Abstract
FoxO3 is a member of FoxO family transcription factors that mediate cellular functions downstream of AKT. FoxO3 phosphorylation by AKT generates binding sites for 14-3-3, which in-turn regulates FoxO3 transcriptional activity and localization. We examine here the functional significance of AKT-FoxO3 interaction and further detail the mechanistic aspects of FoxO3 regulation by AKT and 14-3-3. Our data show that AKT overexpression increases the steady-state levels of FoxO3 protein in a manner dependent on AKT activity and its ability to bind FoxO3. Characterization of the AKT-FoxO3 interaction shows that the three AKT phosphorylation-site-recognition motifs (RxRxxS/T) present on FoxO3, which are required for FoxO3 phosphorylation, are dispensable for AKT binding, suggesting that AKT has a docking point on FoxO3 distinct from the phosphorylation-recognition motifs. Development of a FoxO3 mutant deficient in 14-3-3 binding (P34A), which can be phosphorylated by AKT, established that 14-3-3 binding and not AKT phosphorylation per se controls FoxO3 transcriptional activity. Intriguingly, 14-3-3 binding was found to stabilize FoxO3 by inhibiting its dephosphorylation and degradation rates. Collectively, our data support a model where both AKT and 14-3-3 positively regulate FoxO3 in addition to their established negative roles and that 14-3-3 availability could dictate the fate of phosphorylated FoxO3 toward degradation or recycling.
Keywords: AKT, 14-3-3, FoxO, forkhead domain, transcription factors, protein phosphorylation
1. Introduction
FoxO transcription factors are a family of Forkhead box proteins that include FoxO1 (designated also FoxO1a or FKHR), FoxO3 (FoxO3a/FKHRL1), FoxO4 (AFX) and FoxO6 (for review see [1]. FoxO proteins are evolutionary conserved transcriptional regulators controlled by insulin and insulin-like growth factor receptors in mammals, C. elegans and Drosophila via the PI3K-AKT pathway [2]. They are similar in sequence and contain a highly conserved winged-helix domain, which mediates the DNA binding. FoxO transcription factors coordinate diverse cellular processes including cell proliferation, apoptosis, reactive oxygen species (ROS) response and longevity. Some of their target genes include the pro-apoptotic proteins Bim [3], Bcl-6 [4] and Fas ligand [5], cell cycle regulators such as p27KIP [3], DNA damage response genes such as Gadd45 [6] and oxidative stress response proteins such as MnSOD [7] caveolin-1 [8] and catalase [9]. Given their role in proliferation and apoptosis and the frequent deregulation of their upstream effectors in cancer, FoxO proteins and their target genes are considered attractive targets for cancer therapy [10–12].
While FoxO transcription factors display functional redundancy, some variations in their function have been noted. In mouse knockout models, the FoxO proteins have functional diversity; FoxO1 knockout mice are non-viable while FoxO3 knockout mice are viable, but display abnormal ovarian development [13, 14]. The distribution of FoxO isoforms also varies: FoxO6 and FoxO4 mRNAs are expressed in select tissues, while FoxO3 and FoxO1 mRNAs are ubiquitously expressed, though at varying levels in different tissues [15]. FoxO regulation by post-translational modifications also differs. For example, all FoxO proteins are phosphorylated by AKT; however, FoxO6 lacks the third phosphorylation site present in the other isoforms [1, 16]. Also, only FoxO3 can be phosphorylated and regulated by IκB kinase at Serine 644 [17]. The majority of the research has been focused so far on FoxO1 and FoxO3 isoforms because of their ubiquitous expression pattern and for being the closest homologues of the C. Elegans FoxO, Daf-16, which was originally shown to mediate signaling downstream of the insulin receptor-PI3K-AKT pathway [1, 2, 18].
FoxO3 is regulated by various post-translational modifications including acetylation, ubiquitination and phosphorylation (reviewed in [1, 19]). Phosphorylation, along with a Nuclear Exclusion Sequence (NES) and a Nuclear Localization Sequence (NLS), dictate the subcellular localization of FoxO proteins. Growth factor responsive kinases such as AKT, SGK, CK1 and DYRK1A phosphorylate FoxO proteins and induce their nuclear exclusion, while oxidative stress-regulated kinases such as MST1 and JNK can promote their localization to the nucleus [20, 21].
