC buffer (25 mm Tris·Cl, 130 mm NaCl, 3.0 mm KCl, 1 mm MgCl2, 1 mm DTT, 0.1% PEG8000, pH = 8.0) containing 0.5 mm NAD+ at 37 °C overnight. Then the membranes were blocked and probed with anti-Kmal antibody and anti-Ksucc antibody, respectively. Western blot for SOD-2 was used as a loading control. The arrows indicate the protein bands whose lysine malonylation or lysine succinylation are changed in response to Sirt5. B, Mice lacking SIRT5 show increased global lysine malonylation and succinylation, but no change in protein acetylation. Livers from a Sirt5 −/− (KO) mouse and its wild type littermate control were homogenized and examined for global lysine malonylation, succinylation, and acetylation by Western blot. Ponceau salt staining as well as Western blot for α-tubulin and ETF (mitochondrial electron transfer flavoprotein) were used as loading controls.
To corroborate the result of the in vitro experiment, we examined the lysine malonylation and lysine succinylation status in HepG2 cells, in which Sirt5 was overexpressed or knockdown by siRNA. These experiments demonstrated that lysine malonylation or lysine succinylation were decreased in response to Sirt5 expression (supplemental Fig. S12). Using the protein lysate from livers derived from wt Sirt5 mouse and Sirt5 knockout mouse, we showed that Sirt5 has significant impact on the global lysine malonylation and lysine succinylation, but has little influence on lysine acetylation (Fig. 7B).
Taken together, we reproducibly demonstrated that Sirt5 can catalyze lysine demalonylation reaction and lysine desuccinylation both in vitro and in vivo.
DISCUSSION
Lysine, with a positive charged side chain at physiological pH, plays an important role in protein functions and folding. And lysine methylation and lysine acetylation have been extensively studied in the past decade, revealing their important roles in transcriptional regulation and energy metabolism. In this study, we reported the identification of lysine malonylation in both mammalian and bacterial cells. This modification was validated by Western blot, competition assay, proteomics screening, and synthetic peptides in combination with HPLC/MS/MS. Lysine malonylation induces more dramatic structural changes than both lysine acetylation and methylation, and therefore is expected to have impacts on the substrate proteins.
The class III HDAC family consists of seven sirtuins (Sirt1–7), three of which, Sirt3, Sirt4, and Sirt5, are known to be located in mitochondrion. CPS1 is the only Sirt5-lysine acetylated substrate identified to date. Sirt5 was shown to deacetylate this protein and increase its activity (19). It was reported that Sirt5 may not be a major deacetylase in mitochondria for two reasons. First, no obvious mitochondrial hyperacetylation was detected in SIRT4 or SIRT5 knockout mice (20). In contrast, SIRT3-knockout mice show striking mitochondrial protein hyperacetylation, indicating that the protein is an important mitochondrial deacetylase. Second, human Sirt5 was reported to have weak deacetylase activity in vitro (21).
In this study, we showed robust demalonylation and desuccinylation activities of Sirt5. The activities were initially identified using fluorometric assays using 18 recombinant HDAC proteins. The in vitro demalonylation and desuccinylation activities were confirmed using peptide and protein substrates. The activities were confirmed in vivo by Western blot using transfection experiments and mouse livers from both wild-type mouse and Sirt5-knockout mouse. We further showed that Sirt5 has much higher activities to lysine malonylation and lysine succinylation than lysine acetylation, both in vitro and in vivo.
Eighteen HDACs were annotated in the literature, some of which do not have obvious acetyllysine protein substrates. Identification of Sirt5 as a demalonylation and desuccinylation enzyme suggests a possibility of nondeacetylation activities of other HDACs, especially those that do not have known acetyllysine substrates. On the other hand, although Sirt5 is the first enzyme we identified that has demalonylation and desuccinylation activities, this result does not exclude the possibility of the existence of such activities in other proteins.
