Abstract
ZBRK1, named after its structure of Zinc finger and BRCA1-interacting protein with KRAB domain-1 (ZBRK1), is a transcriptional repressor modulated by BRCA1. Recent evidence also indicated that ZBRK1 collaborated with BRCA1/CtIP to repress angiopoietin-1 expression in preventing over enlargement of blood vessels in tumors, suggesting that ZBRK1 may exert a critical role during tumor progression. However, a direct role of ZBRK1 in tumorigenesis and tumor progression remains obscure. Here we found that ZBRK1 expression was significantly lower in highly malignant cervical cancer cells than the counterpart normal tissue. Ectopic expression of ZBRK1 in HeLa cells significantly inhibits its neoplastic phenotypes including cell proliferation, soft-agar colony formation and tumor growth in nude mice. To explore its mechanisms, analyses of gene expression patterns of these cells revealed groups of genes not only critical for cell proliferation but also for cell motility being down regulated. Consistently, ectopic expression of ZBRK1 inhibits HeLa cells migration in cell migration and invasion assays in culture and metastatic assay in mice. Importantly, ZBRK1 directly represses transcription of the metastatic gene, MMP9, and the loss of ZBRK1 expression is inversely correlated to the elevated expression of MMP9 in cervical cancer specimens. Taken together, these results indicate that ZBRK1 may have a critical role as a tumor suppressor, especially in metastasis, through directly modulating metastatic genes such as MMP9.
Introduction
Cancer metastasis is the most common cause of death among cancer patients (1). It results from several highly organized sequential steps involving interactions between cancer cells and the host. Metastasis is a multi-step process: invasion of tumor cells into adjacent tissues, entry of tumor cells into the systemic circulation (intravasation), survival in circulation, extravasation to distant organs, and finally growth of cancer cells to produce secondary tumors (2, 3). Recently, gene expression analyses of human breast carcinomas with known clinical outcomes revealed profiles associated with disease progression and identified groups of genes, including cell-migration genes, whose characteristic expression patterns can predict the risk of metastatic recurrence (4–8). However, details of candidate genes involved in other aspects of cancer metastasis/invasion processes remain under-investigated. Precisely how tumor cells become metastatic is still largely unknown, especially in terms of a transcriptional factor which serves as a repressor in metastasis/invasion.
Krupple-associated box-containing zinc-finger (KZF) proteins comprise a group of the most widely distributed transcriptional repression proteins in mammals. They are composed of a Krupple-associated box (KRAB) domain at the N-terminus and tandem C2H2 class zinc fingers at the C-terminus. Several KZF proteins modulate cell growth and survival and are implicated in malignant disorders (9). Although a majority of KZF proteins function to repress RNA polymerase II-mediated transcription, their respective target genes, which underlie transcriptional regulation mechanisms and their cellular activities remain largely unclear. Members of the really interesting new gene (RING) family are found throughout eukaryote cells and play key functions in processes as diverse as development, oncogenesis, viral replication, and apoptosis (10, 11).
The KRAB domain is divided into A and B boxes and is required for repression of transcription by recruiting co-repressors, such as KRAB domain-associated protein 1 (KAP1) (12). KAP1 was initially identified as a transcriptional co-repressor and its N-terminal RING-B boxed-coiled-coildomain serves as a protein-protein interaction regionfor binding to the KRAB domain of KZF proteins (12–14). The tandem C2H2 class zinc fingers at the C-terminus were used to bind specific DNA sequences. It has been suggested that some cofactors may cooperate with those zinc fingers to enhance the DNA binding specificity (15, 16).
ZBRK1, which was first identified in a yeast two-hybrid screening for proteins associated with BRCA1, is a typical Krupple-associated box-containing zinc-finger protein (KRAB-ZFP) that contains a highly conserved KRAB domain at the N-terminus, eight consecutive C2H2 zinc finger motifs, and a CTRD domain for BRCA1 interactions at the C-terminus (17). Its zinc-finger repeats were suggested to recognize a consensus DNA binding element, GGGxxxCAGxxxTTT, or to be involved in protein interactions (18). Two co-repressors of RING members, BRCA1 and KAP1, were demonstrated to interact with ZBRK1 in coordinating transcriptional regulation of diverse DNA damage-response genes (19). Some of the ZBRK1-affected downstream targets including GADD45 and p21 have been reported (20–22). Recently, ZBRK1 has been identified as cooperating with the CtIP/BRCA1 to repress angiopoietin-1 (ANG1) gene activation (22) and may play a role in tumor angiogenesis, implying it may act as a potential tumor suppressor. However, whether ZBRK1 plays a direct role in tumor progression, especially in metastasis, has yet to be demonstrated.
In this investigation, we found that reduction of ZBRK1 expression was observed in highly malignant cervical cancer cells as compared to the counterpart normal tissue. Ectopic expression of ZBRK1 in HeLa cells significantly inhibits its neoplastic phenotypes. This paper is the first to report on this significant discovery in cervical cancer cells, it appears that a reduction of ZBRK1 allows the growth of cancer cells, while an increase of ZBRK1 has been shown to inhibit cell growth both in vitro and in vivo, suggesting that ZBRK1 can act as a tumor suppressor. Interestingly, analyses of gene expression patterns of these cells revealed groups of genes not only critical for cell proliferation but also for cell motility being down regulated. Consistently, ectopic expression of ZBRK1 inhibits HeLa cells migration, in part, through directly repressing transcription of the metastatic gene, MMP9. This study also presents the first demonstration of the direct negative repression of the transcriptional regulation of the MMP9 gene. This is further validated in cervical cancer specimens, in which loss of ZBRK1 expression is inversely correlated with the elevated expression of MMP9. All things considered, these results suggest that ZBRK1 plays a critical role in tumor progression, especially in metastasis, through directly modulating metastatic genes.
Materials and Methods
Cell culture and stable cell line establishment
Mammalian cells, U2OS, and various cervical cancer cell lines were cultured in complete medium containing DMEM, 10% FBS, 100 ug/ml streptomycin, and 100 U/ml penicillin. U2OS and HeLa cells stably expressing EGFP or EGFP-ZBRK1 were generated byhygromycin B selection.
