SUMMARY
During yeast cell division, aggregates of damaged proteins are segregated asymmetrically between the bud and the mother. It is thought that protein aggregates are cleared from the bud via actin cable-based retrograde transport toward the mother, and that Bni1p formin regulates this transport. Here we examined the dynamics of Hsp104-associated protein aggregates by video microscopy, particle tracking and image correlation analysis. We show that protein aggregates undergo random walk without directional bias. Clearance of heat-induced aggregates from the bud does not depend on formin proteins but occurs mostly through dissolution via Hsp104p chaperon. Aggregates formed naturally in aged cells also exhibit random walk but do not dissolve during observation. Although our data does not disagree with a role for actin or cell polarity in aggregate segregation, modeling suggests that their asymmetric inheritance can be a predictable outcome of aggregates' slow diffusion and the geometry of yeast cells.
INTRODUCTION
Budding yeast cells divide asymmetrically to generate a new cell with full proliferative potential, and an ageing mother cell that can be anywhere along a finite replicative life span (20–30 cell divisions under standard laboratory conditions) (Mortimer and Johnston, 1959). This asymmetry in replicative age is correlated with an unequal segregation of ageing determinants along the axis of cell polarity in a dividing yeast cell (Egilmez and Jazwinski, 1989; Sinclair and Guarente, 1997). One class of such ageing determinants are aggregates formed by damaged proteins. A seminal study demonstrated that aggregates containing carbonylated proteins, which result from oxidative damage in ageing cells, are preferentially retained by the mother during bud formation and cell division (Aguilaniu et al., 2003). Such asymmetric segregation of damaged proteins appears to be dependent on the actin cytoskeleton (Aguilaniu et al., 2003; Knorre et al., 2010; Tessarz et al., 2009). However, the mechanism by which actin regulates asymmetric partitioning of protein aggregates remains elusive.
In a recent study (Liu et al., 2010), segregation of protein aggregates was studied using a model system where heat-induced protein aggregates were labeled with GFP-tagged Hsp104p, a hexameric AAA ATPase-based chaperon known to play a major role in the modification and dissolution of heat denatured protein aggregates (Parsell et al., 1994; Glover and Lindquist, 1998; Bosl et al., 2006; Doyle and Wickner, 2009). This study implicated several proteins that control the assembly of actin cables, most notably the formin family actin nucleating protein Bni1p, in the segregation of Hsp104-containing protein aggregrates. Actin cables are parallel bundles of actin filaments (F-actin), best known for their role in polarized trafficking of membrane and cell wall materials and organelles during polarized growth (Pruyne et al., 2004). During the early stage of bud growth, actin cables are nucleated from Bni1p localized at the tip of the nascent bud (Evangelista et al., 2002; Sagot et al., 2002). Elongation of actin filaments at their barbed ends generates a retrograde flow of actin subunits in the mother-bound direction (Yang and Pon, 2002). It was proposed that the Hsp104p-containing protein aggregates associate with actin cables, possibly through a direct interaction of Hsp104p with actin, and ride along the retrograde flow to clear from the bud prior to cell division (Liu et al., 2010). Supporting this model, time-lapse movies were presented showing movement of single aggregates in the bud across the bud neck into the mother compartment.
A polarized array of actin cables exist during the part of the cell cycle when the cell undergoes rapid polarized growth, but after mitotic entry the actin organization becomes isotropic (Adams and Pringle, 1984; Lew and Reed, 1995; Pruyne and Bretscher, 2000). Thus, an initial question motivating our study was how mitotic cells might rely on the actin retrograde flow to clear protein aggregates from the bud prior to cytokinesis. We used live imaging to observe the dynamics and movement of Hsp104p-containing protein aggregates. Surprisingly, we found a lack of defined directionality in aggregate movement during budding or other cell cycle stages. Instead, heat-induced aggregates exhibit random walk and are largely cleared from the bud via Hsp104p ATPase-dependent dissolution. Interestingly, while actin depolymerization reduced aggregate random movement and the rate of dissolution, formin proteins do not play any detectable positive role in these events. Based on these findings, we propose an alternative model to explain the asymmetric inheritance of protein aggregates during yeast asymmetric cell division.
