Abstract
A variety of manganese-containing coordination compounds, frequently termed superoxide dismutase (SOD) mimics, have been reported to have SOD activity in vitro and to be effective at improving conditions related to increased oxidative stress in multicellular organisms. We tested the effectiveness of several of these compounds in substituting for authentic SOD enzymes in two simple systems – the prokaryote Escherichia coli and the single-celled eukaryote, Saccharomyces cerevisiae—where strains are available that completely lack cytoplasmic SOD activity and are thus significantly impaired in their ability to grow aerobically. Most of the compounds tested, including Euk-8 and Euk-134, manganese salen derivatives developed by Eukarion, M40403, a manganese complex of a bis(cyclohexylpyridine)-substituted macrocyclic ligand developed by Metaphore, and several manganese porphyrin derivatives, were ineffective in both systems. Only the manganese tetrapyridyl porphyrin complex MnTM-2-PyP and two close relatives were effective in rescuing aerobic growth of E. coli lacking SOD, and, in the case of sod1Δ yeast, only MnTM-2-PyP itself was fully effective. Surprisingly, several compounds reported to be beneficial in other in vivo model systems (Euk-8, Euk-134, M40403) were actually toxic to these organisms lacking SOD, although they had no effect on the wild type parent strains. Our results suggest the possibility that the beneficial effects of some of the so-called “SOD mimic drugs” may be due to some property other than in vivo superoxide dismutase activity.
1. Introduction
Oxidative stress has been implicated in a variety of detrimental health conditions including ischemia-reperfusion injury, chronic inflammation, neurodegenerative disease, and aging [1]. The potential medical utility of a compound (drug or supplement) that would reduce oxidative stress has led to the development and testing of a variety of manganese-containing coordination compounds that have been termed superoxide dismutase (SOD) mimics. In some cases, substantial beneficial effects have been reported for in vivo tests of these compounds (see below for references).
The exact molecular mechanisms by which the manganese-containing SOD mimics function in vivo are unknown, and we therefore decided to address this question using two very simple model systems—bacteria (E. coli) and yeast (S. cerevisiae) that completely lack their cytoplasmic SOD enzymes—to screen compounds for their ability to substitute for bona fide SOD enzymes. We chose a prokaryotic system because of its simplicity and lack of intracellular compartmentalization and a eukaryotic single-celled organism because of its more complex subcellular organization and closer resemblance to higher organisms.
Wild type E. coli have iron-containing and manganese-containing cytoplasmic SODs. E. coli lacking both of these cytosolic SODs, termed sodA−sodB−, exhibit several phenotypes that can be directly attributed to the loss of SOD activity, e.g., poor aerobic growth [2] and auxotrophies for branched-chain [3], aromatic [4] and sulfur-containing amino acids [5]. Most eukaryotes, including yeast, have a single abundant, copper- and zinc-containing SOD (CuZnSOD or SOD1) in the cytosol (which is also present in the nucleus and some other cellular locations) and a much less abundant MnSOD in the mitochondrial matrix. Yeast lacking CuZnSOD (sod1Δ) exhibit a variety of aerobic phenotypes including poor growth, lysine and methionine auxotrophies, and decreased stationary phase survival [6–10]. In both of these systems, i.e., sodA−sodB− E. coli and sod1Δ S. cerevisiae, growth under anaerobic conditions reverses these phenotypes.
We recently demonstrated that all known phenotypes of sod1Δ yeast were completely reversed when the growth medium was supplemented with high levels (5 mM) of ionic manganese [11]. Simple manganese salts have also been reported partially to reverse the phenotypes of sodA− sodB− E. coli, including slow aerobic growth rates, lowered aconitase activity, and sensitivity to hydrogen peroxide [2]. Much lower levels of manganese salts (10 – 100 µM) are effective in the latter case. The exact molecular mechanisms by which ionic manganese functionally substitutes for the SOD enzymes in yeast and E. coli lacking SOD enzymes are not yet known, and we wondered whether the mechanisms of action of the manganese-containing SOD mimics could be related.
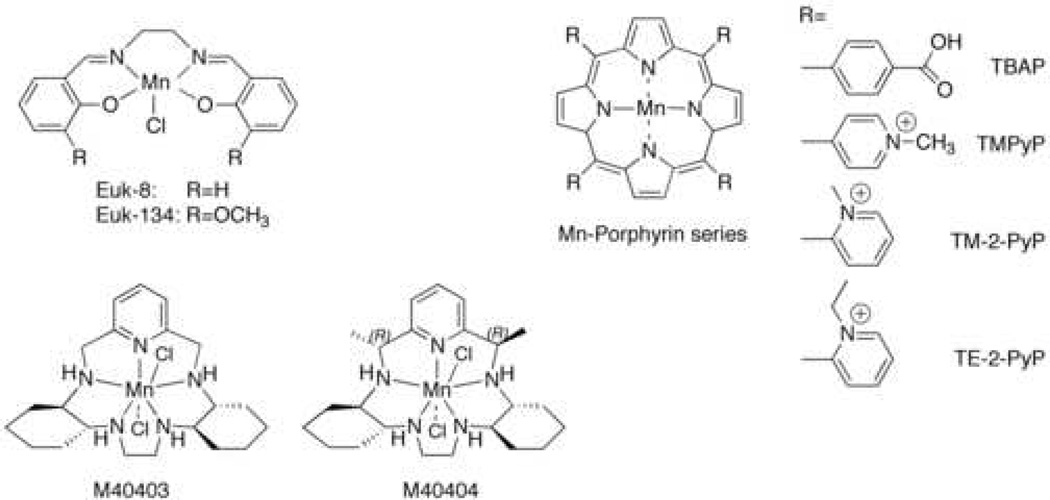
The structures of the SOD mimics that we examined are shown in Scheme 1. The Euk class are manganese salen derivatives that are reported to possess both SOD and catalase activities in vitro, properties which could in theory allow these molecules to protect against a wider range of reactive oxygen species (ROS) and thereby lower oxidative stress [12]. Euk-8 is the prototypical manganese salen SOD mimic; alterations made to the ligand doubled the catalase activity (Euk-134) or improved the solubility of the compound in organic solvents (Euk-189) without significantly affecting the SOD activity. It should be noted, however, that Euk-8 and Euk-134 have been reported to lose activity upon treatment with EDTA [13].
Scheme 1.
The EUK compounds have been shown to be effective treatments for several different types of oxidative stress-related conditions, such as ischemia-reperfusion injury in a rat stroke model [14], ROS-induced apoptosis [15], and paraquat-induced dopaminergic cell death in neurons [16]. They have also been reported to improve survival of sod2 null mice [17], and dose-dependent increases in life span occurred in Caenorhabditis elegans under some growth conditions [18, 19] indicating these compounds entered the cell. Another study of Eukarion compounds in C. elegans found adverse rather than beneficial effects with dose-dependent decreases in life span and fertility [13, 20]. The Eukarion compounds also did not extend life span in the housefly [21].