Assays for Cell proliferation, Focus-formation and Anchorage-independency
For cell proliferation assay, daily cell culture samples were counted by a Neubauer chamber after trypsinization. Viability was assessed by trypan blue exclusion. Assays were performed in triplicates. For focus-forming assay, U2OS and HeLa cells stably expressing EGFP or EGFP-ZBRK1 were plated at low density (1000 cells/plate) onto 100 mm dishes cultured with DMEM containing 5% FBS and the media were changed every 3±4 days. After 2 weeks, the numbers of the colonies were counted with the Sigmascan software program by staining with 2% methylene blue in phosphate buffer saline. For anchorage-independent assay, one ml of 0.3% agarose in complete growth medium containing 2000 HeLa cells stably expressing EGFP or EGFP-ZBRK1 were seeded onto a hard agar base composed of 1.5 ml 10% FBS-DMEM and 0.6% agarose in six-well plates as described (23). After 14 to 21 days, cells were stained with 0.05% crystal violet and the colonies were scored for statistic analysis. All above experiments were performed at least twice in triplicate.
Reverse Transcription Polymerase Chain Reaction (RT-PCR) and Microarray analyses
Total RNA was extracted by using TRIzol RNA extraction reagent (Invitrogen, Carlsbad, CA, USA). The isolated RNAs were subjected to reverse transcription reaction with SuperScript™ III (Invitrogen, Carlsbad, CA, USA) for cDNA synthesis. PCR was performed with the pairs of specific primers as follows: ZBRK1: 5′-GACATATGGAAAGTTGATCATGTGCTG-3 ′ and 5′-ATTCACTGCACACATGATGCTTCTCTA-3 ′ G A P DH: 5′-CCATCACCATCTTCCAGGAG-3′ and 5′-CCTGCTTCACCACCTTCTTG-3′. The total RNA samples harvested from cells stably expressing EGFP or EGFP-ZBRK1 were performed with microarray analysis at WEIGENE Biotech. Comp by Agilent Human whole genome oligo 4 × 44K array.
Tumor growth analysis in mice
Athymic (nu/nu) nude mice (4–6 wk of age) were obtained from the National Laboratory Animal Center and fed with Laboratory Autoclavable Rodent Diet (LabDiet). All animal work was done in accordance with the protocol approved by the Institutional Animal Care. Aliquots of 3 × 106 EGFP and EGFP-ZBRK1 HeLa cells were inoculated subcutaneously into the dorsal rear flanks of nude mice. After injection, tumor size was measured by external caliper, and tumor volumes were calculated using a standard formula as follows: V = height × width × depth. Mice were sacrificed at 11 days after the cell injection.
Experimental metastasis assay
Twelve female 4–6 weeks old NOD-SCID (Non-obese diabetic-severe combined immunodeficiency) mice were injected with 1 × 106 EGFP or EGFP-ZBRK1 HeLa cells. Cells were washed and harvested in PBS and injected intravenously, via the lateral tail vein, in a volume of 0.1 ml. At 14 days after injection, when mice had not died but with significant morbidity, all mice were euthanized and their lungs were removed. The number of surface metastases per lung was counted.
Wound healing assay
After U2OS and HeLa cells were grown to confluence in 6-mm culture plates, an artificial “wound” was created using a P-200 pipette tip to scratch on the confluent cell monolayer. Cell migration into the scratched region was recorded using a Nikon microscopy system (Nikon Instrument) at 0 and 48 h and the number of cell in the scratched region was quantified as the percentage of wound healing.
Cell migration and invasion assays
A migration assay kit (ECM 566 QCM™ 96-well Haptotaxis Cell Migration Assays-Collagen 1, Fluorimetric from Chemicon, Temecula, CA) and an invasion assay kit (ECM 555 QCM™ 96-well cell invasion assay from Chemicon, Temecula, CA) were used according to the manufacturer’s instructions. Cells were seeded at a density of 1.5 × 105 cells per well in 100 ul DMEM onto the upper chambers and DMEM with 1% FBS (150 ul) was added to the lower chambers of the 96 well plates. The plates were incubated at 37°C and 5% CO2 for 6 h to allow migration or invasion through the membrane. Cells that had migrated through to the bottom of the insert membrane were dissociated from the membrane and detected by CyQUANT GR dye. The activity of cell migration and invasion were calculated as the percentage of the fluorescence relative to the controls.
Chromatin immunoprecipitation (ChIP)-PCR assay
The ChIP assay was carried out essentially as described by Furuta et al (22), with a minor modification. Briefly, the sheared chromatin fragments were immunoprecipitated with antibodies specific to GFP, ZBRK1 or control mouse IgG at 4 °C overnight. After dissociating the DNAs from immunoprecipitated chromatin, the DNAs were amplified by PCR amplification. For PCR amplification of specific regions (A and B) of the MMP9 genomic locus, the following sets of primers were used: A fragment of primers, forward: 5′-TGCCCGTAATCCTAGCACTTTGGGA-3′ and reverse: 5′-CCTCACTCCTTTCTTCCTAGCCAGC-3′ B fragment of primers: forward: 5′-AGGCTGTCAGGGAGGGAAAAAGAGG-3′ and reverse: 5′-AGAAAGGGCTTACACCACCTCCTCC-3′. The PCR condition is in a total of 32 cycles of 30 seconds at 94°C, 25 seconds at 56°C, and 1 minute at 72°C.
Reporter plasmids and Luciferase assay
The reporters bearing the different fragments of human MMP9 promoter were generated by PCR with genomic DNA as the template. The primers for PCR reaction as following: MMP9/−1940: 5′-GGGGTACCAGTGACTTGCCCAAGGTCACATAGC-3′, MMP9/−940: 5′-GGGGTACCTACAGGAATGAGCCACCATACCTGG-3′ and MMP9/+113: 5′-CCCAAGCTTTGAGATTGGTTCTCAGGTCTCCAGG-3′. The PCR fragments were cloned into the multi-cloning sites of the promoter-less vector, pGL2-basic vectors and verified by sequencing. For the heterologous reporters, the various copies of repeated ZBRK1- or mutant ZBRK1-binding motifs on the MMP9 promoter were inserted into pGL2-promoter vector. These heterologous reporters were introduced into HeLa cells by Lipofectamine 2000 according to the manufacturer’s instruction, and then the lysates of transfectants were harvested for luciferase assay.
