Abstract
A survey of restriction site polymorphism in the mitochondrial DNA (mtDNA) of the American eel Anguilla rostrata showed no genetic divergence among samples from a 4000-km stretch of North America coastline. Lack of geographic differentiation in mtDNA over such a large area contrasts sharply with results for terrestrial and freshwater vertebrates and is most likely attributable to the extraordinary life history of these catadromous fishes, which involves perhaps a single spawning population in the western tropical mid-Atlantic Ocean and subsequent widespread dispersal of larvae by ocean currents. However, samples of the European eel (nominally Anguilla anguilla) are highly distinct from A. rostrata in mtDNA genotype (distinguishable by 11 of 14 restriction endonucleases), contradicting some previous suggestions that the two forms belong to the same panmictic population. Results of this study emphasize the importance of life history in shaping population genetic structure.
Keywords: Anguilla, population structure, dispersal, panmixia, catadromous fishes
Full text
PDF
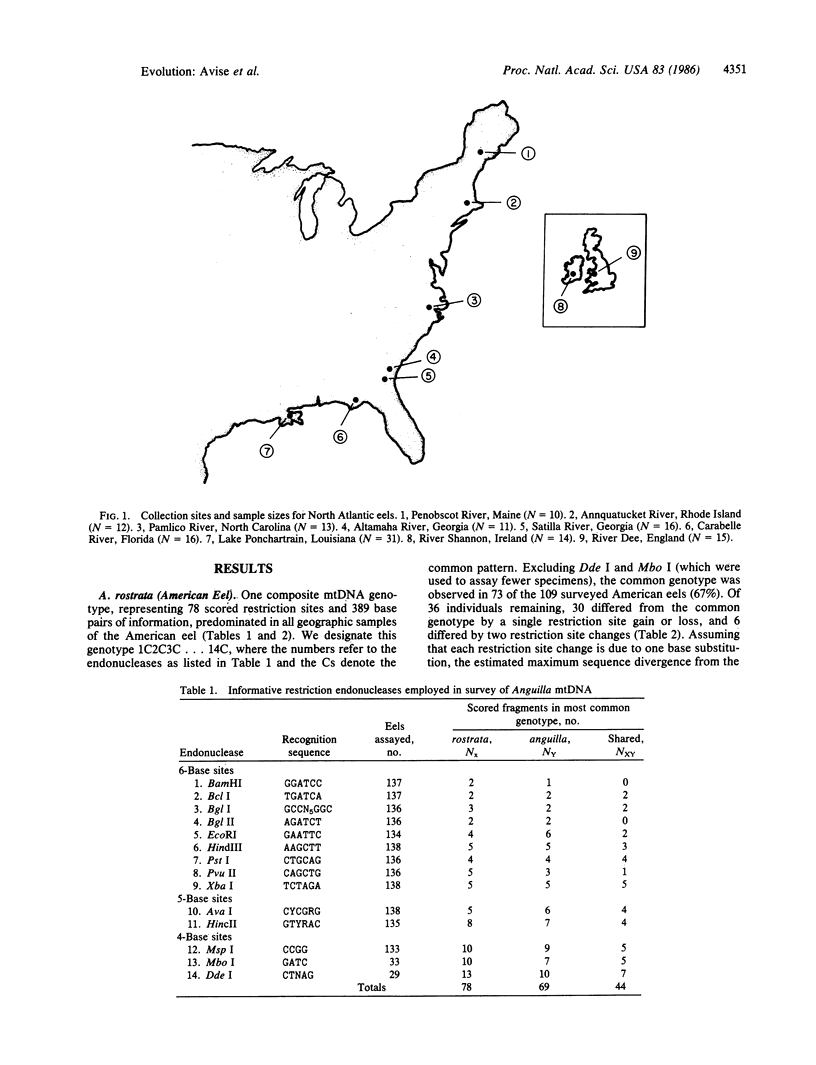
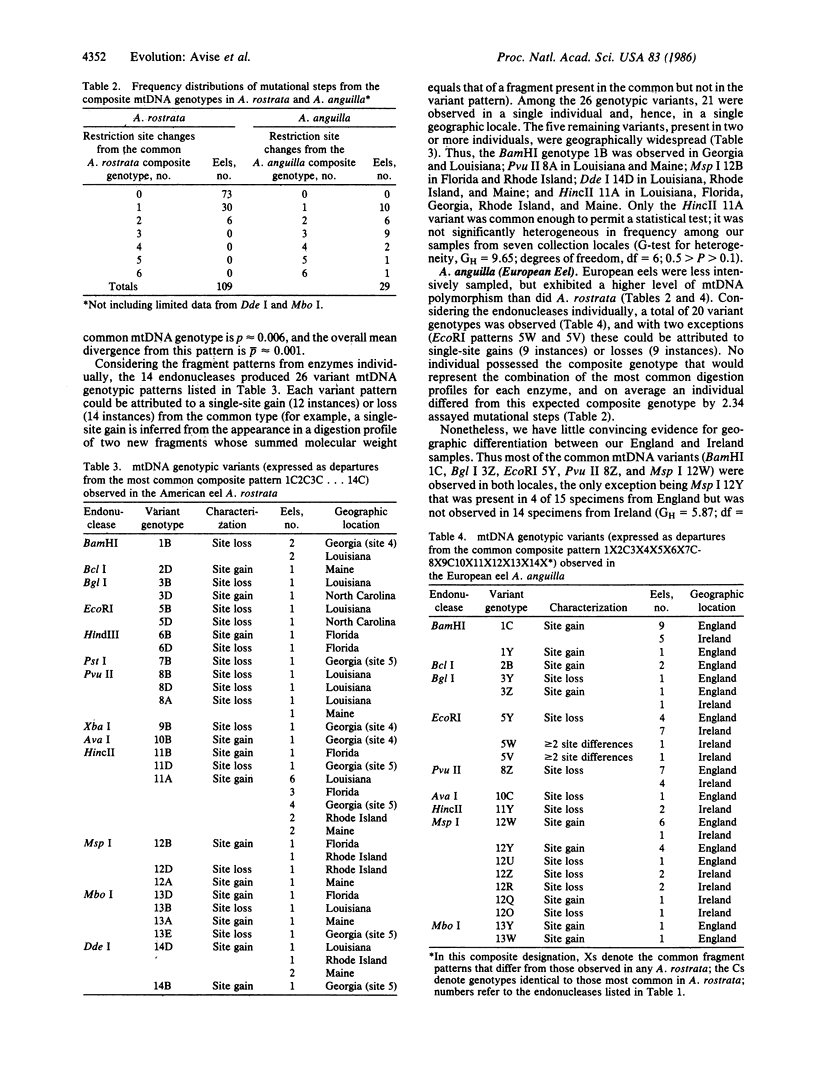
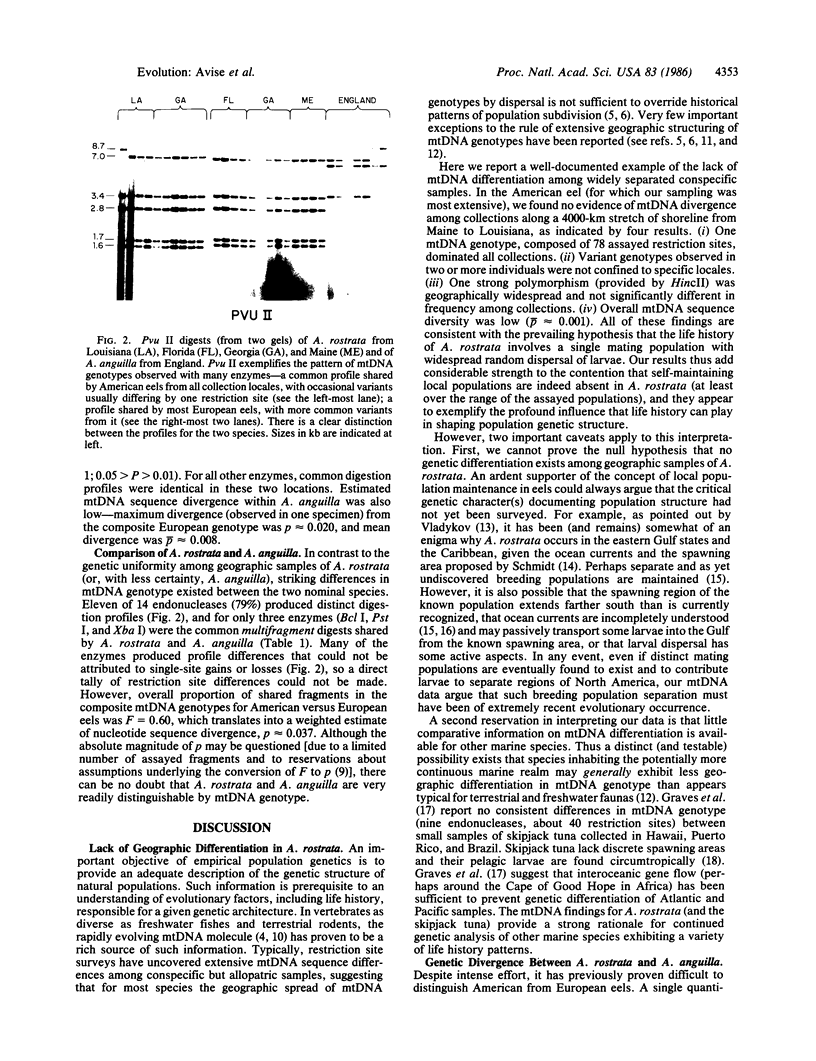

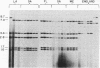
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M. Polymorphism in mitochondrial DNA of humans as revealed by restriction endonuclease analysis. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3605–3609. doi: 10.1073/pnas.77.6.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansman R. A., Shade R. O., Shapira J. F., Avise J. C. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. III. Techniques and potential applications. J Mol Evol. 1981;17(4):214–226. doi: 10.1007/BF01732759. [DOI] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]