The SOD mimics from Metaphore are highly stable manganese complexes of a bis(cyclohexylpyridine)-substituted macrocyclic ligand derivatives that retain SOD activity in the presence of EDTA [22]. They are reported to react specifically with superoxide and not with other ROS or reactive nitrogen species [23, 24]. The SOD-active compound, M40403 (see Scheme 1) was shown to be effective in a variety of model systems, such as ischemia-reperfusion injury in rat heart [25] and inflammation in rat [26, 27]. There is an SOD-inactive, but chemically similar, analog which is quite useful as a control [28]. The chemical basis for this interesting difference in reactivity has been carefully studied by D. Riley and colleagues [29].
Fridovich, Spasojevic, Batinic-Haberle, and coworkers pioneered the use of manganese complexes of water-soluble tetraaryl porphyrin ligands as SOD mimics, all of which are stable in the presence of EDTA. MnTBAP (Scheme 1) exhibited SOD activity in vitro and was reported to improve growth of E. coli lacking superoxide dismutase enzymes [30], to reduce mitochondrial oxidative stress induced by the nucleoside reverse transcriptase inhibitor stavudine [31], and to improve the survival of sod2 deficient neurons [32]. This compound also improved survival of SOD2-deficient mice, although not as well as Euk-8 or Euk-134 [17]. Several related manganese tetrapyridyl porphyrin-based SOD mimics, such as MnTM-2-PyP (Scheme 1) have also been developed, and some are reported to be more effective biologically than MnTBAP [33, 34].
2. Materials and Methods
2.1. Growth conditions for yeast studies
Wild type (EG103; MATα; leu2-3,112; his3Δ1; trp-289a; ura3-52; GAL+) and sod1Δ (EG118; MATα; leu2-3 112; his3Δ1; trp-289a; ura3-52; GAL+; sod1Δ::URA3) strains of S. cerevisiae were streaked from 20% glycerol freezer stocks onto YPD agar plates [35] and incubated under low oxygen conditions (approximately 5% oxygen, Campy Bags, Becton Dickinson Microbiology Systems) at 30 °C for three days [10]. Unless indicated otherwise, experimental yeast cultures and pre-cultures were grown in synthetic complete media (SC) containing 2% glucose (SDC) [35] with a four-fold increase in the concentrations of Leu, His, Trp, Met, Ura, and Ade. Pre-cultures were prepared by inoculating single colonies in 5 ml of SDC in 16 ml culture tubes and growing them overnight at 30 °C and 220 RPM. Experimental cultures were then inoculated at an OD600 of 0.05 (~5 × 105 cells/ml) in 10 mL liquid medium in a 50 mL flask and incubated in air at 30 °C and 220 RPM. To monitor growth, cells were diluted and optical density at 600 nm (OD600) was measured; for yeast, an OD600 of 1 represents approximately 1 × 107 cells per ml. Each experiment included two or three independent colonies for each yeast strain, and was repeated at least twice on different days. Error bars represent standard deviation.
2.2. Growth conditions for E. coli
E. coli strains used were as follows: Wild type (AB1157; F- thr-1; leuB6; proA2; his-4; thi-1; argE2; lacY1; galK2; rpsL; supE44; ara-14; xyl-15; mtl-1; tsx-33) and sodA− sodB− (PN134; same as wild type, except with (sodA::Mu d PR13)25 (sodB−kan)1-Δ2). Cells were streaked from freezer stocks onto LB agar plates supplemented with 0.2% sucrose and incubated in air for 24 hours at 37 °C [36]. Four independent colonies for each strain were each inoculated into three mL LB-sucrose and shaken at 220 RPM in air at 37 °C overnight. These overnights were used to inoculate starter cultures as follows. In 50 mL culture flasks, wild-type E. coli were inoculated into 15 mL LB-sucrose at an OD600 of 0.01, or approximately 1 × 107 cells per ml, and sodA−sodB− E. coli was inoculated into 30 mL LB-sucrose at an OD600 of 0.02. Both were incubated for 2 – 3 hours while shaking in air at 220 RPM at 37 °C. (Sucrose was included to support the growth of sodA−sodB− mutants and minimize the likelihood of suppressors overtaking the cultures during the pregrowth period [37].) Cells were then washed three times with minimal medium (adapted from Miller [38] by addition of 1 mM MgSO4•7 H2O, 16.6 µM thiamine, 0.2% glucose, and 0.5 mM of the L-amino acids Leu, Pro, His, Arg and Thr). For experimental cultures, 96-well microtiter plates with 100 µL of minimal medium per well were inoculated with 10 µl cell suspension per well. Each plate contained vehicle-treated controls, 5.0-µM manganese chloride as a positive control, and a range of concentrations of the test compound(s). Samples containing 5.0-µM manganese chloride and 30.0 µM of the test compound were sometimes included to look for synergistic effects; none were observed (data not shown). The microtiter plates were secured in a 37 °C incubator and shaken at 220 RPM. Growth was measured by recording well turbidity at 600 nm hourly for ten hours using a Molecular Dynamics microtiter plate reader. For experiments with manganese porphyrin complexes, the well turbidity was monitored at 700 nm to reduce interference from the absorbance of the compounds [30]. Doubling times calculated from either 700 nm or 600 nm measurements were comparable to each other (data not shown). Experiments always included four independent colonies for each strain and were repeated on at least two different days. Where n is mentioned, it represents the total number of independent colonies tested. Error bars represent standard deviation.
2.3. Manganese mimic compounds
The structures of the manganese complexes are shown in Scheme 1. Euk-8 and Euk-134 were generous gifts from Dr. Susan Doctrow, Eukarion Inc., Bedford, MA. The Metaphore compounds were generously supplied by Dr. Dennis Riley, Metaphore Pharmaceuticals, Fort Lee, NJ, or synthesized as described [39]. MnTBAP (manganese tetrakis (4-benzoic acid) porphyrin) was purchased from Oxis International Inc., Portland, OR or Calbiochem, San Diego, CA. MnTM-PyP (manganese 5,10,15,20-tetrakis (N-methylpyridinium-4-yl) porphyrin), MnTM-2-PyP (manganese 5,10,15,20-tetrakis (N-methylpyridinium-2-yl) porphyrin), and MnTE-2-PyP (manganese 5,10,15,20-tetrakis (N-ethylpyridinium-2-yl) porphyrin) were from Calbiochem, San Diego, CA. Stock solutions were prepared in water at 10 mM for Euk-8 and Euk-134, at 1.0 mM for M40403 and M40404, and at 10 mM for the Mn porphyrins, and filter sterilized with a 0.22-µM filter (Whatman, Middlesex, UK). MnTBAP was dissolved in DMSO or alkaline water. Mn(II)EDTA2− was prepared by adding a molar equivalent of MnCl2•4H2O to 0.01 M EDTA, pH 8.0.