Malonyl-CoA is an important intermediate in the pathways of cellular metabolism. Malonyl-CoA is known to be synthesized by three major routes: (1) the carboxylation of acetyl-CoA catalyzed by acetyl-Coenzyme A carboxylase (ACC), (2) the carboxylation of acetyl-CoA catalyzed by propionyl-CoA carboxylase, and (3) β-oxidation of old-chain-length dicarboxylic acids. Malonyl-CoA can be disposed in cells through fatty acid synthesis, a process catalyzed by fatty acid synthase and through conversation back to acetyl-CoA by malonyl-CoA decarboxylase. In addition to being an important metabolite, malonyl-CoA was also shown to have regulatory function, e.g. inhibition of mitochondrial carnitine palmitoyltransferase (CPT1) (22). As an important metabolite and regulatory molecule, dysregulation of malonyl-CoA homeostasis affects cellular physiology. For example, malonic aciduria, an inborn error of metabolism is caused by deficient MCD activity (23). ACC2 deficient mice show increased fatty acid oxidation, reduced body fat mass and body weight, despite consuming more food (hyperphagia). They are also protected against diabetes and obesity induced by high fat/high carbohydrate diets (24). These observations suggest ACC2 an attractive therapeutic agents against obesity and diabetes (25). The critical roles of malonyl-CoA in energy homeostasis suggest that lysine malonylation may provide another layer of cellular regulations in response to the dynamic changes of cellular malonyl-CoA concentration.
Although structurally similar, succinyl-CoA and malonyl-CoA have very different metabolic pathways. Succinyl-CoA is a metabolic intermediate in TCA cycle, porphyrin synthesis, and catabolism of odd-chain fatty acids and some branched-chain amino acids. It is possible that cells may use either one of the two PTM pathways (lysine succinylation and lysine malonylation), depending on the available CoA metabolites, to reprogram its cellular networks. It is also possible that some regulatory enzymes are different between the two PTM pathways, as in the case of lysine acetylation and lysine methylation. Identification of lysine malonylation and Sirt5 as the first regulatory enzyme for lysine malonylation is likely to be the first step for the exciting search for other regulatory enzymes and for characterization of biological functions of lysine malonylation. Therefore, lysine malonylation may provide a new paradigm in the cellular regulation, such as energy metabolism, in diverse organisms.
Footnotes
* This work was supported by National Institutes of Health grants to T.C. He, E. V., D.L., and Y.Z. D.X.T. was supported by a National Institute on Aging training grant (T32-AG000114). D.B.L. is a New Scholar in Aging of the Ellison Medical Foundation (AG-NS-0583-09). Work in the Lombard laboratory is also supported by the Ellison Medical Foundation as well as research grants from the Elsa U. Pardee Foundation and the American Foundation for Aging Research; the Richard D. and Katherine M. O'Connor Research Fund of the University of Michigan Comprehensive Care Center.
 This article contains supplemental Figs. S1 to S12, Data Sets S1 to S3 and supplemental Methods.
This article contains supplemental Figs. S1 to S12, Data Sets S1 to S3 and supplemental Methods.
1 The abbreviations used are:
- PTM
- Protein post-translational modification
- KAT
- lysine acetyltransferase
- HDAC
- Histone lysine deacetylase
- MS/MS
- tandem MS
- HPLC
- high performance liquid chromatography.