RESULTS
Heat-induced protein aggregates undergo random walk
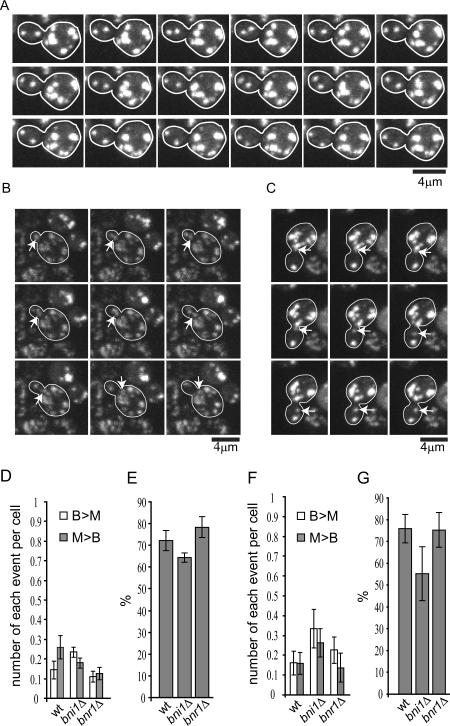
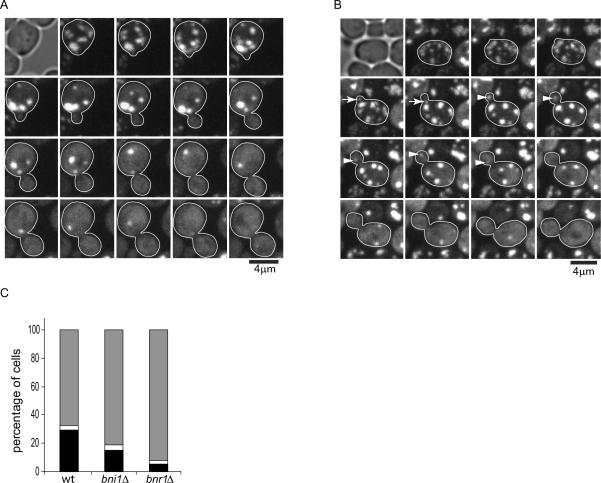
We used the same approach as described (Liu et al., 2010) to observe the dynamics of protein aggregates in live yeast cells. Briefly, protein aggregates were induced with a short (30 min) heat shock at 42 °C and visualized with Hsp104-GFP. As in Liu et al, we used the HSP104-GFP strain from the yeast genome-wide GFP-tagged strain collection (Huh et al., 2003), where GFP was introduced at the 3' end of the HSP104 open reading frame in the genome of the S288c strain BY4741. After a brief recovery at 30 °C following the heat shock, the cells were imaged by acquiring 3-dimensional (3D) time-lapse confocal movies (see Experimental Procedures). As shown in Fig.1A and Movie S1, the majority of protein aggregates were motile in both the mother and the bud at all cell cycle stages. Most aggregate movements were confined within the bud or the mother, but bud-to-mother (B>M) or mother-to-bud (M>B) translocation of the aggregates could be observed at low frequencies with a slight bias in the M>B direction (Fig.1B, C, D, E, Movies S2A,B). Quantification of B>M and M>B movements in small-budded cells (bud diameter equal to or less than 1/3 of that of mother, indicating the stage of polarized growth) revealed no directional bias in aggregate motility (Fig.1F,G).
Figure 1. 3D time-lapse imaging of heat-induced Hsp104p-containing protein aggregates (associated with Movie S1 and Movie S2).
A. Observation of Hsp104-GFP-containing aggregate movement in the wild-type BY4741 strain in a representative 3D time-lapse movie (Movie S1A). Image stacks were collected and shown at 20s intervals, and are shown as maximum projections.
B. Example of an aggregate (arrow) moving from bud to mother in wild type (Movie S2A). Image stacks were collected and shown at 1 min intervals, and are shown as maximum projections.
C. Example of an aggregate (arrow) moving from mother to bud in wild type (Movie S2B). Image stacks were collected at 1 min intervals, and a maximum projection is shown.
D. The frequency of B>M or M>B movement per cell in an exponentially population was calculated by counting the totally number of each type of event divided by the totally number of budded cells observed in each movie. The bar graphs show mean and standard error of the mean (SEM).
E. Percentage of cells in the population in (D) in which no trans-bud neck movement was observed during the 1 hour time lapse movies. Shown are mean and SEM. For both D and E, on average 40 cells/movie from 4–6 movies were quantified for each strain.
F. The frequency of B>M or M>B movement per cell in small budded cells was calculated by counting the totally number of each type of event divided by the totally number of small budded cells observed in each movie. Shown are mean and SEM.
G. Percentage of cells in the population in (F) in which no trans-bud neck movement was observed during the 1 hour time lapse movies. Shown are mean and SEM. For F and G, on average 14 cells/movie from 4–6 movies were quantified for each strain.
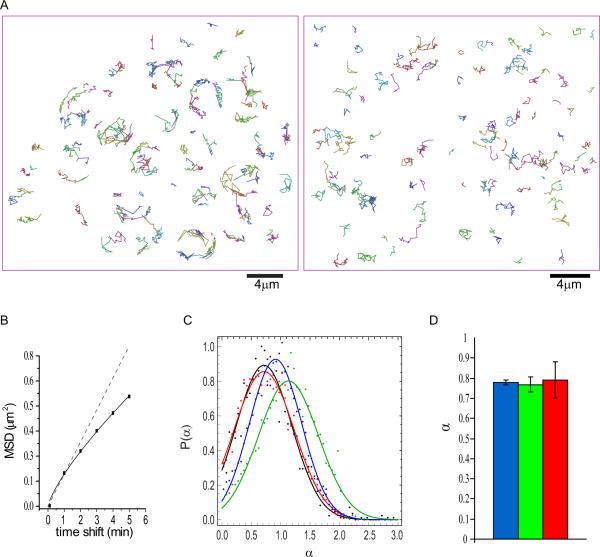
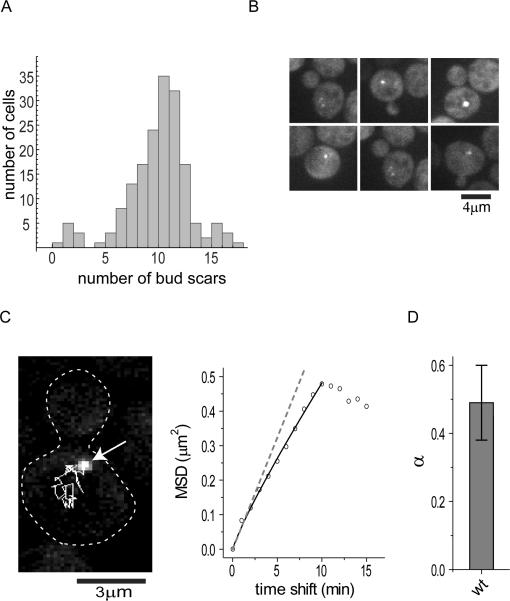
To quantitatively characterize aggregate movement, we performed automated tracking of individual aggregates (Experimental Procedures) (Fig.2A- left panel, Movie S3). The mean square displacement (MSD) averaged from 1068 traces showed that the aggregate movement can be approximated as random walk with a small amount of confinement (Fig.2B). For pure random walk (diffusion), one expects a linear relationship between MSD and time shift (MSD(τ) = 4Dτα, where α = 1), whereas confinement (sub-diffusion) results in a downward trending relationship (α < 1) and transport (super-diffusion) results in an upward trending relationship (α > 1). Simulations of random walk based on the observed trajectory lengths showed overall similar trajectory profiles as those observed experimentally, including the occasional more linear trajectories (Fig.2A – right panel, compared to left panel). As it is possible that a small fraction of transport (or super-diffusion) may be obscured by averaging, we performed 5000 