3. Results
3.1. Eukarion compounds
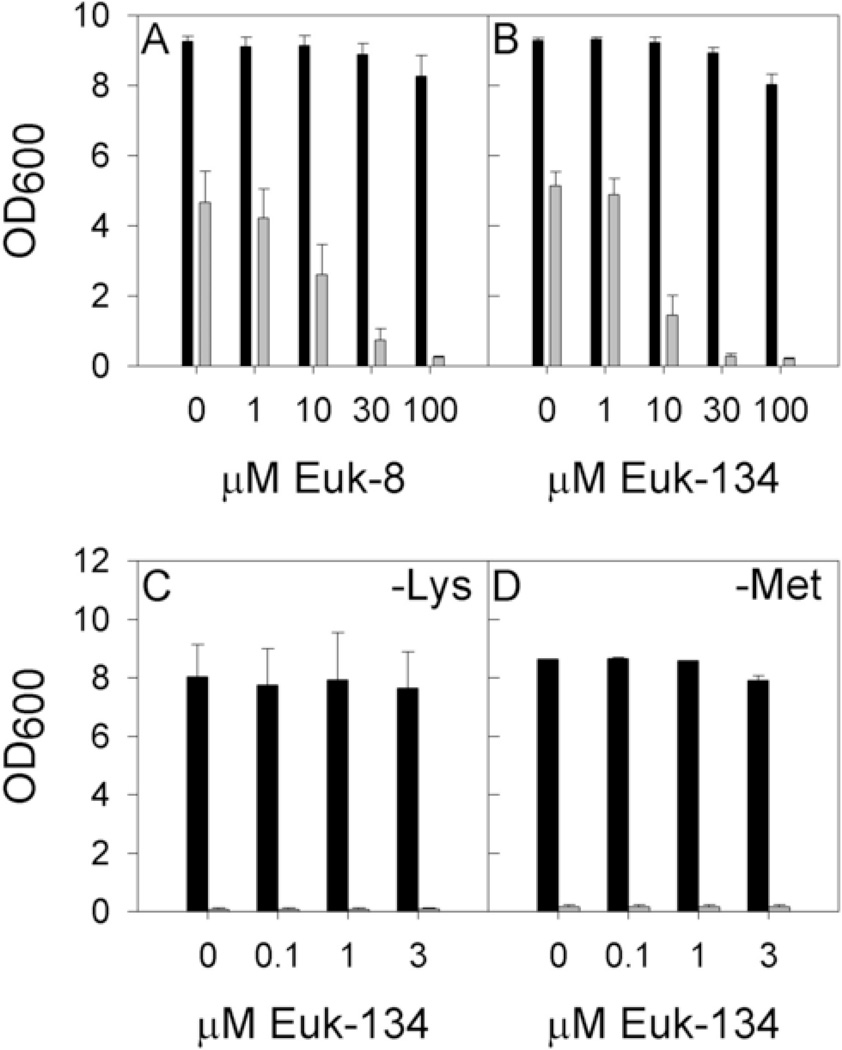
In our initial experiments, wild type and sod1Δ yeast were cultured in SDC, pH 6.0, in the presence of Euk-8 or Euk-134, and cell density was recorded after 24 hours (Figs. 1A and 1B). To our surprise, the poor aerobic growth of sod1Δ S. cerevisiae was not improved by treatment with any Eukarion compound tested, and, in fact, at concentrations of 10 µM and above, inhibition of growth was evident. These compounds had no effect, either positive or negative, on growth of wild-type S. cerevisiae.
Fig. 1. Aerobic growth of wild type and sod1Δ S. cerevisiae with Eukarion compounds.
Wild type (black bars) and sod1Δ (grey bars) S. cerevisiae were grown aerobically for 24 hours in the presence of Eukarion SOD/catalase mimics as described in Materials and Methods. Growth was measured by recording culture turbidity at 600 nm (OD600). Yeast cultures were grown under the following conditions: (A) in SDC pH 6.0 with Euk-8, (B) in SDC pH 6.0 with Euk-134, (C) in SD-Lys pH 6.0 with Euk-134, and (D) in SD-Met pH 6.0 with Euk-134. Error bars represent standard deviation.
In order for sod1Δ yeast to grow under aerobic conditions, both lysine and methionine must be present in the medium [7, 8]. We wondered whether any beneficial effects of the Eukarion compounds might be more evident under these more stringent conditions. However, as shown in Figs. 1C and 1D, Euk-134 at non-toxic levels did not rescue the lysine (Fig. 1C) or methionine (Fig. 1D) auxotrophies. Again, the growth of the wild type was not affected by the presence of Euk-134. Similar results were obtained for Euk-8 (data not shown).
In the case of the bacterial system, we developed a new assay method using microtiter plates, which allowed us to carry out growth assays with many replicates very quickly using minimal amounts of material. By using no more than 110 µl in each well of a 96-well plate and shaking the plates at 220 rpm, we were able to achieve an oxidative environment high enough to trigger the phenotype of sodA−sodB− E. coli. The assay is described in detail in Materials and Methods.
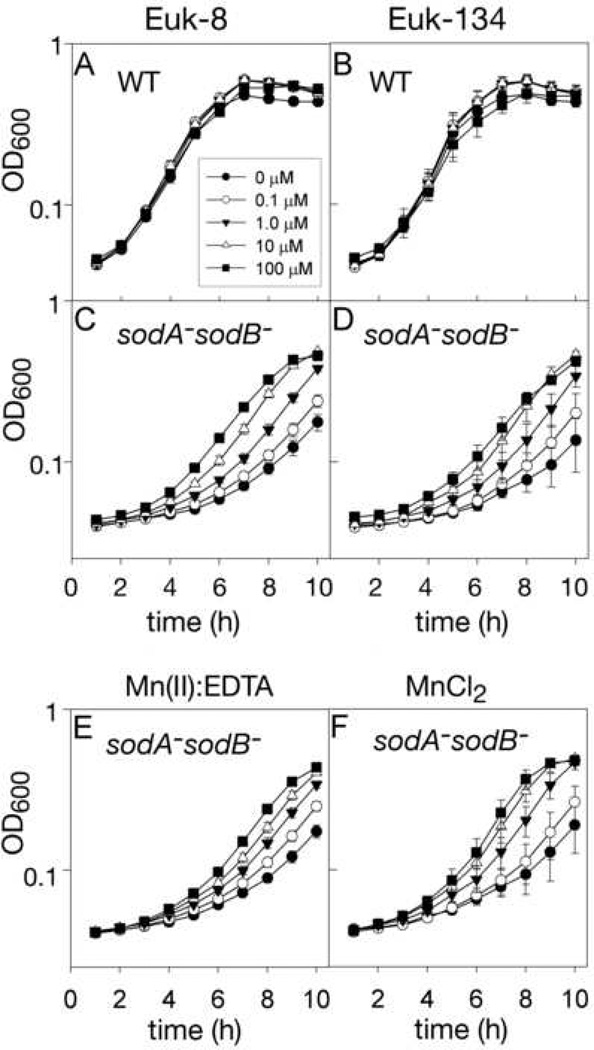
Euk-8 and Euk-134 were tested for their ability to improve the slow growth rate of sodA−sodB− E. coli in air using the new assay system described above. Neither compound, at concentrations up to 100 µM, significantly altered the aerobic growth rate of wild-type E. coli (Fig.s 2A and 2B), but, the growth of the sodA−sodB− strain was significantly improved by treatment with up to 100 µM Euk-8 or Euk-134, as shown in Figs. 2C and 2D. At concentrations of 300 µM or greater, both Eukarion compounds impaired the growth of wild type and sodA−sodB− E. coli (data not shown).