Many experimental studies have focused on the regulation of FoxO proteins by the PI3K-AKT pathway [5, 22–25]. These studies demonstrated that AKT phosphorylates FoxO proteins on three residues, Threonine 32, Serine 253 and Serine 315 (for human FoxO3). This phosphorylation leads to negative FoxO regulation by causing its nuclear exclusion, which is mediated by 14-3-3 proteins and results in inhibition of FoxO transcriptional output. Additional details of this mechanism were discovered through studying the C. elegans homologue, Daf-16, showing that binding of a 14-3-3 dimer to the transcription factor blocked its DNA binding [26]. This effect of 14-3-3 was confirmed later with human FoxOs [27, 28], showing that 14-3-3 binding to the aminoterminal sites blocks FoxO-DNA interactions.
Following phosphorylation of FoxO by AKT and cytoplasmic retention by 14-3-3, FoxO can be degraded by the proteasome [29]. This negative regulation of FoxO can be inhibited by the addition of the PI3K inhibitor, LY294002. Later studies established Skp2 as the probable E3 ligase for FoxO1 [30] and that FoxO1 could be rescued from degradation by dephosphorylation by PP2a, or a PP2a-like enzyme [31]. Besides the negative regulation by the PI3K-AKT pathway, the mitogen-activated protein kinase, ERK, was shown to phosphorylate FoxO3 at residues S294, S344 and S425, leading to its degradation by an MDM2-ubiquitin-proteosome pathway [32–34].
As noted above, AKT phosphorylates FoxO3 on three residues (Fig. 1A). These residues are located at an AKT phosphorylation-recognition motif, RxRxxS/T [35, 36], with the first site (T32) also containing the 14-3-3 consensus-binding motif RSxpS/TxP [37, 38]. Mutation of all three AKT phosphorylation sites of FoxO3 has been shown to give rise to a constitutively active form, which is primarily located in the nucleus and can reverse and/or suppress cellular transformation [5, 39–41]. The individual AKT phosphorylation sites have been further analyzed by single mutations, suggesting that the S256 site on FoxO1 (S253 in FoxO3) is necessary for the phosphorylation of the T24 site (T32 in FoxO3) [42, 43]. Since the AKT-phosphorylation site mutants show dominant negative effects on AKT activity, it is not clear which one of the observed functions of the mutants is the result of FoxO transcriptional activity and which one stems from AKT inhibition.
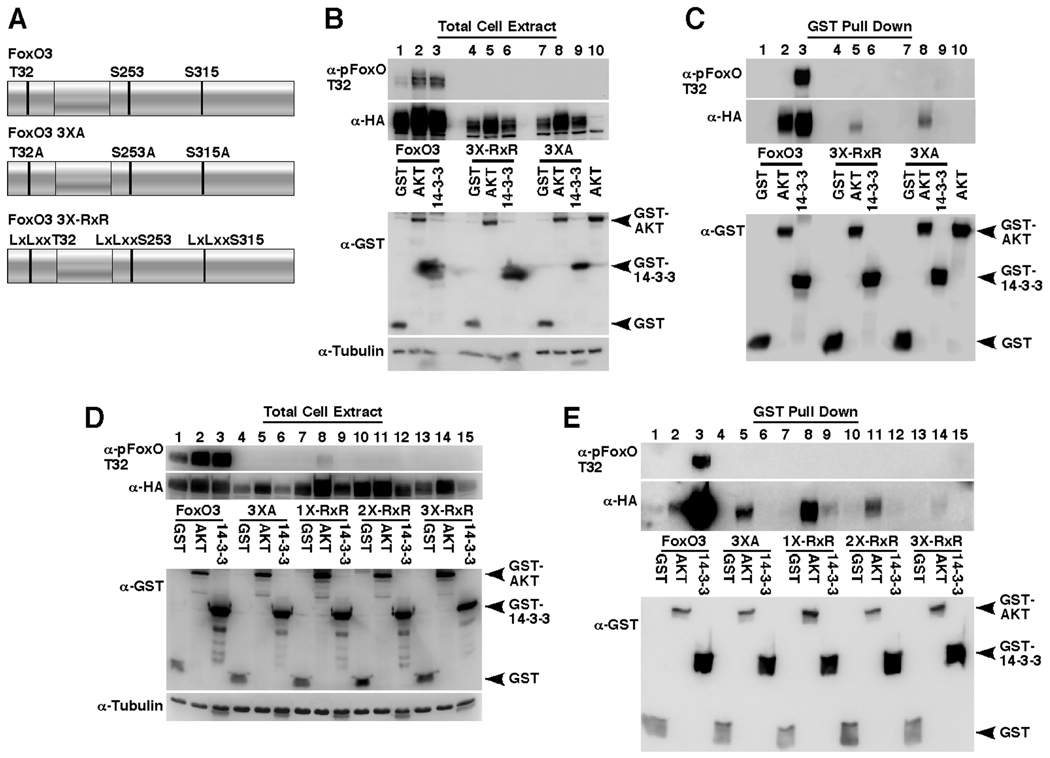
Fig. 1. AKT and 14-3-3ζ stably bind FoxO3 and increase its steady-state protein levels.