Gelatin zymography
The presence of MMP9 in supernatants of EGFP and EGFP-ZBRK1 HeLa cells was analyzed with gelatin zymograms as described previously (24). Briefly, the cells were incubated in serum-free DMEM with or without 100 ng/ml of EGF, and the supernatants were collected after stimulation for 24 h, clarified by centrifugation, normalized to cell number, mixed with non reducing Laemmli sample buffer and separated by electrophoresis in 10% sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGE) containing 1 mg/ml gelatin (Sigma, St. Louis, MO, USA). After electrophoresis the gels were renatured by washing in 2.5% Triton X-100 solution (Sigma, St. Louis, MO, USA) twice for 30 minutes to remove all SDS. The gels were then incubated in 50 mM Tris-HCl (pH 7.4), 5 mM CaCl2, 1% Triton X-100 and 0.02% NaN3 at 37°C overnight. After incubation, the gels were stained with 0.05% Coomassie Brilliant Blue R-250 for 1 h at room temperature and then destained in distilled water. The MMP9 activities were visible as clear bands on a blue background where the gelatin substrate had been hydrolyzed by enzyme activity.
Tissue samples
Patients of 12 cases of cervical cancer were surgically resected at the National Cheng Kung University Hospital. Total RNA samples were extracted from tumor tissues and adjacent unaffected cervical or colon tissues. All tumor specimens of patients were obtained from surgically resected tissues that had previously been pathologically assessed at the National Cheng Kung University Hospital. The fresh tissue samples were immediately cut into small pieces, snap-frozen in liquid nitrogen, and stored in a deep freezer. Total RNA was extracted from the tumorous and paired non-tumorous tissue using the TRIzol RNA extraction reagent.
Lentiviral shRNA
The lentiviral expression vectors p L K O. 1-shLuc, containing 5′-CTTCGAAATGTCCGTTCGGTT-3′, and pLKO.1-shZBRK1, containing 5′-GCTAACCATGAACGACTTCAT-3′, were obtained from the National RNAi Core Facility located at the Genomic Research Center of Institute of Molecular Biology, Academia Sinica. Virus was produced as described using Lipofectamine 2000 cotransfection of Phoenix cells with the pLKO.1-shLuc or pLKO.1-shZBRK1 vector together with pMD2.G and psPAX2.
Results
Elevated expression of ZBRK1 retards cancer cell proliferation, soft-agar colony formation and tumor growth in mice model
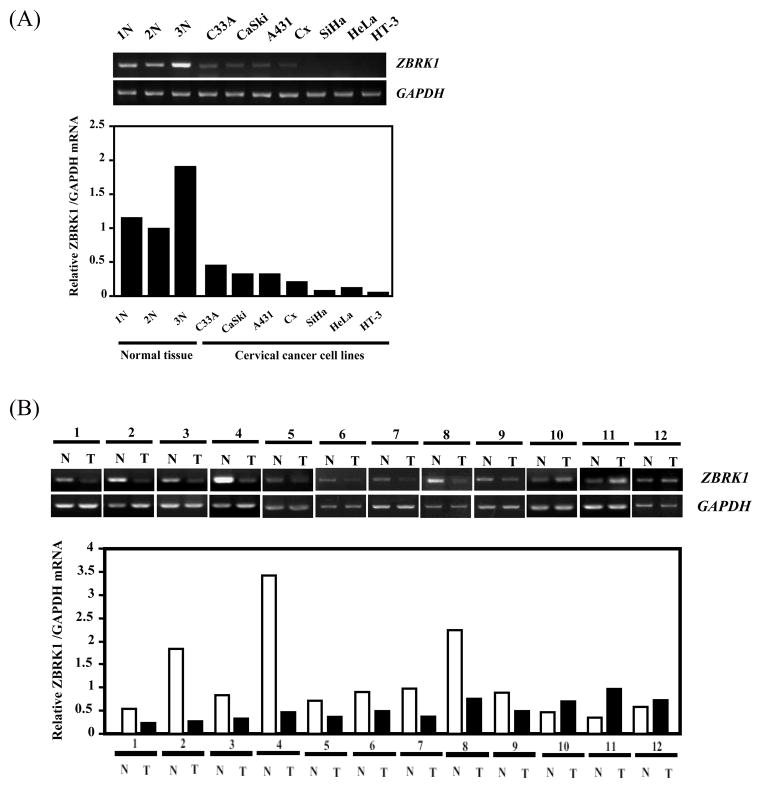
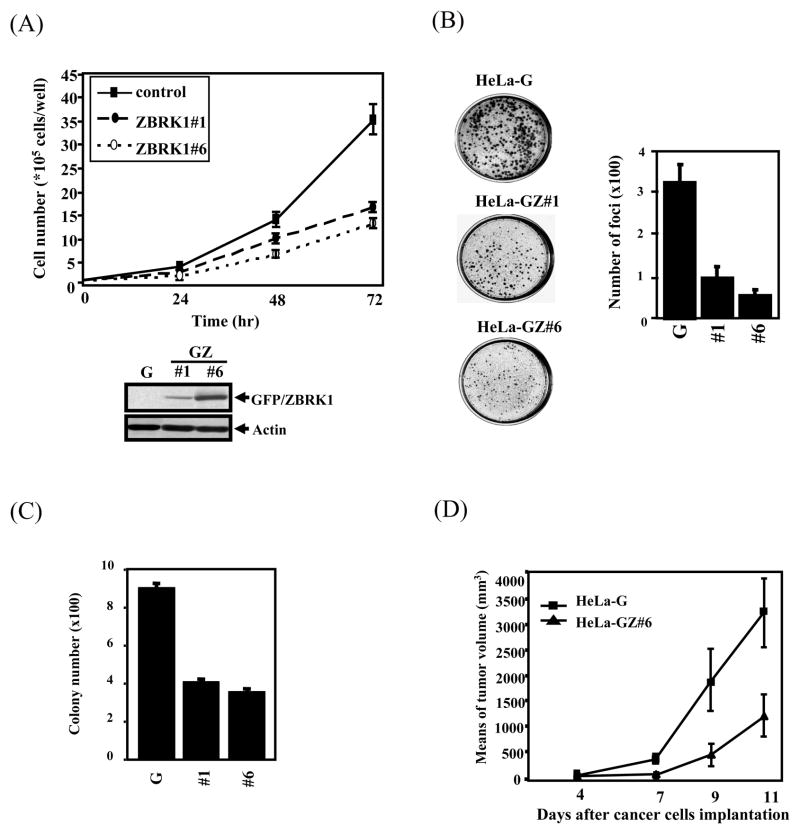
ZBRK1 is a transcriptional repressor and a potential tumor suppressor since it cooperates with BRCA1/CtIP in modulating ANG1 expression (22). It has not been known whether its expression level has any relationship with tumor malignancy. To test this possibility, we examined the mRNA expression level of ZBRK1 in a panel of cervical cancer cells and clinical cervical cancer specimens. It found that 7 out of 7 cervical cancer cell lines and 9 out of 12 cervical cancer specimens expressed significantly reduced amounts of ZBRK1 RNA when compared with the normal cervical tissues (Fig. 1A and 1B). This result suggests that reduction of ZBRK1 expression may be involved in tumorigenesis. To further corroborate this observation, stable cell lines bearing ectopically expressed EGFP-tagged ZBRK1 in HeLa and U2OS cells were established for comparison with the parental cells in growth rate, plating colony formation efficiency, soft agar colony formation and tumor formation in mice. As shown in Fig 2, HeLa cells constitutively expressing ZBRK1 either clone 1 or 6, which expressed different levels of ZBRK1, grew slower (Fig. 2A), formed less colonies in plating efficiency (Fig. 2B), less colonies in soft agar (Fig. 2C) and much smaller tumors in nude mice (Fig. 2D). These results suggest that ZBRK1 expression inversely correlates with the malignancy of tumor cells and ZBRK1 acts as a tumor suppressor to inhibit tumor progression.