Fig. 2. Aerobic growth of wild type and sodA− sodB− E. coli with Eukarion compounds or Mn(II).
Cultures of E. coli were grown aerobically in 96-well microtiter plates as described in the Methods section with treatments as indicated. Growth was recorded hourly by measuring well turbidity (as OD600) using a microtiter plate reader. Error bars represent standard deviation. (A) Wild type with various concentrations of Euk-8 (n=16); (B) wild type with Euk-134 (n=8); (C) sodA− sodB− with Euk-8 (n=16); (D) sodA− sodB− with Euk-134 (n=8); (E) sodA− sodB− grown with Mn(II):EDTA (n=24); (F) sodA− sodB− E. coli were grown with MnCl2 (n=8). Solid circles, 0 µM; open circles 0.1 µM; solid triangles, 1.0 µM; open triangles 10 µM; solid squares, 100 µM.
Previously published results indicated that manganese chloride can itself improve growth of sodA− sodB− E. coli [2] and that Mn(II)EDTA2− possesses some superoxide scavenging activity in vitro [2, 40]. For comparative purposes, therefore, we also tested these two manganese compounds in our assay. As can be seen in Figs. 2E and 2F, both showed a degree of rescue similar to that seen for Euk-8 and Euk-134 [41]. In contrast to the case with the SOD mimics, MnCl2 and Mn(II)EDTA2− at up to 1 mM or more showed no sign of toxicity, although uncomplexed EDTA strongly inhibited growth at low concentrations (data not shown).
3.2. Metaphore compounds
The SOD mimic from Metaphore, M40403, is reported to have SOD but not catalase activity in vitro, and it is quite stable chemically and highly unlikely to dissociate manganese ions under the conditions of our experiments [22–24]. The structurally similar but SOD-inactive compound, M40404, provided a useful control.
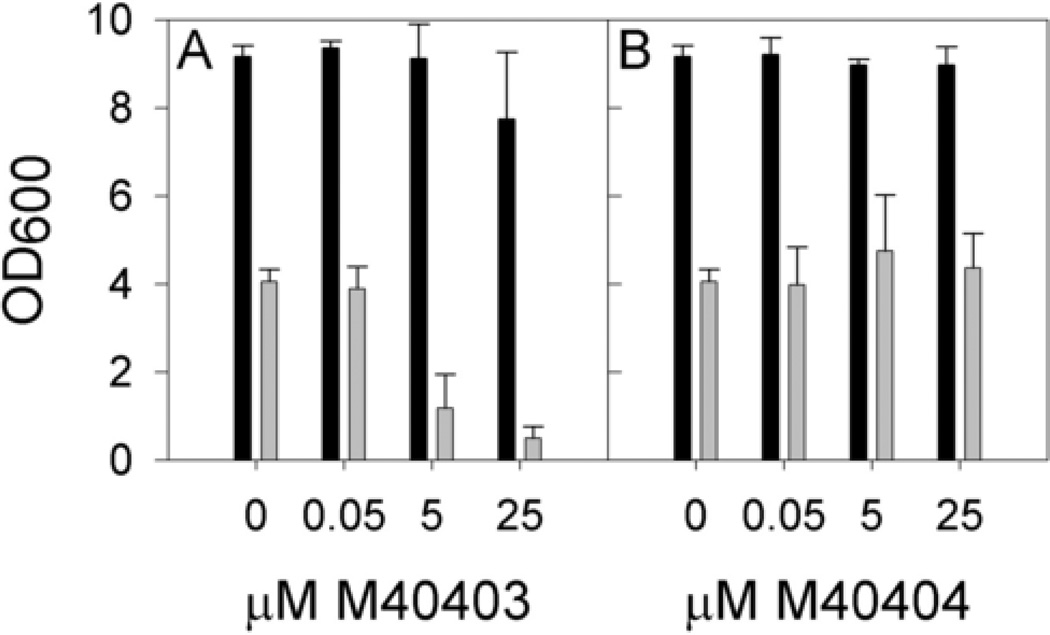
Wild type and sod1Δ yeast were grown with varying levels of the SOD-active compound, M40403, or the SOD-inactive compound, M40404, and culture density was measured at 24 hours (Fig. 3). Growth of wild-type S. cerevisiae was unaffected by treatment with either compound. However, concentrations of 5 µM or above of M40403 reduced the final cell density of the sod1Δ S. cerevisiae cultures. These results are in stark contrast to those observed for the SOD-inactive compound, M40404, which had no effect on growth of the sod1Δ strain. In conclusion, the SOD-active compound was growth inhibitory to sod1Δ yeast, but the SOD-inactive compound was not. Because the two compounds are quite similar chemically, it is unlikely that one is taken up by the cells while the other is not, but we do not have definitive proof that this is the case.
Fig. 3. Aerobic growth of wild type and sod1Δ S. cerevisiae with Metaphore compounds.
Wild type (black bars) and sod1Δ S. cerevisiae (grey bars) were grown in flasks as described in Materials and Methods in the presence of (A) M40403, the active compound, or (B) M40404, the inactive compound. Growth was measured via culture turbidity at 600 nm (OD600), and values shown are averages of 4 independent colonies grown on two different days with error bars representing standard deviation.