(A) Depiction of mammalian and C. elegans FoxO isoforms and corresponding AKT phosphorylation sites.
(B) HEK-293T cells were transfected for 24 hours with HA-FoxO3 together with GST (lane 1), GST-AKT (lane 2) or GST-14-3-3ζ (lane 3). Immunoblotting showing expression of HA-FoxO3, T32-phosphorylated FoxO3 and the GST-fusion proteins in total cell extracts is presented in the left panel and the recovery of the same proteins following GST-pull-down is presented in the right panel.
(C and D) HEK-293T cells were transfected as in B with wildtype FoxO3 (C) or FoxO3 3XA (T32A/S253A/S315A, D) for 24 (lanes 1–3) or 48 hours (lanes 4–6) and expression of the transfected proteins in total cell extracts was examined by immunoblotting.
AKT was shown to have stable interactions with some of its substrates, including FoxO3 [44–46]. Functionally, AKT binding was shown to have both negative and positive effects on its substrates, depending on the target. For example, AKT binding results in inhibition of Par-4 and Merlin activities [47, 48], while increasing the activities of VCP and actin [49, 50]. The mechanistic aspects of AKT interaction with its substrates is, however, largely unknown [44, 51, 52]. The RxRxxS/T phosphorylation-recognition motif was determined by defining the substrate specificity of AKT [35] and by screening peptide and protein libraries [36] and was shown to be necessary for the phosphorylation of all of its known substrates. A peptide containing this motif, derived from GSK3, was shown to bind to the AKT kinase pocket in co-crystallization studies [53]. However a critical question remains regarding the role of this motif in mediating AKT-substrate interactions [52]. In this respect, it appears that the Ser/Thr residue itself is not obligatory for the binding since many substrates with alanine substitution at the phosphorylation site still bind to AKT. The role of the arginines in the binding has not been established. This question is of special significance since defining the binding requirements may offer new ways for targeting the pathway by blocking AKT-substrate interactions, allowing the identification of more substrate-specific inhibitors than the currently available inhibitors that primarily target AKT kinase activity.
The complex nature of AKT and 14-3-3 interactions with their substrates as well as the functional aspects of these interactions on the activity of their substrates, led us to further examine the roles of AKT and 14-3-3 in the regulation of FoxO3. We provide here a more detailed characterization of AKT-FoxO3 interaction by establishing that the AKT phosphorylation-recognition motif, RxRxxS, while critical for the ability of AKT to phosphorylate FoxO3, is negligible for AKT-FoxO3 binding. This finding points to the existence of a docking point on Foxo3 for AKT binding, distinct form the established phosphorylation recognition motif. Furthermore, our data suggest a positive regulation of FoxO3 by both 14-3-3 and AKT, with AKT acting via increasing FoxO3 steady-state protein levels and 14-3-3 via protecting FoxO3 from dephosphorylation and degradation. Thus, our findings propose a more complex mode of regulation of FoxO3 by AKT and 14-3-3 indicating on positive roles in addition to the reported negative roles.
2. Materials and Methods
2.1. Cell Lines, Tissue Culture and Antibodies
293T cells and HepG2 cells were maintained in high-glucose DMEM (Invitrogen) with either 10% Newborn Calf Serum (293T) or 10% Fetal Bovine Serum (HepG2) in a humidified incubator with 5% CO2. Cells for Luciferase Reporter Assays were cultured in 60mm Nunc plates. All other experiments were performed in 100mm Corning plates. The following antibodies were purchased from Cell Signaling Technologies: FoxO3 (#9467), pT32FoxO3 (#9464), pS256FoxO3 (#9466), AKT (#9272), pS473AKT (#9271), GST (#2624). Tubulin monoclonal antibody was purchased from Sigma Aldrich. The HA mouse monoclonal antibody was produced using the 12CA5 hybridoma. Secondary horseradish-conjugated anti-mouse and anti-rabbit antibodies were purchased from Jackson ImmunoResearch Laboratories.