Figure 1. Expression level of ZBRK1 is attenuated in various cervical cancer cell lines and clinical cervical cancer patients.
(A) ZBRK1 expression was examined in various cervical cancer cell lines by RT-PCR. GAPDH serves as an internal control. (B) ZBRK1 expression was determined in normal and tumor areas of surgical biopsies from 12 cervical cancer patients by RT-PCR. GAPDH transcript level was used as the load control. “N” and “T” respectively denote “normal” and “tumor” areas of the same patients. The bottom panels show the amount of ZBRK1 mRNA, relative to GAPDH by RT-PCR.
Figure 2. Overexpression of ZBRK1 inhibits cell proliferation and anchorage-independent activity of cancer cells.
(A) ZBRK1 attenuated the growth rate of cells. Equal amounts of HeLa cells stably expressing EGFP (G) or EGFP-ZBRK1 (GZ#1 and GZ#6) were seeded, and then the viable cell number was determined at the indicated times. Stably expressed levels of ZBRK1 in HeLa cells were analyzed by Western blot. (B) An increase in ZBRK1 expression attenuated cell proliferation. A focal formation assay was performed with the stable cell lines of HeLa cells. Values are the relative cell number ± S.E.M. (C) Overexpression of ZBRK1 attenuated focal formation of cancer cells. A soft agar assay was performed with HeLa cell lines stably expressing EGFP (G) or EGFP-ZBRK1 (#1 and #6). Values are the relative cell number ± S.E.M. (D) Overexpression of ZBRK1 reduced the formation of tumors in nude mice. Tumor volumes were measured every day after a subcutaneous injection of EGFP or EGFP-ZBRK1 HeLa cells. Data from six mice in each group are presented as the mean ± SD.
Ectopic expression of ZBRK1 inhibits expression of groups of genes essential for cell proliferation and migration
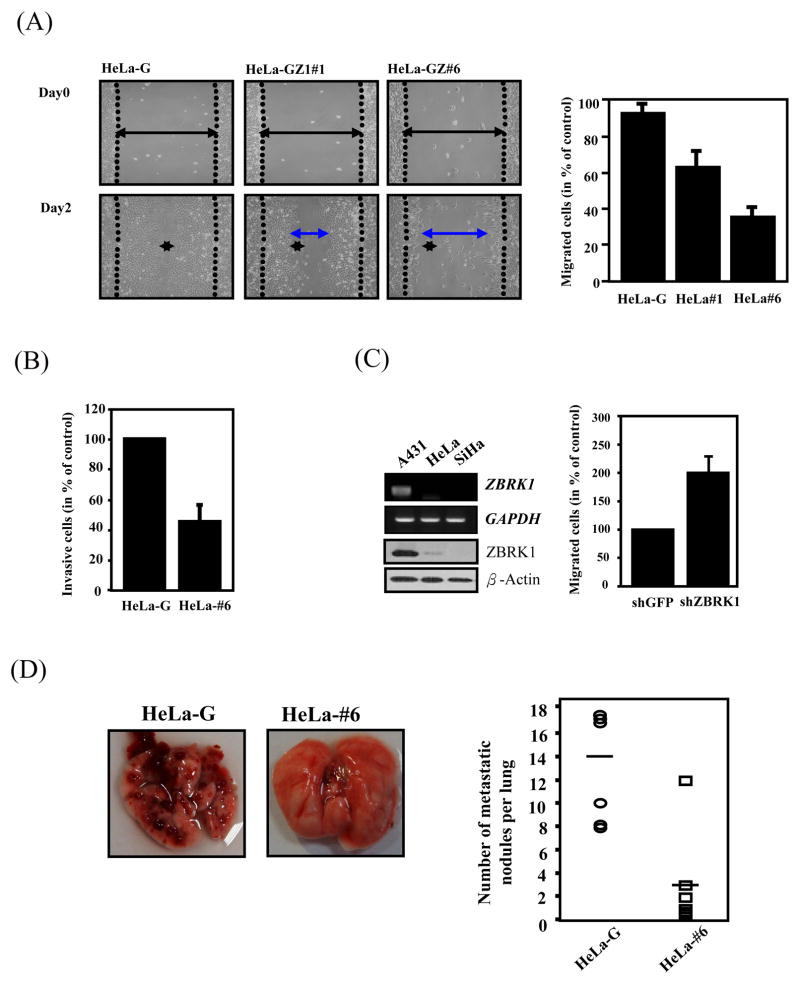
To search the genes regulated by ZBRK1, two sets of microarray analyses using stable cell lines, EGFP-ZBRK1 and EGFP HeLa cells, and EGFP-ZBRK1 and EGFP U2OS cells, were respectively performed. About 304 and 196 upregulated genes, and 473 and 341 downregulated genes, were respectively identified from these two sets of expression profiles of U2OS and HeLa cells (SFig. 2A, B). Interestingly, 36 ZBRK1-downregulated genes and 23 ZBRK1-upregulated genes were commonly shared between these two ZBRK1 ectopically expressed cells (Supplemental table 1). The expression of some of these commonly regulated genes was further validated by RT-PCR (SFig. 1D and E). These ZBRK1-regulated genes were divided into groups by the Ingenuity program according to their known biological functions (Supplemental table 2). Based on this analysis, it appeared that the major functions of these genes affected by ZBRK1 could be categorized into cell proliferation, which is consistent with the observation as described in Fig. 2, and cell migration. Subsequently, to verify the potential role of ZBRK1 in regulating cell migration, we then performed a wound-healing assay and showed a slow sealing effect in EGFP-ZBRK1 HeLa cells in a dose-dependent manner when compared with the parental cells (Fig. 3A). Similar results were also observed in U2OS cells (30%~40% reduction) (SFig. 3A). The migration and invasion assays were performed individually, which showed coincident results with wound-healing assay (SFig. 3B and Fig. 3B). To further validate this notion, a poorly metastatic/invasive cell line A431 with higher expression levels of ZBRK1 mRNA (Fig. 1) as well as protein (Fig. 3C), was employed for depletion of ZBRK1 via lentiviral-mediated shRNA knockdown approach. As shown in Fig. 3C, knockdown of ZBRK1 expression in A431 cells promoted cell migration in the QCM™ Haptotaxis Cell Migration Assay, suggesting that the reduction of ZBRK1 has the advantage of promoting cancer cell migration. To further evaluate the effects of ZBRK1 on metastasis/invasion, EGFP-ZBRK1 and EGFP HeLa cells were injected into the lateral tail vein of scid mice, and their metastasis and growth in lung tissues were measured. After 4 weeks, the injected control tumor cells had formed 8~20 metastatic nodules per lung in all 6 mice. In contrast, mice injected with the same amount of EGFP-ZBRK1 HeLa cells formed 0~3 nodules (Fig. 3D). These results demonstrate that ectopic expression of ZBRK1 reduced or eliminated the metastasis/invasion ability of cancer cells.