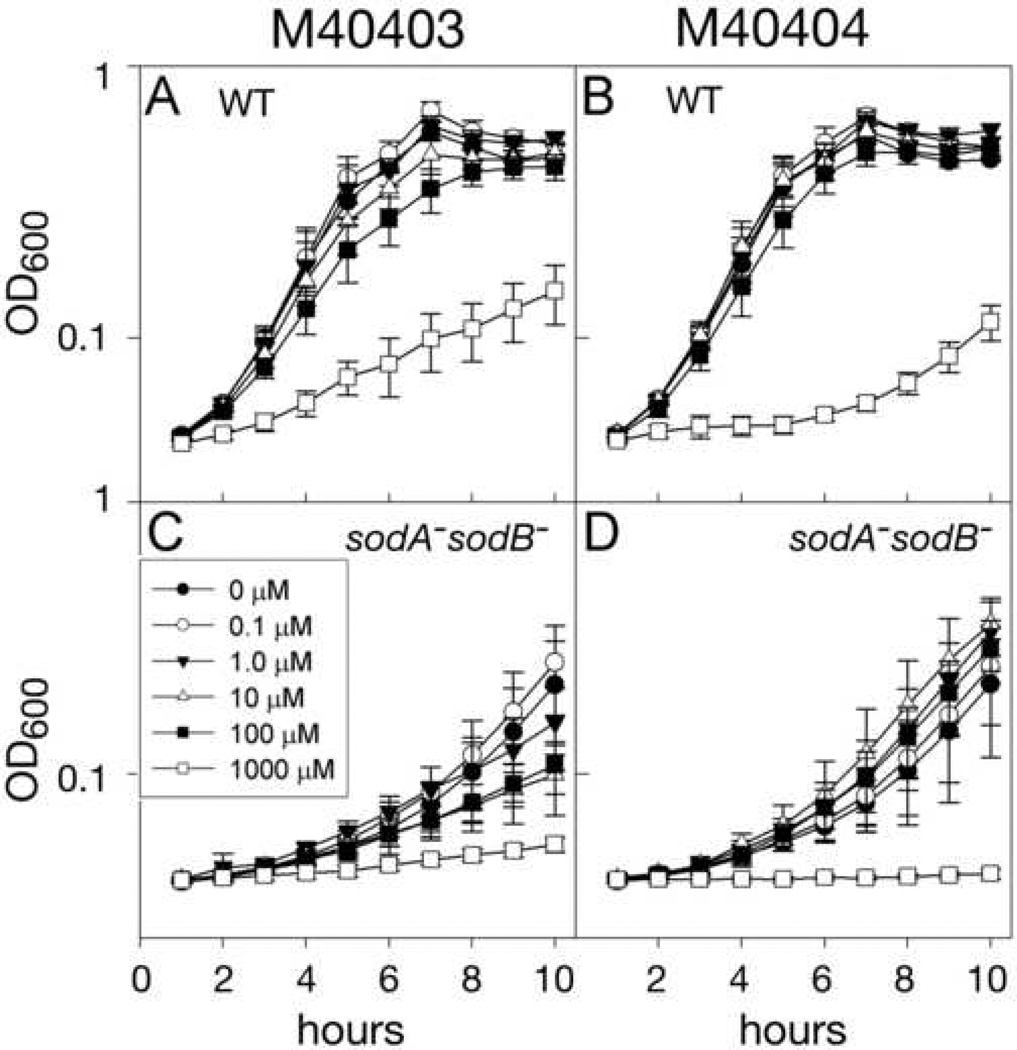
In the E. coli system, using the microtiter plate assay system described above, low levels of the SOD-active compound M40403 did not affect growth of either wild type or sodA−sodB−, but higher levels reduced growth relative to the untreated control (Figs. 4A and 4C). This reduction in growth rate was more pronounced in the sodA−sodB− strain, but was evident in the wild type as well. M40404, the inactive analog, had little effect on growth of wild-type E. coli until a concentration of 1 mM was reached, at which point it was highly growth-inhibitory (Fig. 4B). sodA− sodB− E. coli cultures treated with ≤ 100 µM M40404 grew marginally better than the control culture but the effect was less pronounced than that of MnCl2 alone, and, once again, 1 mM M40404 completely inhibited growth (Fig. 4D).
Fig. 4. Aerobic growth of wild type and sodA− sodB− E. coli with Metaphore compounds.
E. coli were cultured as described in Materials and Methods, in 100 µL minimal medium supplemented as indicated in 96-well microtiter plates. Growth was recorded hourly as turbidity (OD600) using a microtiter plate reader. Results shown are averages of eight cultures for each condition (n=8). Wild type (A) and sodA− sodB− E. coli (C) with the SOD-active compound M40403; Wild type (B) and sodA− sodB− E. coli (D) with the SOD-inactive compound M40404.
3.3 Manganese porphyrins
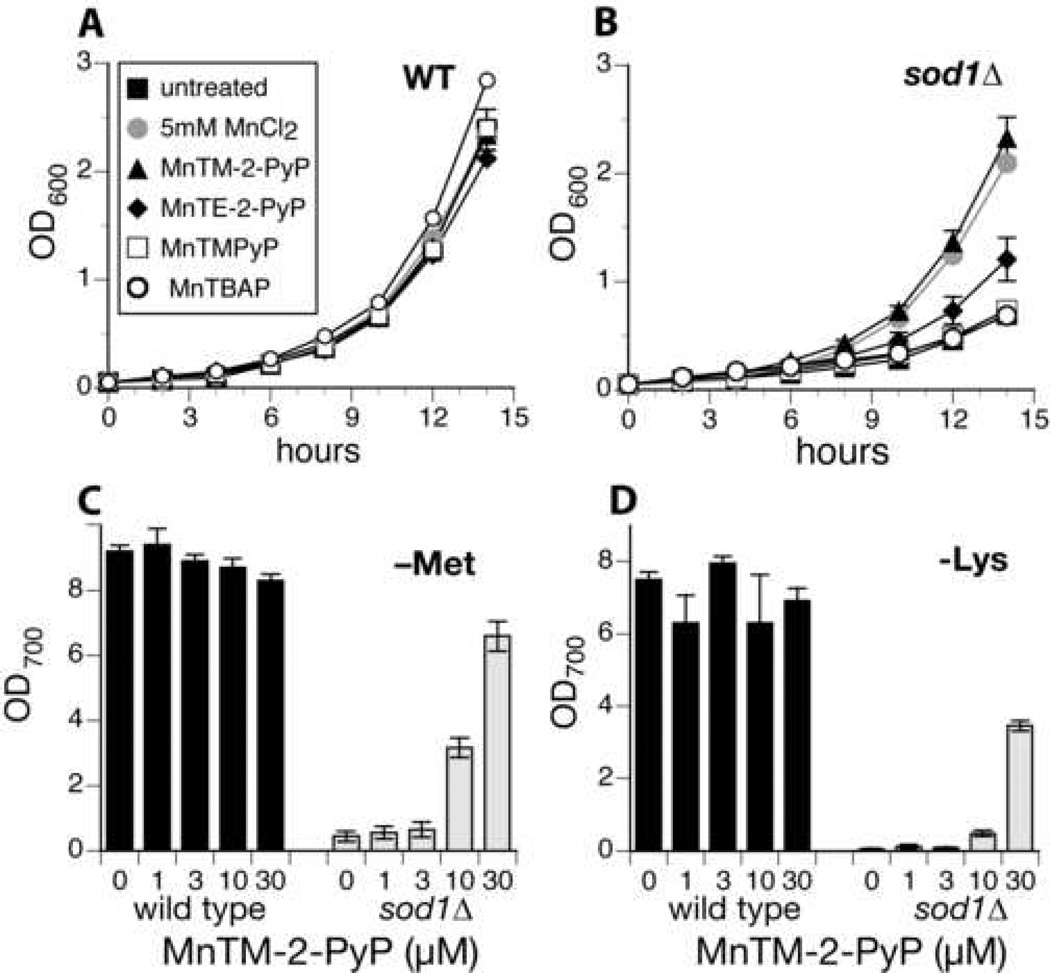
Several manganese porphyrins, including MnTBAP, MnTM-2-PyP, MnTM-PyP, and MnTE-2-PyP (see Scheme 1) have been studied as SOD mimics, and some have previously been shown to be beneficial in the E. coli model system [30, 33]. We assayed these compounds in our wild type and sod1Δ yeast strains. MnTBAP and MnTM-PyP had no effect on the 24 hour growth of sod1Δ in SDC at either pH 4 or pH 6 (Figs. 5A and 5B and data not shown), nor did they enable sod1Δ S. cerevisiae to grow without lysine or methionine at either pH 4.0 or pH 6.0. (Data not shown.)