simulations of aggregate movement with the diffusion coefficient and trajectory length distribution similar to those observed in yeast cells for three scenarios: 1) pure random walk; 2) random walk with 10% super-diffusion; 3) random walk with 30% sub-diffusion. The distribution of α values for each population was compared to the experimentally observed distributions (Fig.2C). This analysis found that the experimentally observed distribution matches the shape of the predicted distribution from scenario (3), indicating that the experimental data is consistent with a model that does not include transport. Quantification of α values in small budded cells showed that the mode of aggregate motility at this cell cycle stage is indistinguishable from that of the entire population (Fig.2D).
Figure 2. Hsp104p-containing protein aggregates undergo random walk (associated with Movie S3).
A. Left panel: A representative field of trajectories from tracking of protein aggregates from 1 hr time-lapse movies. For comparison, the right panel shows a field of simulated trajectories of random walk with observed distribution of trajectory lengths and diffusion coefficient.
B. A plot of MSD vs time shift, showing that aggregate movement can be characterized as random walk with a small amount of confinement (solid line). Dotted line shows plot expected for pure random walk.
C. α value distributions from 5000 simulations of aggregate movement with the diffusion coefficient and trajectory length distribution similar to those observed in yeast cells for each of the three scenarios: 1) pure random walk (blue); 2) random walk with 10% super-diffusion (green); 3) random walk with 30% sub-diffusion (red). The distribution of α values for each population was compared to the experimentally observed distribution (black), showing the observed is consistent with scenario (3).
D. Comparison of average α values (mean and SEM) from aggregates in different population of cells in 60min 3D time-lapse movie. Blue: aggregates in all the cells in the movies (N=3542); green: all aggregates in the small budded S/G2 cells (N=182); red: aggregates in the buds of the S/G2 cells (N=31). N refers to the number of aggregates. See also Movie S3.
To ensure that the Hsp104-GFP-expressing strain in our collection had not accumulated random mutations that might disrupt the normal behavior of protein aggregates, we obtained and tested the Hsp104-GFP strain used in the previous work (Liu et al., 2010). This strain showed the same aggregate behavior as our Hsp104-GFP strain (Movie S4, Fig.3A,B). We also reconstructed a new Hsp104-GFP strain by genomic tagging of HSP104 in the BY4741 parental wild-type strain. Observation of protein aggregate movement using this strain was again consistent with the above two strains (data not shown).
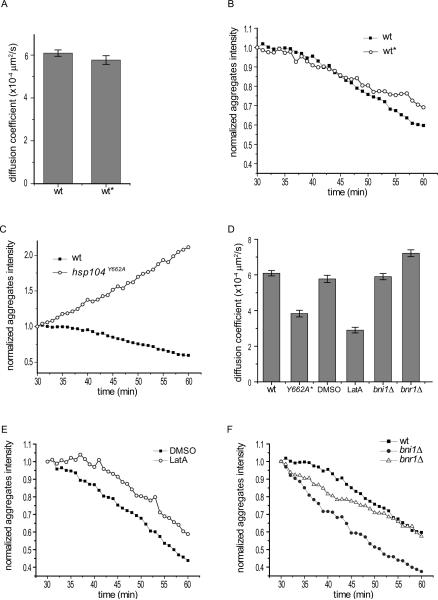
Figure 3. Comparison of dissolution kinetics and diffusion coefficients among different strains and conditions (associated with Figure S1, S2, Movie S4, S6, S7, S8).
A. Comparison of diffusion coefficients calculated from aggregate tracking between the two wild type strains. Shown are mean and SEM. More than 1000 aggregates from at least 2 movies/strain were tracked. wt, RLY7110; wt*, YBD401 (Liu et al., 2010).
B. Quantification of aggregate dissolution (see Experimental Procedures) in the two strains in (A). Shown are plots of total aggregate intensity as a function of time from time-lapse movies of a field of cells (50–100 cells per field) starting from the 30 min frame when aggregates in wild type no longer grew in brightness. Each plot is an average from two movies. Black square: RLY7110; open circle: YBD401.
C. Quantification of aggregate dissolution following the same method as in (B) in wild-type (wt) cells and hsp104Y662A cells.
D. Diffusion coefficient of each strain and condition as indicated quantified from aggregate tracking data (see Experimental Procedures). Shown are mean and SEM. *The quantification for hsp104Y662A cells was done from the 3.5 min heat shock experiment to mimic wild-type aggregate density. For each strain or condition, more than 450 (and in many cases over 1000) aggregates from 2–4 movies were tracked and quantified.