Figure 3. ZBRK1 inhibits cell migration.
(A) Wound-healing migration was performed with EGFP- and EGFP-ZBRK1-expressing cells. Representative images of wound sealing were taken on the day of the laceration and day 2 after the wound scratch. The level of cell migration into the wound scratch was quantified as the percentage of wound sealing. Values represent the average ±S.E.M. of three independent measurements. (B) Overexpression of ZBRK1 inhibits invasion of cancer cells. The cells were seeded in ECMatrix layer and the level of cell invasion was determined using QCM™ 96-well cell invasion assay as described in Materials and Methods. (C) Inactivation of ZBRK1 increased migration of cancer cells. Left panel: mRNA and protein levels of ZBRK1 in A431, HeLa, and SiHa cervical cancer cell lines. Right panel: A431 cells were treated with lentiviral shZBRK1 or shGFP in the QCM™ 96-well cell migration assay. The top panel shows the amounts of ZBRK1 and GAPDH. (D) Equal amounts of EGFP- and EGFP-ZBRK1-#6 HeLa cells were injected into the tail vein of scid mice. The experimental mice were sacrificed to calculate the metastatic nodes on lung tissues after 4 weeks. Data from six mice in each group are presented as the mean ± SD. Representative pictures taken at the time of sacrifice are shown (left panel).
ZBRK1 directly regulates cell metastasis-related gene MMP9
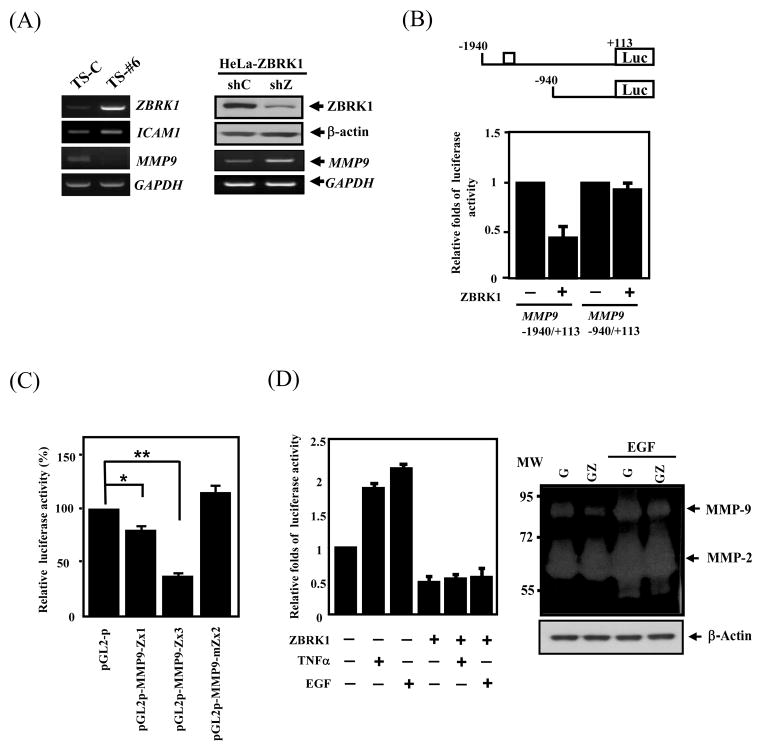
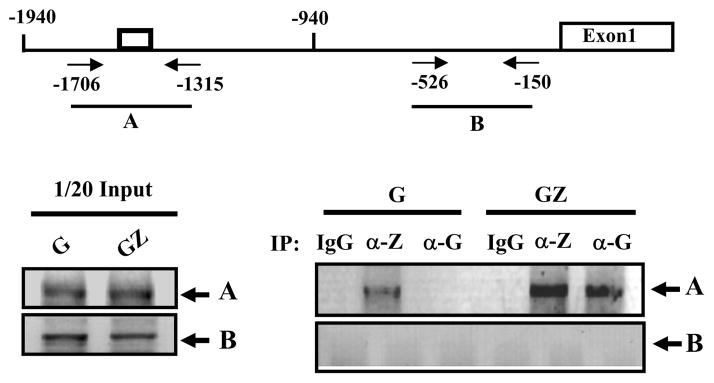
Among those cell motility genes potentially regulated by ZBRK1 (Supplemental table 2), MMP9 is of particular interest because MMP9 expression was the most significantly reduced in tumor specimens derived from HeLa clone 6 grown in mice when compared with HeLa-C tumors (Fig. 4A, left panel). Similarly, using shRNA to knockdown ZBRK1 mRNA in HeLa cells, which ectopically expressed ZBRK1, expression of MMP9 mRNA was elevated in these cells when compared with the control shRNA (Fig. 4A, right panel). These results strongly suggest that ZBRK1 down modulates MMP9 expression. A putative ZBRK1-binding motif, −1504/−1518, was identified on MMP9 promoter. Therefore, we performed an EMSA by using in vitro translated ZBRK1 to incubate with the fragment, −1524 to −1497, on MMP9 promoter region. As shown in SFig. 4, ZBRK1 can bind to the putative ZBRK1-binding motif. Next, we cloned the MMP9 promoter to verify whether ZBRK1 regulates MMP9 transcription through its promoter region. Ectopic expression of ZBRK1 specifically reduced the MMP9 reporter activity containing the putative ZBRK1-binding motif, MMP9-1940/+113, but not the MMP9-940/+113 reporter, with no putative ZBRK1-binding motif (Fig. 4B). To further confirm the repressive effect of the ZBRK1-binding motif on the MMP9 promoter, heterologous reporters generated by inserting a wild-type ZBRK1-binding motif, −1524/−1497 oligonucleotides, or a mutated ZBRK1-binding motif, −1524/−1497 oligonucleotides into the pGL2-promoter vector, were used for the reporter assay. The MMP9-1524/−1497 reporters, pMMP9-Zx1 and pMMP9Zx3, showed repressive activity in dose-dependent manners, but the reporter bearing a mutant ZBRK1-binding motif, pMMP9-mZx2, showed no repressive activity (Fig. 4C). These results suggest that ZBRK1 can inhibit MMP9 promoter activity through the ZBRK1-binding motif, −1524/−1497. Importantly, ectopic expression of ZBRK1 overcame the transcriptional activation by TNF-α or EGF, which is known to induce MMP9 reporter activity (Fig. 4D, left panel) (25, 26). Consequentially, MMP9 enzymatic activity induced by EGF was also reduced in cells ectopically expressing ZBRK1 (Fig. 4D, right panel). To determine whether ZBRK1 indeed binds to the specific region containing the ZBRK1-binding motif of the MMP9 promoter in vivo, chromatin immunoprecipitation assay was performed and showed that the A fragment, a PCR product of −1706/−1315, but not the B fragment, a PCR product of −526/−150, of the MMP9 promoter region was detected following immunoprecipitation by ZBRK1 antibodies or GFP antibodies in EGFP or EGFP-ZBRK1 cells (Fig. 5). These results strongly suggest that ZBRK1 directly down regulates MMP9 expression through specifically binding to the MMP9 promoter region containing the ZBRK1-binding motif.