2.2. Plasmid Constructs and Transient Transfections
pcDNA3.1-FoxO3-HA, a gift form Dr. Karen Arden (UCSD), pEBG-GST-14-3-3ζ and pEBG-GST-AKT, a gift from Dr. Jim Woodgett (Samuel Lunenfeld Research Institute), have been previously described [26, 54, 55]. pcDNA3.1-FoxO3-HA and pEBG-GST-AKT mutants and pcDNA3.1-FoxO3 fragments used in this study were created using QuikChange site-directed or multi-site-directed mutagenesis kits (Stratagene) and were confirmed by full-length sequencing. FHBE-Luciferase reporter for FoxO was obtained from Addgene (submitted by Michael Greenberg’s lab [5]). pTK/Renilla Luc reporter was a gift from Dr. Arun Rishi (Wayne State University). Transient transfections were performed using FuGENE HD (Roche) according to the manufacturer’s instructions.
2.3. Cell Extraction, Fractionation and Western Blot Analysis
Cells were lysed in buffer containing 50mM Tris-Cl, pH 7.5, 100mM NaCl, 1% Triton, 1mM EDTA, 1mM EGTA, 1mM DTT, 50mM β-glycerolphosphate, 2mM Na2 VO4 and protease inhibitors and cleared by centrifugation. For nuclear/cytoplasmic fractionation, we used the NE-PER fractionation kit from Pierce according to the manufacturer’s instructions. Protein extracts were separated using SDS-PAGE, transferred to PVDF membranes (0.2uM Immun-Blot, BioRad) and were immuno-blotted using the indicated antibodies followed by ECL. The blots were analyzed using ChemiDoc digital imaging system (BioRad).
2.4. Protein-Protein Interaction Studies
Cells expressing the indicated vectors were lysed and proteins were extracted as detailed above. Equal amounts of protein (1–2mg) were incubated with GSH beads (Amersham/GE Healthcare) for 90 minutes followed by 2X washes with lysis buffer, 1X wash with lysis buffer containing 0.5M LiCl and 2X washes with buffer containing 40mM Tris-Cl, pH 7.5, 0.1mM EDTA and 5mM MgCl2. For the last wash, the GSH beads were transferred to a new tube and the proteins were eluted using SDS-PAGE sample buffer at 95°C for 5 minutes. Total cell extracts were saved from each sample to verify protein expressions.
2.5. FoxO Transcription Reporter Assay
HepG2 cells were co-transfected with the FHBE-Luciferase vector containing FoxO binding sites and the pTK/Renilla Luc control vector together with the indicated pcDNA3.1 control or FoxO3 expression vectors in 60mm plates for 48 hours. Cells were lysed in 200µL lysis buffer and 50µl protein extracts were distributed to a white flat-bottom 96-well plate. Equal amounts of the Dual-Glo Luciferase Assay System (Promega, E2920) buffer A was added to each well and incubated for 15 minutes before reading. The Stop and Glo reagent was added and incubated for 15 minutes before reading Renilla activity. Luminescence was read with a BioTek Synergy 2 using Gen5 software. Expression of FoxO3 variants in each experiment was verified by immunoblotting the cell extracts.
3. Results
3.1. 14-3-3ζ and AKT bind FoxO3 and increase its steady-state protein levels
The binding of FoxO proteins to 14-3-3 has been well established [5, 26, 27] and FoxO binding to AKT also has been reported [45]. To confirm the interaction with AKT, HA-FoxO3 was co-transfected with GST control or with GST-tagged AKT or 14-3-3ζ in 293T cells for 24 hours and a GST pull-down was performed (Fig. 1B). These experiments confirmed that both AKT and 14-3-3ζ were capable of stably binding FoxO3. Interestingly, while 14-3-3ζ was able to pull-down phosphorylated FoxO3, no FoxO3 phosphorylation was detected in AKT pull-downs (Fig. 1B and Fig. 2C, p-FoxO blot, compare lanes 2 and 3). In addition, examination of FoxO3 expression levels in total cell extracts demonstrated that co-expression of AKT increased FoxO3 steady-state protein levels when compared to the GST control (Fig. 1B, total cell extract blot, compare lanes 1 and 2). To further explore this observation, FoxO3 was co-expressed with AKT or 14-3-3ζ for 24 or 48-hours (Fig. 1C). While longer co-expression with AKT did not notably increase FoxO3 levels (Fig. 1C compare lanes 2 and 5), the longer co-expression with 14-3-3ζ significantly increased the steady-state levels of both phosphorylated and total FoxO3 proteins (Fig. 1C compare lanes 3 and 6). Importantly, while AKT was able to bind and increase the steady-state levels of both, wildtype FoxO3 and a mutant lacking the three AKT phosphorylation sites, FoxO3 3XA, 14-3-3ζ was only able to bind and stabilize the wildtype form (Fig. 1D).