Fig. 5. Aerobic growth of wild type and sod1Δ S. cerevisiae with Mn-porphyrin compounds.
Yeast were grown aerobically in flasks as described in Materials and Methods. A & B. Wild type (A) and sod1Δ (B) were grown in SDC either untreated (solid squares), treated with 5 mM MnCl2 (grey solid circles), or treated with 30 µM MnTM-PyP (open squares), MnTBAP (open circles), MnTM-2-PyP (solid triangles), or MnTE-2-PyP (solid diamonds). C & D. wild type (black bars) and sod1Δ (grey bars) yeast were grown in SD-methionine (C) or SD-lysine (D) in the presence of the indicated concentration of MnTM-2-PyP. Growth was measured after 24 hours via culture turbidity at 700 nm (OD700). Data shown are averages of results from 2 or 3 independent cultures done on the same day; each experiment was repeated at least twice. In addition, similar results were obtained when yeast were grown in microtiter plates (not shown).
By contrast, the compounds with ortho substituents, MnTM-2-PyP and MnTE-2-PyP, were able to rescue the phenotype of sod1Δ yeast. MnTM-2-PyP in particular very effectively restored growth to aerobic cultures of sod1Δ (Fig. 5A), and was able to rescue the lysine and methionine auxotrophies (Figs. 5C and 5D). This is the only SOD mimic we have found to rescue the rate of growth of sod1Δ yeast anywhere near as well as MnCl2 or to restore growth in the absence of methionine or lysine, and it is interesting to note that this mimic is effective at far lower concentrations than is MnCl2.
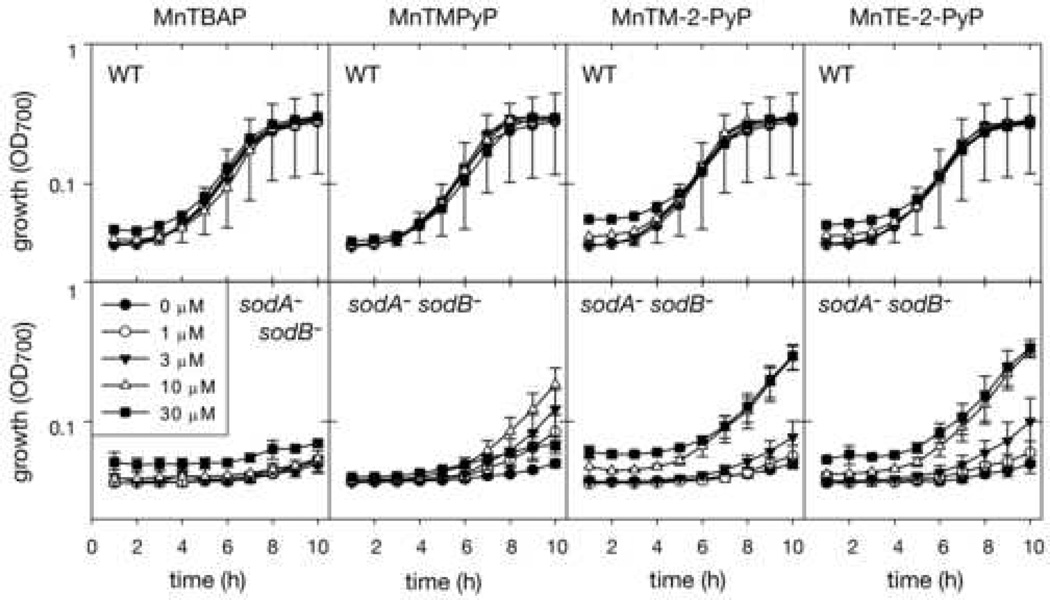
We tested these same Mn-porphyrin compounds for their effect on growth of wild type and sodA−sodB− E. coli (Fig. 6). Under our conditions, treatment with MnTBAP caused no change in aerobic growth in either strain, despite earlier reports that it functions in vitro and in vivo as an SOD mimic [30]. The other three Mn-porphyrins were more biologically active in our experimental system, as would be expected based on previously reported results [33]. MnTE-2-PyP and MnTM-2-PyP were the most effective, restoring near wild type growth rates at 10 µM concentrations. MnTM-PyP was very active, restoring growth more effectively at 3 µM than the others, but it also exhibited toxicity, such that the sod1A− sod1B− samples treated with 30 µM MnTM-PyP did not grow. MnTE-2-PyP and MnTM-2-PyP did not show such toxicity, and growth still occurred in WT and sodA−sodB− cultures treated with 100 µM of these two compounds (data not shown). (It should be noted that it cannot be concluded that a lower saturation density was reached in samples treated with higher levels of compound, because the colored SOD mimic compounds that cause the higher starting OD were removed from the medium as growth proceeded; thus it is not valid to subtract the starting OD700 from the later points.)
Fig. 6. Aerobic growth of wild type and sodA− sodB− E. coli with Mn-porphyrin compounds.
E. coli cells were grown aerobically for ten hours in 100 µL minimal medium supplemented as indicated in 96-well microtiter plates at 37 °C shaking at 220 RPM. Growth was recorded hourly as turbidity using a microtiter plate reader. In this case the turbidity measurement was done at 700 nm (OD700) to minimize interference due to absorbance by the Mn-porphyrins themselves. Error bars represent standard deviation; for clarity in the wild type samples only error bars for the untreated sample are shown; the treated samples showed similar error.