E–F: Quantification of aggregate dissolution following the same method as in (B) for the same conditions or strains examined in (D). See also Figure S1, S2, Movie S4, S6, S7, S8.
Hsp104p chaperon-mediated dissolution is the main mechanism of heat-induced aggregate clearance
Aside from the random motion of the protein aggregates described above, another apparent behavior of the aggregates is their dissolution: during the initial segment of the movies (up to ~20 min), the aggregates grew slightly brighter but then underwent gradual reduction in brightness and number during the rest of the 1–1.5 hr movies (Fig.4A, arrow, MovieS5). Near the end of the movies, most buds, which had fewer aggregates initially, were cleared of aggregates by this means while the mothers had drastically reduced number of aggregates compared to the initial amounts. By acquiring serial confocal slices spanning the entire Z dimension of cells in these movies, we could rule out the possibility that aggregate disappearance was due to movement out of the focal plane. Concomitant with aggregate dissolution, the diffuse cytosolic GFP level rose likely as a result of release of free Hsp104-GFP (Fig.4A). This observation is consistent with previously reported role for Hsp104 in dissolving heat-induced aggregates (Parsell et al., 1994; Glover and Lindquist, 1998; Bosl et al., 2006; Doyle and Wickner, 2009).
Figure 4. Heat-induced aggregates are cleared from the bud through Hsp104p chaperon-mediated dissolution (associated with Movie S5 and Movie S6).
A. Hsp104-GFP-associated aggregates show gradual clearance through dissolution. The arrow indicates an example of an aggregate dissolving; arrowheads indicate two aggregates fusing and then dissolving. Images stacks were collected at 1 min intervals; the montage shows maximum projections at 6min intervals from a 90 min movie (Movie S5).
B. Comparison of the percentage of cells where the last aggregate was cleared from the bud by dissolution (white bar) to that by bud-to-mother movement (gray bar) in the wild type. On average 47 cells/movie from 5 movies were quantified. Shown are mean and SEM. C. Aggregate dissolution depends on chaperon activity of Hsp104p. In hsp104Y662A mutant cells, the aggregates not only did not dissolve over a representative 3hr movie (Movie S6A) but also increased in brightness over time. Montage starts from 18 min of the movie and frames (maximum projections) are shown at 18min intervals.
To evaluate the contribution of dissolution to aggregate clearance from the bud relative to that from B>M movement, we quantified the likelihood of these events from the time-lapse movies. As shown in Fig.4B, in ~96% of the cells observed, aggregate clearance (defined as disappearance of the last aggregate) from the bud occurred via dissolution (white bar), while only ~4% cells lost their last aggregate by B>M movement (gray bar). Furthermore, in nearly all cases of the latter, the observed B>M movement was a single such event per cell, before which most of the aggregates in the bud were cleared already via dissolution. As previously reported (Liu et al., 2010), we also observed fusion of colliding aggregates in both mother and the bud (Fig.4A, arrowhead, Movie S5), and large aggregates could sometimes be observed to split into smaller aggregates (not shown).
We next tested the possibility that the observed aggregate dissolution is a result of the Hsp104p chaperon activity by using a mutation, hsp104Y662A, which does not affect Hsp104p binding to protein aggregates but disrupts the refolding activity of Hsp104p (Lum et al., 2004). A strain was constructed where the genomic copy of HSP104 was replaced with hsp104Y662A-GFP. This mutant strain exhibited more aggregates than the wild type immediately after heat shock, and the aggregates in the mutant did not show any dissolution throughout the 3 hour time-lapse movie (Fig.4C, Movie S6A). In fact, quantification showed that the aggregates in hsp104Y662A-GFP strain continued to increase in their intensity (Fig.3C). This observation is consistent with the notion that the Hsp104p chaperon activity plays a major role in protein disaggregation in yeast (Parsell et al., 1994; Glover and Lindquist, 1998; Bosl et al., 2006; Doyle and Wickner, 2009).
Hsp104p and F-Actin, but not Bni1, play a role in the random movement and dissolution of heat-induced aggregates
We observed that, in addition to a lack of dissolution, the movement of protein aggregates was largely abolished in hsp104Y662A-GFP cells (Movie S6A). As this reduced movement could possibly be due to over-crowding of the aggregates in the mutant cells, we performed another experiment by subjecting the mutant cells to a much shorter (3.5 min) heat shock, which results in an aggregate density similar to that in wild-type cells. In these cells the aggregates still did not dissolve but did exhibit some motility (Movie S6B). Quantification from particle tracking data confirmed that the diffusion coefficient (D) for aggregates in the hsp104Y662A mutant was reduced compared to the wild type (Fig.3D). This observation suggests that the Hsp104p ATPase activity plays a role in the movement of the aggregates. We next examined the involvement of F-actin in aggregate movement and dissolution by treating cells with latrunculin A (LatA), an inhibitor of actin polymerization. Cells were first subjected to the aforementioned protocol of 30 min heat shock and a brief recovery and then incubated with 100 μM LatA. Time-lapse imaging started 10 min after LatA addition, at which time most of the cellular actin filaments were undetectable by fluorescent phalloidin staining (Fig.S1), and the drug was present throughout the subsequent time-lapse imaging. LatA treatment significantly reduced, though did not completely abolish, the diffusion coefficient (D) of protein aggregates (Fig.3D, Movie S7). Interestingly, the rate of aggregate dissolution was also visibly reduced (Fig.3E), suggesting that actin contributes to but is not absolutely required for the movement and dissolution of heat-induced aggregates.