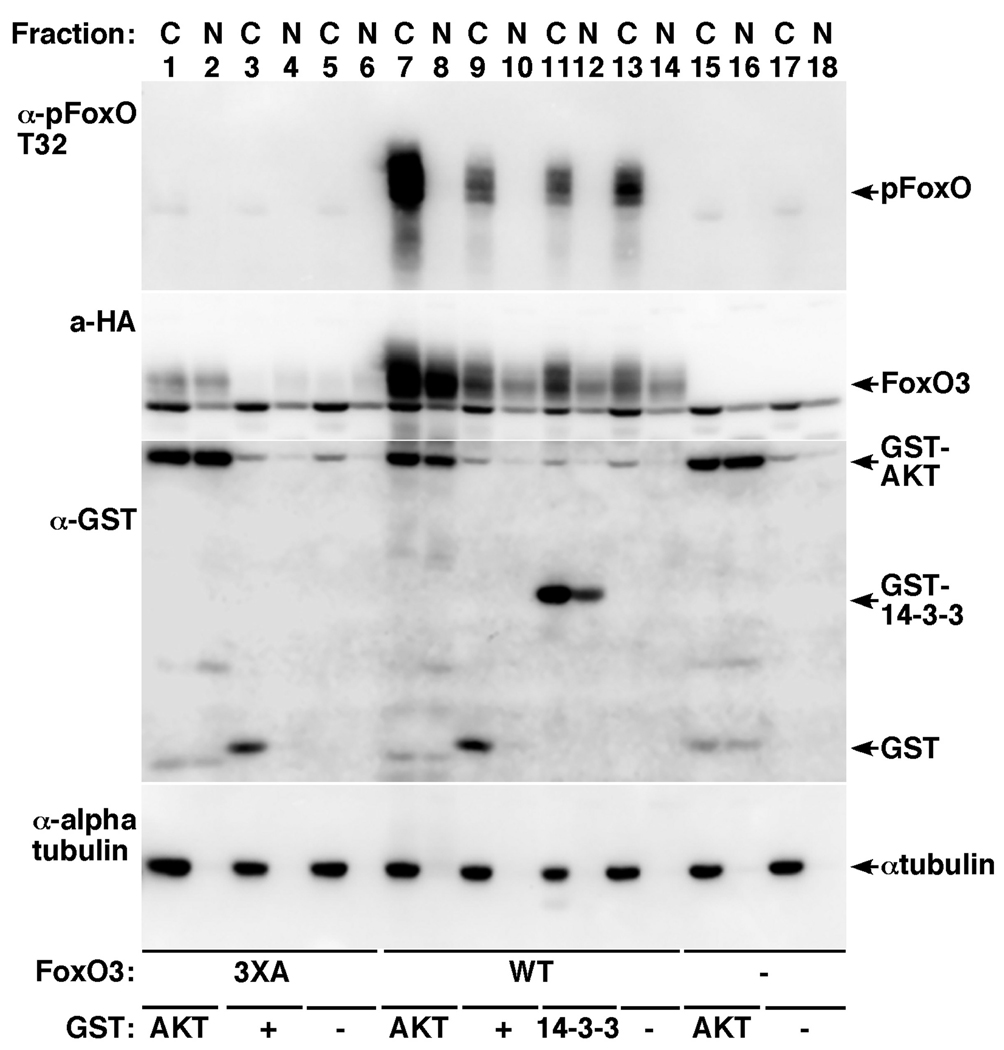
Fig. 2. AKT stabilizes both the nuclear and cytoplasmic fractions of FoxO3.