4. Discussion
This study provides a comparison of the several classes of manganese-containing small molecule SOD mimicking compounds and manganous chloride with respect to their abilities to substitute functionally for the SOD enzymes in vivo. Two simple unicellular model systems, SOD gene deletion strains of E. coli and S. cerevisiae. were used for the assay, which measured the ability of the compounds to restore wild type-like growth in air to these normally oxygen-sensitive strains. The manganese-containing SOD mimics assayed were two Eukarion mimics, one Metaphore mimic, and a series of manganese porphyrin mimics, each of which had demonstrated beneficial antioxidant properties in other in vivo systems.
In the sodA−sodB− E. coli system, all of the compounds except MnTBAP were found to be capable of rescuing to approximately the same degree. The main differences between the compounds were the concentration at which toxicity (or growth inhibition) occurred. The active compounds include the three Eukarion compounds, Euk-8, Euk-134, and Euk-189, the Metaphore compounds M40403 (reported to be SOD active in vitro) and M40404 (reported to be SOD inactive in vitro), and three manganese porphyrins, MnTE-2-PyP, MnTM-2-PyP, and MnTM-PyP. Although all of these compounds were effective in lowering the doubling time of sodA−sodB− E.coli, none of them was more effective than an equimolar amount of MnCl2, and some of them, in particular M40403, showed toxicity at relatively low levels.
In the case of the Eukarion compounds, it is possible that manganese ion is dissociating from the ligands and providing the beneficial effect, since these complexes have been shown to lose manganese ions in the presence of a strong chelator [13], but for the more stable Metaphore compound and manganese porphyrins, this possibility is less likely. We conclude therefore that, in the case of the sodA−sodB− E.coli system, the manganese-containing SOD mimic compounds are acting either on their own as antioxidants or as a source of manganese ion, which itself has a beneficial antioxidant effect, but in no case was any antioxidant effect observed greater than that provided by an equimolar amount of manganous chloride.
The same series of compounds was tested in the sod1Δ yeast system with quite different results. The three Eukarion compounds, Euk-8, Euk-134, and Euk-189 showed no beneficial effect at concentrations up to 100 µM, and in fact were growth inhibitory starting at 10 µM. Likewise, the SOD-active Metaphore compound M40403 was inhibitory starting at 5 µM, but the SOD-inactive Metaphore compound M40404 and two of the the Mn-porphyrins had no effect in either direction. We do not at this time know the explanation for the unexpected growth inhibitory effects of the Eukarion compounds and M40403 (see below) but the fact that they are observed argues that the compounds are getting into the cells. The other two manganese porphyrins—MnTE-2-PyP and MnTM-2-PyP—did show activity as discussed further below. The growth inhibitory effects of the Eukarion compounds and M40403 on both the sodA−sodB− E. coli and sod1Δ yeast systems are interesting and unexpected, especially in light of the observation that the SOD-inactive Metaphore compound M40404, which is very similar structurally to the SOD-active compound M40403, showed no adverse effects. We cannot rule out that the lack of toxicity of M40404 is associated with a failure of the cells to take up this compound, because we have no way to detect it in vivo at the low levels used. Nevertheless, we tentatively conclude that observed growth inhibition is related to their SOD activities. One interesting speculations is that superoxide may be involved in growth-signaling [42] and that reducing it to too low levels may therefore interfere with cellular growth signaling processes in already compromised SOD-deficient cells, but this possibility remains untested.
The most interesting results, from our perspective, were with the manganese porphyrin, MnTM-2-PyP (and the closely related but less potent MnTE-2-PyP), which were the only SOD mimics to show a high degree of rescue in the yeast system. The two other manganese porphyrin complexes, MnTBAP and MnTM-PyP were not effective at any concentration tested and provided neither rescue nor toxicity in the yeast system. It is also interesting that in E. coli, MnTM-2-PyP and MnTE-2-PyP showed good rescue activity at the relatively low concentration of 10 µM, although it was not more effective than a similar concentration of MnCl2. The growth rescue in yeast provided by this compound occurs at µM concentrations unlike the rescue due to simple manganese salts, for which mM levels are required in yeast. It is possible that in order to be effective these SOD mimics or manganese ions have to be in a particular cellular location(s) at sufficient concentration and be in a suitable chemical form to scavenge superoxide. It is interesting in this regard to note that MnTE-2-PyP has recently been demonstrated to target mouse heart mitochondria [43]. The differences in the efficacy observed for some SOD mimics, such as EUK compounds, that work in E. coli and not in yeast could be due to mislocalization or due to lack of sufficient concentration buildup of active species of these compounds in compartments where the radicals are present. Measuring cellular distribution of these SOD mimics correlated with radical levels, SOD and catalase activity levels, along with gene expression profiles may provide insight into the mode of action of various SOD mimics. These findings also send a cautionary note to investigators who declare a compound as an antioxidant based on results obtained in one model system, as it may not replicate universally.
Acknowledgements
This work was supported by grant NIH DK 46828 to JSV.
Abbreviations
- SOD
superoxide dismutase
- ROS
reactive oxygen species
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We dedicate this paper to our colleague, Ed Stiefel, an inspirational researcher, teacher, and friend.