To examine if formin proteins are involved in the dissolution and movement of protein aggregates, we tagged Hsp104 with GFP in the bni1Δ and bnr1Δ strains (BY4741 background) and performed 3D time-lapse imaging of heat-induced aggregates (Movie S8A, B). Neither mutant showed reduced frequency of B>M movement of aggregates (Fig.1D). In fact, we observed a slightly higher frequency of B>M in bni1Δ compared to that in wild type (Fig.1D). Quantification of the rates of aggregate movement and dissolution showed that neither mutant is defective in these processes (Fig. 3D, F). The aggregates showed slightly faster movement in bnr1Δ cells and faster dissolution in bni1Δ than those processes in wild-type cells (Fig. 3D, F), suggesting that Bni1 and Bnr1 may contribute slightly negatively to aggregate dissolution or movement, respectively. Thus, the roles of F-actin in aggregate movement and dissolution appear to be largely independent of the formin proteins. We note that the calculation of aggregate diffusion coefficients in various strains or under the specified conditions was confirmed by the method of spatial temporal image correlations spectroscopy (STICS) analysis (Berland et al., 1995; Hebert et al., 2005; Kolin et al., 2006) (Fig.S2), which is independent of the particle tracking approaches. The rates calculated using both methods were consistent.
Asymmetric inheritance of protein aggregates during bud formation
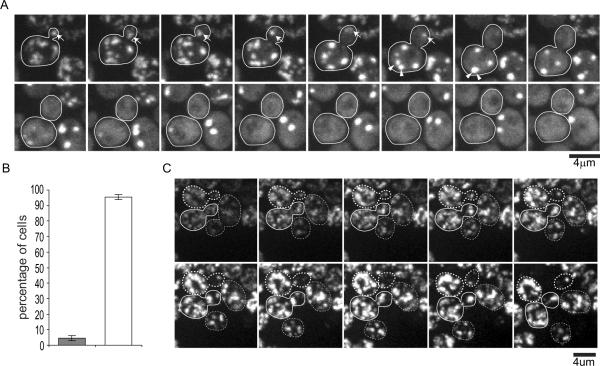
During our time-lapse confocal movies, new buds could be observed to form from a number of mother cells in each experiment. In the majority (68%) of such wild-type cells observed, the aggregates were successfully retained in the mother while the bud grew and the new bud was devoid of aggregates throughout the movies (Fig. 5A, Movie S10A). In the reminder of the population, 1 (2 in rare cases) aggregate moved into the bud from the mother (Fig. 5B, Movie S10B). In the majority of these cases, the aggregate dissolved after entry into the bud (Fig.5B,C, Movie S10B, yellow arrow), while in a small fraction the aggregate moved back to the mother (Fig.5B,C, Movie S10B, red arrow). Formin mutant (bniΔ and bnrΔ) cells undergoing budding during the time-lapse imaging essentially showed similar behavior, that is, the majority of aggregates were retained in the mother (Fig.5C, Movie S9), and those occasionally entering the bud most often dissolved (Fig.5C).
Figure 5. Retention of heat-induced protein aggregates in the mother during bud formation (associated with Movie S8, Movie S9 and Movie S10).
A. An example of retention of protein aggregates in a wild-type mother cell undergoing budding. Image stacks were collected 1 min intervals; montage is shown as maximum projections every 4 min (see also Movie S10A).
B. Time-lapse images of a cell showing two different fates of aggregates that leaked into the bud from the mother during bud formation. Image stacks were collected 1 min intervals; montage is shown as maximum projections every 4 min. Arrows point to an aggregate that returned to the mother after leakage into the bud; arrowhead points to an aggregate that dissolved after leakage into the bud. See movie S10B to observe these events at a higher time resolutions.
C. Percentages of cells displaying three different types aggregate behavior during new bud formation and growth in three different genetic backgrounds as indicated. Cells either with a tiny bud initially devoid of any aggregates or giving birth to a new bud during the first 30min of 1 hr long 3D time-lapse movies were scored. Gray bar: cells with no aggregates leakage into the bud; black bar: cells in which aggregates leaked into the bud were subsequent cleared by dissolution; white bar: cells in which aggregates leaked into the bud and subsequent moved back to mother. N=108 cells (wt); 74 cells (bni1Δ), 133 cells (bnr1Δ). See also Movie S8, S9.
We next performed time-lapse movies to observe the dynamics and distribution of naturally accumulated Hsp104-positive protein aggregates. Previous work showed that Hsp104p associates with protein aggregates containing carbonylated proteins resulting from oxidative damage in cells of older replicative age (Erjavec et al., 2007). In an exponentially growing population, where most cells are young, Hsp104p-containing aggregates were rarely observed (data not shown). We hence used a magnetic beads-based sorting protocol (Chen et al., 2003) to obtain populations of wild-type and sirΔ enriched for older cells (see bud scar staining in Fig.6A).
Figure 6. Observation of motility and distribution of Hsp104p-containing aggregates in aged wild-type cells (associated with Movie S11).
A. Age distribution of magnetic-beads sorted old wild-type cells determined by bud scar counting (> 170 cells were counted).
B. Representative images showing asymmetric distribution of Hsp104-associated protein aggregates in aged wild-type from the populations in (A). Shown are maximum projections of 3D image stacks.