HEK-293T cells transfected for 24 hours with control vector (lanes 15–18), HA-FoxO3 (lanes 7–14) or HA-FoxO3 3XA mutant (lanes 1–6) together with empty control vector (lanes 5, 6, 13, 14, 17, 18) GST control (lanes 3, 4, 9, 10), GST-AKT (lanes 1, 2, 7, 8, 15, 16) or GST-14-3-3ζ (11, 12) were fractionated to nuclear (N) and cytoplasmic (C) fractions. Immunoblot showing total FoxO3, pT32-FoxO3 and GST-fusion protein expression are presented. Alpha tubulin immunoblot served to verify the purity of the fractionations.
To examine which pool of FoxO3 was stabilized by AKT, cells expressing FoxO3 or the FoxO3 3XA mutant, alone or together with GST control, GST-AKT or GST-14-3-3ζ were fractionated 24h after transfection to cytoplasmic and nuclear fractions and examined for total FoxO3 and pFoxO3 levels (Fig. 2). These experiments showed that AKT expression increased the steady-state protein levels of FoxO3 and the 3XA mutant in both nuclear and cytoplasmic fractions. pFoxO3, however, was only increased in the cytoplasm, corroborating previous observations that pFoxO is being shuttled rapidly out of the nucleus.
3.2. AKT binding to FoxO3 and its ability to enhance FoxO3 steady-state protein levels are not dependent on the RxRxxS/T motif
After confirming that AKT stably binds FoxO3, we wanted to further characterize the AKT-FoxO3 interaction. To examine the role of the three AKT phosphorylation recognition motifs of FoxO3, we substituted the arginines at positions –3 and –5 of the phosphorylation residue with leucines, generating a set of FoxO3 mutants at the RxR motif (illustrated in Fig. 3A). These arginines were shown to be critical for the ability of AKT to phosphorylate peptide substrates [35, 36]. As presented in Fig. 3, co-immunoprecipitation experiments demonstrated that mutations at the RxRxxS/T motifs of FoxO3, abolish binding to 14-3-3ζ but have little effect on AKT binding (Fig. 3C, compare lanes 3, 6 and 9 for 14-3-3ζ binding and lanes 2, 5 and 8 for AKT binding; note that the expression levels of the 3XA and 3X-RxR mutants is significantly lower than wildtype FoxO3, Fig. 3B, compare lanes 1–3, 4–6 and 7–9). In addition, AKT was still able to increase the steady-state protein levels of the RxR and 3XA mutants, while the effect of 14-3-3ζ was abolished (Fig. 3B). These results suggest that the RxRxxS/T motifs do not play a central role in AKT-FoxO3 interaction or the ability of AKT to increase FoxO3 steady-state protein levels.
Fig. 3. The ability of AKT to bind and increase FoxO3 steady-state protein levels is not dependent on the three FoxO3 RxRxxS/T motifs.
(A) A depiction of FoxO3 variants used in this figure: wildtype FoxO3; indicated are the three AKT phosphorylation sites T32, S253 and S315. 3XA, alanine substitutions at the three AKT phosphorylation sites. 3X-RxR, leucine substitutions at the three RxR AKT-phosphorylation-recognition motifs.
(B and C) HEK-293T cells were transfected for 24 hours with HA-FoxO3 (lanes 1–3), HA-FoxO3 3X-RxR (lanes 4–6) or HA-FoxO3 3XA (lanes 7–9) mutant together with GST, GST-AKT or GST-14-3-3ζ. Immunoblots showing expression of HA-FoxO3, T32-phosphorylated FoxO3, tubulin and the GST-fusion proteins in total cell extracts are presented in B and the recoveries of the same proteins following GST-pull-down are presented in C.
(D and E) HEK-293T cells were transfected as in B with HA-FoxO3 (lanes 1–3), HA-FoxO3 3XA (lanes 4–6), HA-FoxO3 1X-RxR (R27/29L, lanes 7–9), HA-FoxO3 2X-RxR (R27/29L, R248/250L, lanes 10–12) or HA-FoxO3 3X-RxR mutant (lanes 13–15) and expression of the transfected proteins in total cell extracts (D) and in GST-pull-downs (E) are presented.