Figure 4. ZBRK1 represses MMP9 promoter activation.
(A) Left panel: ZBRK1 inhibited the MMP9 transcripts in xenogenic tumor. Expression levels of multiple genes of tumor sample from mice resulting after subcutaneously injected with EGFP (TS-C) or EGFP-ZBRK1#6 (TS-#6) HeLa cells were analyzed by RT-PCR. The transcripts of human GAPDH served as a control. Right panel: The loss-of-function ZBRK1 enhanced MMP9 transcripts. Stable ZBRK1-expressing cells were incubated with lentiviral shRNA of ZBRK1 or the control. The lysates and total RNA of infected cells were harvested for Western blot and RT-PCR analyses, respectively. (B) ZBRK1 inhibited the MMP9 reporter. EGFP-ZBRK1-expressing HeLa cells transfected with the −1940/+113 and −940/+113 MMP9 reporters contained wild-type or loss-of-function ZBRK1 responsive element, respectively. Lysates of the transfectants were harvested after 12 h of transfection for luciferase assay. (C) ZBRK1 regulates MMP9 transcription through binding to ZBRK1 motifs. The heterologous reporters bearing the wild-type or mutant ZBRK1 motifs of the MMP9 promoters were transfected in EGFP-ZBRK1-expressing HeLa cells. The results shown are averages from three independent transfection assays and are plotted as relative activities to cells transfected by the backbone reporter, pGL2-promoter (pGL2-p). The latter activity was considered to be 100. Data are shown as the mean ± SEM. * p < 0.05, by Student’s t-test. (D) Left panel: ZBRK1 suppresses TNFα- and EGF-induced MMP9 transcription. HeLa cells transfected the region of −1940/+113 of the MMP9 promoter and stimulated with or without TNF-α or EGF. Right panel: An increase in ZBRK1 attenuated EGF-induced enzyme activity of MMP9. The concentrated lysates for gelatin zymography were performed by the supernatants harvested from the cells stably expressing EGF or EGF-ZBRK1 upon EGF treatment or not.
Figure 5. ZBRK1 binds to the MMP9 promoter in vivo.
The sheared formaldehyde-cross-linked chromatins, extracted from HeLa cells stably expressing EGFP (G) or EGFP-ZBRK1 (GZ), were immunoprecipitated with the ZBRK1 or GFP antibody. The figure represents PCR products obtained using specific primers on the MMP9 promoter region as shown in the upper panel. Very similar results were obtained from two independent experiments.
Expression of ZBRK1 is inversely correlated to the expression of MMP9 in cervical cancer specimens
Next, to test whether repression of MMP9 expression by ZBRK1 also occurs in clinical cancer specimens, we then collected 12 pairs of cervical cancer and non-cancer tissues from the same patient for comparison of their expression pattern. Since we have previously reported that SUZ12, a polycomb protein overexpressed in specimens of cervical cancer (23), we then analyzed the three genes ZBRK1, SUZ12 and MMP9 using the collected specimens. As shown in SFig. 5 and Table 1, reduction of the ZBRK1 expression was observed in 9 out of 12 pairs that inversely correlated with the up-regulation of MMP9 with 100% coincidence. While expression of SUZ12 was also elevated in the majority of cervical cancer samples (9 out of 12), the inverse correlation with ZBRK1 was not completely concordant (Table 1). This data further supports the notion that ZBRK1 down regulates MMP9 in cervical cancer specimens.
Table 1. The correlation among ZBRK1, SUZ12 and MMP9 in cervical cancer specimens.
By Chi-square analysis, ZBRK1 repression was significantly associated with upregulation of MMP9 (p=0.007) but not SUZ12.
| Increase of ZBRK1 in CC (n=3, 25%) | Repression of ZBRK1 in CC (n=9, 75%) | P-value | ||
|---|---|---|---|---|
| SUZ12 | Downregulation | 1 (50%) | 1 (11%) | 0.197 |
| Upregulation | 1 (50%) | 8 (89%) | ||
| MMP9 | Downregulation | 2 (66%) | 0 (0%) | 0.007* |
| Upregulation | 1 (33%) | 9 (100%) | ||
statistically significant.