C. Trajectory (white line in left panel) and MSD (right panel) analysis of an Hsp104-GFP-associated aggregate (arrow) in an old wild-type cell from a 60 min 3D time-lapse movie.
D. Hsp104-GFP-containing protein aggregates in old wild-type cells exhibit a similar type of movement. Shown are mean and SEM of α values calculated from long aggregate tracks. N=21 tracks for wild-type aggregates. See also Movie S11-.
The naturally formed aggregates (Fig.6B) do not dissolve during the duration of our observation by time-lapse movies, allowing long tracking of aggregate movement (Fig.6C, Movie S11). The individual long traces show nearly linear fitting of MSD with time with a slight downward trend, suggesting that this movement can be described as random walk with slight confinement (Fig.6C, as an example). The distributions of α values from tracking many aggregates in older wild-type cells were similar to that of heat-induced aggregates (Fig.6D, compared to Fig.2C). These observations suggest that the dynamics of the Hsp104-associated protein aggregates formed in different ways exhibit the same type of movement.
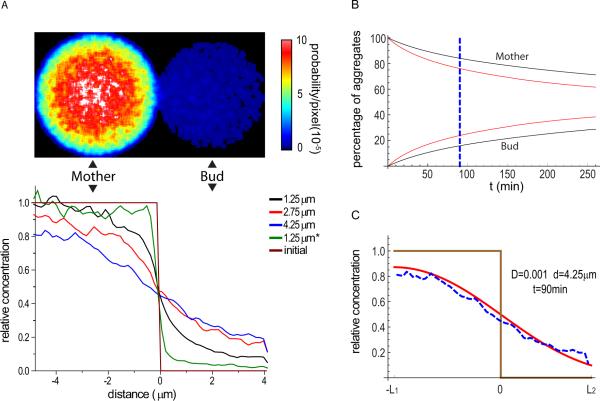
A recent study investigating the mechanism of asymmetric partitioning of episomal DNA circles reported free diffusive motion of episomes in the nucleoplasm (Gehlen et al., 2011). Using a numerical simulation, they demonstrated that the retention of episomes in the mother can be explained simply by the geometry of the dividing nucleus and the brief window of time in the cell cycle when nuclear division occurs. As the protein aggregates also undergo random motion, a similar explanation may be applied to the preferential retention of aggregates in the mother during cell division. To test this idea, we performed a 3D numerical simulation of pure random walk with the diffusion rate D = 1.0×10−3 μm2/s computed from particle tracking of natural Hsp104-GFP-containing aggregates in older cells. A yeast cell was represented as two spheres with radius of R1=2.5 μm (mother) and R2=0.85×R1 (bud) connected by a narrow neck with width d=1.25 μm. Starting from an initial distribution of aggregates entirely in the mother, the probability density map after 90 min (a cell cycle) showed maintenance of a biased distribution of aggregates in the mother (Fig.7A, top panel). To evaluate the impact of the geometry of yeast cells on this partitioning, the bud neck size was varied in the simulations. Figure 7A (bottom panel) shows that increasing bud neck size reduces the asymmetry in aggregate partitioning. Addition of confinement (α=0.50, approximating the average value in Fig. 6D) also enhanced the segregation asymmetry as expected (Fig. 7A). Aggregate asymmetric segregation can also be described using a 1D (equivalent to wide open neck) analytical model, which again shows preferential retention of aggregates in the mother even with no confinement (α=1) (Fig.7B, C).
Figure 7. 3D numerical simulation and 1D analytical model of aggregate partitioning between mother and bud during a yeast cell cycle.
A. Probability density map of protein aggregates after a cell cycle period (90 min) from a 3D numerical simulation of aggregate retention assuming random walk (α = 1) (see Experimental Procedures) as a 2D sum projection (top) and lateral profiles of central plane (bottom). Each colored line corresponding to simulation with a particular neck size (diameter) as indicated. The brown line shows the initial distribution at the start of the simulation and the green line shows the 1.25 μm neck size simulation with confined diffusion (α = 0.50, as measured in aged cells).
B. Analysis of protein aggregate retention using a 1D analytical solution (Formula S12, see Supplementary Information) for D=0.0005 μm2/s (black) and D=0.001μm2/s (red). The dashed blue line denotes 90 min time point.
C. A comparison of aggregates distribution after 90min in 3D simulation for D=0.001 μm2/s and neck size of 4.25 μm (mimicking open neck, blue dashed curve) to the 1D analytical solution (Formula S7, using 20 terms, see Extended Experimental Procedures) (red solid curve). The brown curve represents the initial aggregates distribution. 0 on x axis is the position of bud neck and mother-bud orientation as in A; L1 and L2 is the length of mother and bud cell, respectively.
Discussion
Based on the analyses described above, we conclude that the clearance of protein aggregates from the bud does not depend on actin cable-mediated retrograde transport. Our conclusion is based on quantification of aggregates movement from time-lapse recording of a large number of cells. Although linear bud-to-mother movement was observed occasionally, mother-to-bud movement of aggregates was observed at a similar frequency, and thus there was no net directionality in aggregate movement along the mother-bud axis. Particle tracking and trajectory analysis showed that the movement of protein aggregates can be described as random walk with a small amount of confinement. Even though some apparently linear trajectories can be observed, these can be recapitulated by random walk simulations using the same diffusion coefficient and trajectory length. We emphasize that although our data does not support a direct role for actin-based retrograde transport in aggregate segregation, the data does not necessarily argue against an involvement for actin cytoskeleton or cell polarity proteins in this process. It is
conceivable, for example, that the abnormally wide bud neck in some of the cell polarity mutants could increase the leaking of protein aggregates into the bud, although such effect is likely to be too subtle to observe reliably in our experiments.