References
- 1.Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. Oxford, UK: Oxford University Press; 1999. [Google Scholar]
- 2.Al-Maghrebi M, Fridovich I, Benov L. Arch. Biochem. Biophys. 2002;402:104–109. doi: 10.1016/S0003-9861(02)00065-6. [DOI] [PubMed] [Google Scholar]
- 3.Brown OR, Smykrandall E, Draczynskalusiak B, Fee JA. Arch. Biochem. Biophys. 1995;319:10–22. doi: 10.1006/abbi.1995.1262. [DOI] [PubMed] [Google Scholar]
- 4.Benov L, Fridovich I. J. Biol. Chem. 1999;274:4202–4206. doi: 10.1074/jbc.274.7.4202. [DOI] [PubMed] [Google Scholar]
- 5.Benov L, Kredich NM, Fridovich I. J. Biol. Chem. 1996;271:21037–21040. doi: 10.1074/jbc.271.35.21037. [DOI] [PubMed] [Google Scholar]
- 6.Liu XF, Elashvili I, Gralla EB, Valentine JS, Lapinskas P, Culotta VC. J. Biol. Chem. 1992;267:18298–18302. [PubMed] [Google Scholar]
- 7.Chang EC, Kosman DJ. J. Bacteriol. 1990;172:1840–1845. doi: 10.1128/jb.172.4.1840-1845.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gralla EB, Kosman DJ. Adv Genet. 1992;30:251–319. doi: 10.1016/s0065-2660(08)60322-3. [DOI] [PubMed] [Google Scholar]
- 9.Bilinski T, Krawiec Z, Liczmanski A, Litwinska J. Biochem. Biophys. Res. Commun. 1985;130:533–539. doi: 10.1016/0006-291x(85)90449-8. [DOI] [PubMed] [Google Scholar]
- 10.Longo VD, Gralla EB, Valentine JS. J. Biol. Chem. 1996;271:12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez RJ, Srinivasan C, Munroe WH, Wallace MA, Martins J, Kao TY, Le K, Gralla EB, Valentine JS. J. Biol. Inorg. Chem. 2005;10:913–923. doi: 10.1007/s00775-005-0044-y. [DOI] [PubMed] [Google Scholar]
- 12.Baudry M, Etienne S, Bruce A, Palucki M, Jacobsen E, Malfroy B. Biochem. Biophys. Res. Commun. 1993;192:964–968. doi: 10.1006/bbrc.1993.1509. [DOI] [PubMed] [Google Scholar]
- 13.Keaney M, Matthijssens F, Sharpe M, Vanfleteren J, Gems D. Free Radic. Biol. Med. 2004;37:239–250. doi: 10.1016/j.freeradbiomed.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Baker K, Marcus CB, Huffman K, Kruk H, Malfroy B, Doctrow SR. J. Pharmacol. Exp. Ther. 1998;284:215–221. [PubMed] [Google Scholar]
- 15.Pong K, Doctrow SR, Huffman K, Adinolfi CA, Baudry M. Exp. Neurol. 2001;171:84–97. doi: 10.1006/exnr.2001.7747. [DOI] [PubMed] [Google Scholar]
- 16.Peng J, Stevenson FF, Doctrow SR, Andersen JK. J. Biol. Chem. 2005;280:29194–29198. doi: 10.1074/jbc.M500984200. [DOI] [PubMed] [Google Scholar]
- 17.Melov S, Doctrow SR, Schneider JA, Haberson J, Patel M, Coskun PE, Huffman K, Wallace DC, Malfroy B. J. Neurosci. 2001;21:8348–8353. doi: 10.1523/JNEUROSCI.21-21-08348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melov S, Ravenscroft J, Malik S, Gill MS, Walker DW, Clayton PE, Wallace DC, Malfroy B, Doctrow SR, Lithgow GJ. Science. 2000;289:1567–1569. doi: 10.1126/science.289.5484.1567. [DOI] [PubMed] [Google Scholar]
- 19.Sampayo JN, Gill MS, Lithgow GJ. Biochem. Soc. Trans. 2003;31:1305–1307. doi: 10.1042/bst0311305. [DOI] [PubMed] [Google Scholar]
- 20.Collins JJ, Evason K, Kornfeld K. Exp. Gerontol. 2006;41:1032–1039. doi: 10.1016/j.exger.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 21.Bayne AC, Sohal RS. Free Radic. Biol. Med. 2002;32:1229–1234. doi: 10.1016/s0891-5849(02)00849-3. [DOI] [PubMed] [Google Scholar]
- 22.Riley DP, Henke SL, Lennon PJ, Aston K. Inorg. Chem. 1999;38:1908–1917. doi: 10.1021/ic981319b. [DOI] [PubMed] [Google Scholar]
- 23.Riley DP. Chem. Rev. 1999;99:2573–2588. doi: 10.1021/cr980432g. [DOI] [PubMed] [Google Scholar]
- 24.Riley DP, Weiss RH. J. Am. Chem. Soc. 1994;116:387–388. [Google Scholar]
- 25.Masini E, Cuzzocrea S, Mazzon E, Marzocca C, Mannaioni PF, Salvemini D. Br. J. Pharmacol. 2002;136:905–917. doi: 10.1038/sj.bjp.0704774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udipi K, Ornberg RL, Thurmond KB, 2nd, Settle SL, Forster D, Riley D. J. Biomed. Mater. Res. 2000;51:549–560. doi: 10.1002/1097-4636(20000915)51:4<549::aid-jbm2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 27.Wang ZQ, Porreca F, Cuzzocrea S, Galen K, Lightfoot R, Masini E, Muscoli C, Mollace V, Ndengele M, Ischiropoulos H, Salvemini D. J. Pharmacol. Exp. Ther. 2004;309:869–878. doi: 10.1124/jpet.103.064154. [DOI] [PubMed] [Google Scholar]
- 28.Salvemini D, Wang ZQ, Zweier JL, Samouilov A, Macarthur H, Misko TP, Currie MG, Cuzzocrea S, Sikorski JA, Riley DP. Science. 1999;286:304–306. doi: 10.1126/science.286.5438.304. [DOI] [PubMed] [Google Scholar]
- 29.Riley DP, Lennon PJ, Neumann WL, Weiss RH. J. Am. Chem. Soc. 1997;119:6522–6528. [Google Scholar]
- 30.Faulkner KM, Liochev SI, Fridovich I. J. Biol. Chem. 1994;269:23471–23476. [PubMed] [Google Scholar]
- 31.Velsor LW, Kovacevic M, Goldstein M, Leitner HM, Lewis W, Day BJ. Toxicol. Appl. Pharmacol. 2004;199:10–19. doi: 10.1016/j.taap.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Patel MN. Aging Cell. 2003;2:219–222. doi: 10.1046/j.1474-9728.2003.00055.x. [DOI] [PubMed] [Google Scholar]
- 33.Batiniac-Haberle I, Benov L, Spasojeviac I, Fridovich I. J. Biol. Chem. 1998;273:24521–24528. doi: 10.1074/jbc.273.38.24521. [DOI] [PubMed] [Google Scholar]
- 34.Okado-Matsumoto A, Batinic-Haberle I, Fridovich I. Free Radic. Biol. Med. 2004;37:401–410. doi: 10.1016/j.freeradbiomed.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 36.Imlay JA, Linn S. J. Bacteriol. 1987;169:2967–2976. doi: 10.1128/jb.169.7.2967-2976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imlay JA, Fridovich I. J. Bacteriol. 1992;174:953–961. doi: 10.1128/jb.174.3.953-961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 39.Aston K, Rath N, Naik A, Slomczynska U, Schall OF, Riley DP. Inorg. Chem. 2001;40:1779–1789. doi: 10.1021/ic000958v. [DOI] [PubMed] [Google Scholar]
- 40.Cheton PL, Archibald FS. Free Radic. Biol. Med. 1988;5:325–333. doi: 10.1016/0891-5849(88)90104-9. [DOI] [PubMed] [Google Scholar]
- 41.Doctrow SR, Huffman K, Marcus CB, Tocco G, Malfroy E, Adinolfi CA, Kruk H, Baker K, Lazarowych N, Mascarenhas J, Malfroy B. J. Med. Chem. 2002;45:4549–4558. doi: 10.1021/jm020207y. [DOI] [PubMed] [Google Scholar]
- 42.Moraitis C, Curran BP. Yeast. 2004;21:313–323. doi: 10.1002/yea.1078. [DOI] [PubMed] [Google Scholar]
- 43.Spasojevic I, Chen Y, Noel TJ, Yu Y, Cole MP, Zhang L, Zhao Y, St. Clair DK, Batinic-Haberle I. Free Radic. Biol. Med. 2007;42:1193–1200. doi: 10.1016/j.freeradbiomed.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]