Discussion
ZBRK1 is a transcriptional repressor with potential activity in tumor suppression because of its association with BRCA1, a bona fide tumor suppressor. In this communication, our results indicate that ZBRK1 plays an important role in suppressing tumor progression including metastasis. First, ZBRK1 expression was significantly lower in highly malignant cervical cancer cells and clinical cervical cancer specimens than in normal tissue counterpart, while ectopic expression of ZBRK1 in HeLa cells significantly inhibits its neoplastic phenotypes including tumor growth in nude mice. Second, the metastasis suppressing activity is demonstrated by ectopic expression of ZBRK1 that inhibits HeLa cells metastasis shown in both cell mobility assay in culture and experimental metastatic assay in mice. Third, ZBRK1 directly represses transcription of the metastatic gene, MMP9, and loss of ZBRK1 expression is inversely correlated to the elevated expression of MMP9 in cervical cancer specimens. These results indicate that ZBRK1 has a critical role in tumor progression, notably in metastasis.
Reduction of the expression of ZBRK1 in cervical tumors and cell lines provides a clue suggesting that ZBRK1 may behave as a tumor suppressor. Down regulation of ZBRK1 was observed in many cervical cancer cell lines and 75% of cervical cancer specimens (n=12) (Fig. 1 and Table 1). Interestingly, our preliminary data indicates that the reduction of ZBRK1 expression is also observed in other cancers including 50% of hepatocellular carcinomas (n=8) and 37.5% of colorectal cancers (n=8) (CC Liao and JM Wang unpublished data). These results suggest that ZBRK1 may have a broad role in diverse human cancer tumorigenesis.
Intriguingly, overexpression of ZBRK1 has the potential to inhibit cancer cell migration and suppress metastasis activity (Fig. 3), suggesting that ZBRK1 may behave as a metastasis suppressor. It was reported that metastasis was inhibited in an in vivo assay, while tumorigenicity was not significantly affected following re-expression of a metastasis-suppressor gene in a tumor cell line (27, 28). This is an essential criterion as a metastasis-suppressor gene, which does not affect the growth of the primary tumor. However, the proliferation assay (Fig. 2A) and downstream targets, including EGR1 and HMGA2, showed in profiling suggested that ZBRK1 also could be a proliferation suppressor. Therefore, we could not rule out the potential that the involvement of proliferation in these experimental migration assays. Even so, these current results suggested that ZBRK1 serves as an upstream regulator integrating the proliferation and cell migration in tumor progression. Therefore, ZBRK1 may be a useful therapeutic target by reactivation of ZBRK1 expression to inhibit metastasis of tumor cells through blocking the metastatic cascade. Further pursuit of this possibility is warranted.
Further, expression of ZBRK1 inversely correlated with the expression of a group of genes participating in cell proliferation and mobility, particularly a well characterized metastatic gene, MMP9 (Supplemental table 2), in both cell lines as well as in clinical specimens. MMP9 is thought to play important roles in invasion and metastasis and has been shown to have multiple effects on cell motility (29). Although extrinsic stimulators in this activation include EGF, PMA, and TNF-α, other transcriptional activators, including AP-1, and CBP/p300 have been reported to be involved in the transcriptional activation of the MMP9 gene (30–32). Negative regulators of the MMP9 transcription such as interferon-γ and interleukin (IL)-10, have also been reported to inactivate MMP9 transcription (33, 34). However, the precise mediator for MMP9 was not elucidated until this report revealed that ZBRK1 directly binds to the promoter and serves as a transcriptional repressor of MMP9. Interestingly, how these external stimulators and negative regulators integrate to modulate the de-repression of ZBRK1 and subsequently activate the transcription of MMP9 will be a complicated but exciting subject to resolve.
Based on the global profiling regulated by ZBRK1, several other interesting candidates involved in two processes of migration and invasion were identified (SFig. 2 and Supplemental table 1), which accounting for the function of ZBRK1 as a tumor suppressor, especially in repression of metastasis/invasion. For instance, ZBRK1 inhibits MMP3, LAMA1, FGF18, EGR1, and HMGA2, in addition to MMP9 but activates ICAM1, ANK1, NCAM1 and VIM. Therefore, the inactivation of ZBRK1 which results in the increase of MMP3 and MMP9, and the decrease of ICAM1 and NCAM1 may benefit the cancer cells migrate from extracellular matrix of origin for invasion. ZBRK1 directly represses the MMP9 expression through binding to its promoter, similarly to other ZBRK1 regulated genes including ANG1 (22). Although the detailed mechanism of transcriptional repression remains to be unraveled, one possibility is through its partner proteins such as BRCA1 and KAP1 to differentially regulate these genes. It was reported that ZBRK1 interacts with BRCA1 through its C- terminus to negatively regulate the transcription of GADD45 and ANG1 (35), while p21 transcription was repressed by ZBRK1 through KAP1 recruitment (21). It is likely that either BRCA1 or KAP1 will work with ZBRK1 to repress MMP9. For those genes repressed by ZBRK1, it will be interesting to classify which one is going through BRCA1 or KAP1 or both. Furthermore, how ZBRK1 activates the expression of ICAM1 and other genes is another unknown process. ZBRK1 apparently can directly bind to the putative ZBRK1 motif on the ICAM1 promoter based on our EMSA. However, it is not known which co-activator will interact with ZBRK1 to turn on the expression of ICAM1. Elucidating each specific regulation mode of ZBRK1 will provide important information to explain its tumor-suppressive effects.
ZBRK1 mutation rarely occurs in tumor cells (35, 36). This current study clearly showed that reduction of ZBRK1 transcription in 75% of cervical cancer specimens is crucial for cancer progression. Therefore, elucidation of ZBRK inactivation in cancer cells would be an interesting issue. The ZBRK1 gene locates on chromosome 19q13.41. A recent publication demonstrated a high frequency of 19q12 deletion (37), yet the frequent deleted locus is 19p13.11-q12 (15% cases), which does not include the ZBRK1 gene. This suggests that the transcriptional regulation or the loss of heterozygosity may explain the inactivation of ZBRK1 gene, which will be the focus of the next pursuit in order to fully elucidate the contribution of ZBRK1 during cancer progression.
Supplementary Material
Acknowledgments
This work was supported in part by the grants to YMW (NSC 98-2320-B-006-036-MY3 and the NCKU landmark grant of C007, and WHL (NIH RO1 94170. Funding to pay the Open Access publication charges for this article was provided by the grant of NSC 98-2320-B-006-036-MY3 To YMW. Thanks are due to Miss Christine C. Hsieh and Dr. Kazi Ahmed for review of this manuscript.