Disassembly of actin filaments following LatA treatment partially abrogated aggregates movement and slowed their dissolution. Although it is possible that residual actin filaments remained and were below detection in LatA treated cells, this observation suggests that actin cables, which were no longer present in LatA-treated cells, are not required for the movement or dissolution of heat-induced aggregates. The effect of LatA on aggregate movement or dissolution may also be indirect if the loss of actin filaments induces a high-stress state interfering with aggregate dissolution or motility. Regardless of the mechanism, our observed effect of LatA on aggregate motility does not help explain a positive role for actin in aggregate asymmetric segregation, as based on our model, reduced aggregate diffusive motion would predict their better retention in the mother. The inhibitory effect of actin depolymerization on the Hsp104-dependent aggregate dissolution is consistent with a functional linkage between actin and Hsp104 reported in recent studies (Erjavec et al., 2007; Tessarz et al., 2009).
We did not observe any apparent requirement for the formin protein Bni1p or Bnr1p in the motility or retention of heat-induced protein aggregates in the mother. The experiments performed previously on the retention of protein aggregates relied on quantification of nonsynchronized cell populations at different time points during a continuous incubation at high temperature (Liu et al., 2010). Such an assay would not be sufficient for distinguishing between the possibilities of aggregate clearance by dissolution versus by movement. Because the dynamics of new aggregate formation during the prolonged heat exposure were unknown, buds displaying no aggregates might be new buds that formed after the cells had adapted to the heat stress and ceased the formation of new aggregates, rather than reflecting active aggregate clearance from the bud. Additionally, the unbudded cells observed in this assay might not be the new G1 cells after cytokinesis but instead might be arrested cells failing to adapt to the heat stress. By contrast, we directly observed retention of pre-formed aggregates during new bud formation by time-lapse movies, and we did not observe any defect in this process in the formin mutants.
Finally, based on the observed diffusion rates of protein aggregates, and borrowing the insight from a recent study explaining the asymmetric inheritance of episomal DNA during yeast nuclear division (Gehlen et al., 2011), we propose that the limited mobility and the narrowness of the bud neck ensure that the vast majority of the proteins aggregates accumulated with ageing are retained in the mother during the limited time of a cell cycle. This idea is supported by our 3D simulation as well as a 1D analytical model. This model implies that no additional mechanism may be needed to confer the asymmetric segregation of protein aggregates during yeast cell divisions.
Experimental procedures
Yeast strains
Yeast strains used in this study are based on the BY4741 S288c strain background as listed in Extended Experimental Procedures. Gene deletion or GFP tagging were performed with PCR mediated homologous recombination (Longtine et al., Yeast 1998) and correct integrations were confirmed by PCR. To create the hsp104Y662A mutant (RLY7200), site directed mutagenesis was performed by PCR using the primers described previously (Lum et al., 2004). hsp104Y662A mutant DNA was sequenced and correct integration and introduction of the mutation into the hsp104Δ background was confirmed by PCR. All mutant strains were analyzed by FACS and qPCR (Pavelka et al., 2010) to ensure no abnormalities in karyotype.
3D fluorescence time-lapse imaging of Hsp104-GFP-containing protein aggregates
Yeast cells were grown in synthetic complete (SC) media containing 2% dextrose overnight at 30 °C. The overnight culture was diluted to OD600=0.3 and allowed to grow another 2hr at 30 °C to reach an OD600 of roughly 0.5. 4 ml of this mid-log culture was transferred to 42 °C for 30 min (unless indicated otherwise) and then recovered at 30 °C for 10min and 23 °C for 5min as described in Liu et al (2010). 1.5mL cultures were used when heat shock was 3.5min. The cells were placed on a thin SC (2% dextrose) agarose gel pad to allow for prolonged imaging (Tran et al., 2004). Confocal movies of Hsp104-GFP-associated aggregates were acquired using a Yokagawa CSU-10 spinning disc on the side port of a Zeiss 200m inverted microscope. 488 nm excitation was used to excite GFP, and emission was collected through a 500–550 nm filter onto a Hamamatsu C9100-13 EMCCD. 3D image stacks were acquired every minute for 60–90 min or every 20 sec for 20min. The latter movies were performed to insure no motility behavior was missed due to insufficient time resolution. In these movies time-lapse recording started 20 min later than the former movies after heat shock recovery in order for better observation of aggregate movement). As we did not observed any apparent difference using two time resolutions, all data analysis presented came from movies with frames 1 minute apart. Each z-series was acquired with 0.5 micron step size and 15 total steps. Visual inspection confirmed that the yeast cells did not leave the focus of the z-stack during the progression of the movie. Max projections were applied for each z-stack to generate the final 2D data set. All images processing was performed in Image J software (NIH).
Particle tracking and calculation of diffusion coefficient
Tracking of Hsp104-GFP-associated protein aggregates was accomplished using MOSAIC ParticleTracker software (Sbalzarini and Koumoutsakos, 2005) as a plugin to ImageJ. Prior to tracking, max-projection time-series were smoothened and a rolling-background subtraction with a width of 5 pixels was applied. Inside the ParticleTracker software, a radius of 1 pixel and a percentile of 1.5% were selected (meaning the top 1.5% of most intense pixels were considered for aggregate tracking). As visual inspection verified that motion spanning 10 pixels in x or y were extremely rare, an interframe linking range of 1 and max displacement per step of 10 pixels were used. In order to calculate a MSD plot, only trajectories lasting longer than 5 time points were considered. The MSD was calculated using the equation, MSD(τ) = 〈|r(t + τ) − r(t)|2)〉, in Mathematica (Wolfram Research, Champaign, IL). The first 5 points were fit to MSD(τ) = 4Dτα using non-linear least squares fit in Mathematica. Here D the diffusion coefficient and α is a factor indicating non-random diffusion.