REFERENCES
- 1. Witze E. S., Old W. M., Resing K. A., Ahn N. G. (2007) Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798–806 [DOI] [PubMed] [Google Scholar]
- 2. Walsh C. T., Garneau-Tsodikova S., Gatto G. J., Jr. (2005) Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem. Int. Ed Engl. 44, 7342–7372 [DOI] [PubMed] [Google Scholar]
- 3. Johnson S. A., Hunter T. (2005) Kinomics: methods for deciphering the kinome. Nat. Methods 2, 17–25 [DOI] [PubMed] [Google Scholar]
- 4. Denu J. M., Dixon J. E. (1998) Protein tyrosine phosphatases: mechanisms of catalysis and regulation. Curr. Opin. Chem. Biol. 2, 633–641 [DOI] [PubMed] [Google Scholar]
- 5. Guarente L. (2007) Sirtuins in aging and disease. Cold Spring Harb. Symp Quant Biol. 72, 483–488 [DOI] [PubMed] [Google Scholar]
- 6. Martin C., Zhang Y. (2005) The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 [DOI] [PubMed] [Google Scholar]
- 7. Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. (2011) Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 9. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 10. Chen Y., Kwon S. W., Kim S. C., Zhao Y. (2005) An integrated approach for manual verification of peptides identified by searching protein sequence databases with tandem mass spectra. J. Proteome Res. 4, 998–1005 [DOI] [PubMed] [Google Scholar]
- 11. Allis C. D., Chicoine L. G., Richman R., Schulman I. G. (1985) Deposition-related histone acetylation in micronuclei of conjugating Tetrahymena. Proc. Natl. Acad. Sci. U.S.A. 82, 8048–8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mitzen E. J., Koeppen A. H. (1984) Malonate, malonyl-coenzyme A, and acetyl-coenzyme A in developing rat brain. J. Neurochem 43, 499–506 [DOI] [PubMed] [Google Scholar]
- 13. Koeppen A. H., Papandrea J. D., Mitzen E. J. (1979) Fatty acid biosynthesis in Wallerian degeneration of rat sciatic nerve. Muscle Nerve 2, 369–375 [DOI] [PubMed] [Google Scholar]
- 14. DeGnore J. P., Qin J. (1998) Fragmentation of phosphopeptides in an ion trap mass spectrometer. J. Am. Soc. Mass Spectrom 9, 1175–1188 [DOI] [PubMed] [Google Scholar]
- 15. Carr S. A., Huddleston M. J., Annan R. S. (1996) Selective detection and sequencing of phosphopeptides at the femtomole level by mass spectrometry. Anal. Biochem. 239, 180–192 [DOI] [PubMed] [Google Scholar]
- 16. Darula Z., Chalkley R. J., Baker P., Burlingame A. L., Medzihradszky K. F. (2010) Mass spectrometric analysis, automated identification and complete annotation of O-linked glycopeptides. Eur. J. Mass Spectrom. 16, 421–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wells L., Vosseller K., Cole R. N., Cronshaw J. M., Matunis M. J., Hart G. W. (2002) Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics 1, 791–804 [DOI] [PubMed] [Google Scholar]
- 18. Brown W. H., Foote C. S., Iverson B. L. (2009) Organic Chemistry. 5th edn, (Brooks/cole Cengage Learning) [Google Scholar]
- 19. Nakagawa T., Lomb D. J., Haigis M. C., Guarente L. (2009) SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137, 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lombard D. B., Alt F. W., Cheng H. L., Bunkenborg J., Streeper R. S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., Yang Y., Chen Y., Hirschey M. D., Bronson R. T., Haigis M., Guarente L. P., Farese R. V., Jr, Weissman S., Verdin E., Schwer B. (2007) Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell Biol. 27, 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. North B. J., Marshall B. L., Borra M. T., Denu J. M., Verdin E. (2003) The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437–444 [DOI] [PubMed] [Google Scholar]
- 22. Wolfgang M. J., Lane M. D. (2011) Hypothalamic malonyl-CoA and CPT1c in the treatment of obesity. FEBS J. 278, 552–558 [DOI] [PubMed] [Google Scholar]
- 23. MacPhee G. B., Logan R. W., Mitchell J. S., Howells D. W., Tsotsis E., Thorburn D. R. (1993) Malonyl coenzyme A decarboxylase deficiency. Arch. Dis. Child 69, 433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abu-Elheiga L., Oh W., Kordari P., Wakil S. J. (2003) Acetyl-CoA carboxylase 2 mutant mice are protected against obesity and diabetes induced by high-fat/high-carbohydrate diets. Proc. Natl. Acad. Sci. U.S.A. 100, 10207–10212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harwood H. J., Jr. (2004) Acetyl-CoA carboxylase inhibition for the treatment of metabolic syndrome. Curr. Opin. Investig. Drugs 5, 283–289 [PubMed] [Google Scholar]