References
- 1.Parker B, Sukumar S. Distant metastasis in breast cancer: molecular mechanisms and therapeutic targets. Cancer Biol Ther. 2003;2:14–21. doi: 10.4161/cbt.188. [DOI] [PubMed] [Google Scholar]
- 2.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–72. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 3.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 4.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 6.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 7.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 9.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome biology. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–52. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 11.Sun Y, Tan M, Duan H, Swaroop M. SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal. 2001;3:635–50. doi: 10.1089/15230860152542989. [DOI] [PubMed] [Google Scholar]
- 12.Friedman JR, Fredericks WJ, Jensen DE, et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–78. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 13.Kim SS, Chen YM, O’Leary E, Witzgall R, Vidal M, Bonventre JV. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:15299–304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moosmann P, Georgiev O, Le Douarin B, Bourquin JP, Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic acids research. 1996;24:4859–67. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 16.Peng H, Zheng L, Lee WH, Rux JJ, Rauscher FJ., 3rd A common DNA-binding site for SZF1 and the BRCA1-associated zinc finger protein, ZBRK1. Cancer Res. 2002;62:3773–81. [PubMed] [Google Scholar]
- 17.Chen CF, Li S, Chen Y, Chen PL, Sharp ZD, Lee WH. The nuclear localization sequences of the BRCA1 protein interact with the importin-alpha subunit of the nuclear transport signal receptor. J Biol Chem. 1996;271:32863–8. doi: 10.1074/jbc.271.51.32863. [DOI] [PubMed] [Google Scholar]
- 18.Bellefroid EJ, Poncelet DA, Lecocq PJ, Revelant O, Martial JA. The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc Natl Acad Sci U S A. 1991;88:3608–12. doi: 10.1073/pnas.88.9.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng L, Pan H, Li S, et al. Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol Cell. 2000;6:757–68. doi: 10.1016/s1097-2765(00)00075-7. [DOI] [PubMed] [Google Scholar]
- 20.Yun J, Lee WH. Degradation of transcription repressor ZBRK1 through the ubiquitin-proteasome pathway relieves repression of Gadd45a upon DNA damage. Mol Cell Biol. 2003;23:7305–14. doi: 10.1128/MCB.23.20.7305-7314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YK, Thomas SN, Yang AJ, Ann DK. Doxorubicin down-regulates Kruppel-associated box domain-associated protein 1 sumoylation that relieves its transcription repression on p21WAF1/CIP1 in breast cancer MCF-7 cells. J Biol Chem. 2007;282:1595–606. doi: 10.1074/jbc.M606306200. [DOI] [PubMed] [Google Scholar]
- 22.Furuta S, Wang JM, Wei S, et al. Removal of BRCA1/CtIP/ZBRK1 repressor complex on ANG1 promoter leads to accelerated mammary tumor growth contributed by prominent vasculature. Cancer Cell. 2006;10:13–24. doi: 10.1016/j.ccr.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Ko CY, Hsu HC, Shen MR, Chang WC, Wang JM. Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. J Biol Chem. 2008;283:30919–32. doi: 10.1074/jbc.M804029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pratap J, Javed A, Languino LR, et al. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol. 2005;25:8581–91. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Liang J, Castrillon DH, DePinho RA, Olson EN, Liu ZP. FoxO4 regulates tumor necrosis factor alpha-directed smooth muscle cell migration by activating matrix metalloproteinase 9 gene transcription. Mol Cell Biol. 2007;27:2676–86. doi: 10.1128/MCB.01748-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uttamsingh S, Bao X, Nguyen KT, et al. Synergistic effect between EGF and TGF-beta1 in inducing oncogenic properties of intestinal epithelial cells. Oncogene. 2008;27:2626–34. doi: 10.1038/sj.onc.1210915. [DOI] [PubMed] [Google Scholar]
- 27.Gobeil S, Zhu X, Doillon CJ, Green MR. A genome-wide shRNA screen identifies GAS1 as a novel melanoma metastasis suppressor gene. Genes Dev. 2008;22:2932–40. doi: 10.1101/gad.1714608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 29.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–74. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 30.Chandrasekar B, Mummidi S, Mahimainathan L, et al. Interleukin-18-induced human coronary artery smooth muscle cell migration is dependent on NF-kappaB-and AP-1-mediated matrix metalloproteinase-9 expression and is inhibited by atorvastatin. J Biol Chem. 2006;281:15099–109. doi: 10.1074/jbc.M600200200. [DOI] [PubMed] [Google Scholar]
- 31.Chou YT, Wang H, Chen Y, Danielpour D, Yang YC. Cited2 modulates TGF-beta-mediated upregulation of MMP9. Oncogene. 2006;25:5547–60. doi: 10.1038/sj.onc.1209552. [DOI] [PubMed] [Google Scholar]
- 32.Song H, Li Y, Lee J, Schwartz AL, Bu G. Low-density lipoprotein receptor-related protein 1 promotes cancer cell migration and invasion by inducing the expression of matrix metalloproteinases 2 and 9. Cancer Res. 2009;69:879–86. doi: 10.1158/0008-5472.CAN-08-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuga H, Morisaki T, Nakamura K, et al. Interferon-gamma suppresses transforming growth factor-beta-induced invasion of gastric carcinoma cells through cross-talk of Smad pathway in a three-dimensional culture model. Oncogene. 2003;22:7838–47. doi: 10.1038/sj.onc.1207046. [DOI] [PubMed] [Google Scholar]
- 34.Krishnamurthy P, Rajasingh J, Lambers E, Qin G, Losordo DW, Kishore R. IL-10 inhibits inflammation and attenuates left ventricular remodeling after myocardial infarction via activation of STAT3 and suppression of HuR. Circ Res. 2009;104:e9–18. doi: 10.1161/CIRCRESAHA.108.188243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia V, Garcia JM, Pena C, et al. The GADD45, ZBRK1 and BRCA1 pathway: quantitative analysis of mRNA expression in colon carcinomas. J Pathol. 2005;206:92–9. doi: 10.1002/path.1751. [DOI] [PubMed] [Google Scholar]
- 36.Ayyanathan K, Lechner MS, Bell P, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–69. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilting SM, Steenbergen RD, Tijssen M, et al. Chromosomal signatures of a subset of high-grade premalignant cervical lesions closely resemble invasive carcinomas. Cancer Res. 2009;69:647–55. doi: 10.1158/0008-5472.CAN-08-2478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.