Dissolution rate measurements
In order to quantitatively compare the dissolution of Hsp104-GFP containing aggregates in various strains, it was necessary to measure the intensity of the aggregates as a function of time. All measurements were performed on background subtracted versions of the maximum projection image for the mask creation and the sum projection image, both performed using ImageJ. A mask of the aggregates was created as follows: the maximum projection image was smoothened with a one pixel radius, followed by background subtraction using the rolling ball background subtraction ImageJ method with a radius of 5 pixels. The image was then thresholded to binary with a threshold of 10000, and finally the binary image was dilated to ensure full coverage of the aggregates. A mask of cell areas was calculated from a smoothened maximum projection image using a threshold of 2500. The cytoplasmic mask was calculated from the aggregates mask and the cell mask using the ImageJ xor function.
Once masks were created, they were used to calculate the average aggregates region intensity (includes cytosolic background), average cytosolic intensity, and total aggregates area. The average cytosolic intensity was subtracted from the average aggregates region intensity to obtain the corrected average aggregates intensity without cytosolic background. Finally, the total aggregates intensity was calculated by multiplying the corrected average aggregates intensity by the aggregates area. Aggregates intensities were measured starting at 30 minutes after the start of acquisition because during the early part of the movies some aggregates grew in intensity. Note the absolute value of aggregates intensity is not important given that they were normalized to the initial intensity for comparison between mutants or conditions.
Supplementary Material
Acknowledgement
The authors thank I. Pinto for assistance with yeast strain construction, J. Zhu and A. Box for help with aged cell sorting, A. Das for assistance in microscopy, and B. Liu and T. Nystrom for kindly providing their Hsp104-GFP strain. This work was supported by NIH grant RO1-GM057063 to RL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION Supplemental information includes Extended Experimental Procedures, two figures, eleven movies and can be found online at ……
References
- Adams AE, Pringle JR. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- Berland K, So PTC, Gratton E. Two-Photon Fluorescence Correlation Spectroscopy: Method and Application to the Intracellular Environment. Biophys J. 1995;68:694–701. doi: 10.1016/S0006-3495(95)80230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosl B, Grimminger V, Walter S. The molecular chaperone Hsp104--a molecular machine for protein disaggregation. J Struct Biol. 2006;156:139–148. doi: 10.1016/j.jsb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Chen C, Dewaele S, Braeckman B, Desmyter L, Verstraelen J, Borgonie G, Vanfleteren J, Contreras R. A high-throughput screening system for genes extending life-span. Exp Gerontol. 2003;38:1051–1063. doi: 10.1016/s0531-5565(03)00186-4. [DOI] [PubMed] [Google Scholar]
- Doyle SM, Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Egilmez NK, Jazwinski SM. Evidence for the involvement of a cytoplasmic factor in the aging of the yeast Saccharomyces cerevisiae. J Bacteriol. 1989;171:37–42. doi: 10.1128/jb.171.1.37-42.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N, Larsson L, Grantham J, Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21:2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4:32–41. doi: 10.1038/ncb718. [DOI] [PubMed] [Google Scholar]
- Gehlen LR, Nagai S, Shimada K, Meister P, Taddei A, Gasser SM. Nuclear geometry and rapid mitosis ensure asymmetric episome segregation in yeast. Curr Biol. 2011;21:25–33. doi: 10.1016/j.cub.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Hebert B, Costantino S, Wiseman PW. Spatiotemporal Image Correlation Spectroscopy (STICS) Theory, Verification, and Application to Protein Velocity Mapping in Living CHO Cells. Biophys J. 2005;88:3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Knorre DA, Kulemzina IA, Sorokin MI, Kochmak SA, Bocharova NA, Sokolov SS, Severin FF. Sir2-dependent daughter-to-mother transport of the damaged proteins in yeast is required to prevent high stress sensitivity of the daughters. Cell Cycle. 2010;9:4501–4505. doi: 10.4161/cc.9.22.13683. [DOI] [PubMed] [Google Scholar]
- Kolin DL, Ronis D, Wiseman PW. k-Space Image Correlation Spectroscopy: A Method for Accurate Transport Measurements Independent of Fluorophore Photophysics. Biophys J. 2006;91:3061–3075. doi: 10.1529/biophysj.106.082768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- Liu B, Larsson L, Caballero A, Hao X, Oling D, Grantham J, Nystrom T. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140:257–267. doi: 10.1016/j.cell.2009.12.031. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279:29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. J Cell Sci. 2000;113(Pt 4):571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Sbalzarini F, Koumoutsakos P. Feature Point Tracking and Trajectory Analysis for Video Imaging in Cell Biology. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P, Schwarz M, Mogk A, Bukau B. The yeast AAA+ chaperone Hsp104 is part of a network that links the actin cytoskeleton with the inheritance of damaged proteins. Mol Cell Biol. 2009;29:3738–3745. doi: 10.1128/MCB.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HC, Pon LA. Actin cable dynamics in budding yeast. Proc Natl Acad Sci U S A. 2002;99:751–756. doi: 10.1073/pnas.022462899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.