The RxR motifs were also examined individually and AKT was found to bind and increase the steady-state protein levels of the examined mutants to a comparable extent, indicating that these sites play little role in AKT binding or its effect on FoxO3 levels (Fig. 3D and E, compare lanes 2, 5, 8 11 and 14; note that in this experiment we reduced the DNA amounts of wildtype FoxO3 during transfection to achieve comparable expression levels of wildtype and mutant FoxO3 forms). In contrast to AKT, 14-3-3ζ exhibited diminished binding to the RxR mutants and was unable to increase their steady-state protein levels (Fig. 3D and E, compare lanes 3, 6, 9 12 and 15), indicating that the RxR motifs are critical for this positive regulatory role of 14-3-3 on FoxO3.
3.3. The ability of AKT to increase FoxO3 steady-state protein levels involves the N-terminal protein segment
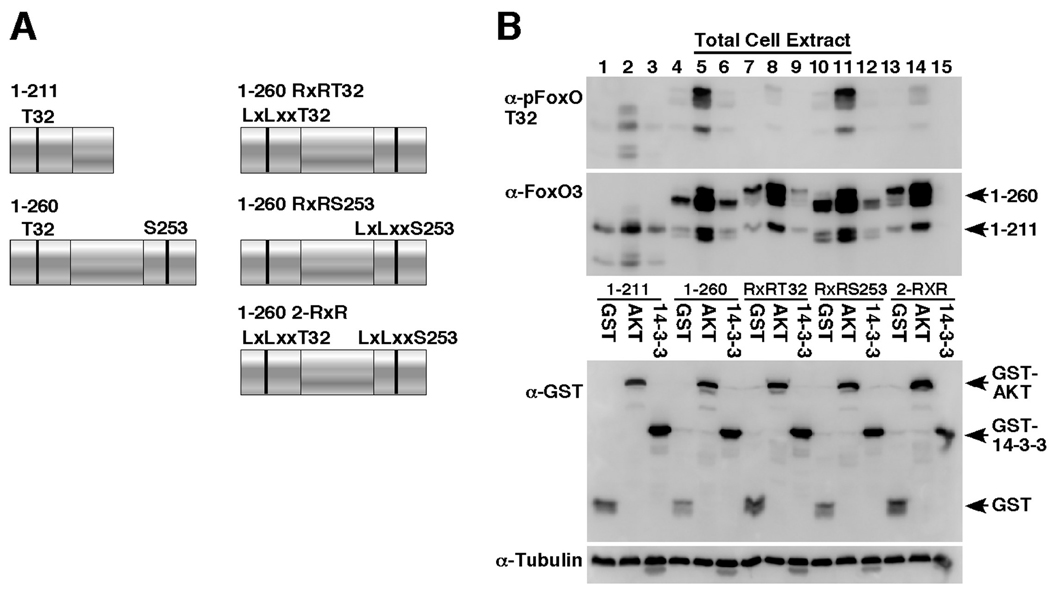
Since the RxR motif mutants were not capable of eliminating the stabilizing effect of AKT on FoxO3, we created several N-terminal FoxO3 deletion variants that included mutations at the RxR motif (illustrated in Fig. 4A). As can be seen in Fig. 4B, the 1–260 N-terminus fragment of FoxO3 is sufficient for the AKT-induced steady-state protein level increase (Fig. 4B, compare lanes 4 and 5). This increase was not dependent on the RxR motif, similarly to the results obtained with full-length FoxO3 (Fig. 4B, compare lanes 7, 10, 13 with lanes 8, 11, 14). The ability of 14-3-3ζ to increase steady-state levels of FoxO3 was eliminated in the fragments, suggesting that 14-3-3ζ needs the full-length FoxO3 protein for this effect (Fig. 4B, compare lanes 4, 7, 10, 13 with lanes 6, 9, 12, 15).
Fig. 4. The ability of AKT to increase FoxO3 steady-state protein levels involves the N-terminal segment.
(A) Depiction of FoxO3 variants used in this figure: 1–211, 1–260 and 1–260 fragment containing leucine substitutions at the RxR AKT-phosphorylation recognition motifs.
(B) HEK-293T cells were transfected for 24 hours with FoxO3 1–211 (lanes 1–3), FoxO3 1–260 (lanes 4–6), FoxO3 1–260 RxRT32 (R27/29L, lanes 7–9), FoxO3 1–260 RxRS253 (R248/250L, lanes 10–12) or FoxO3 1–260 2X-RxR fragment (R27/29L, R248/250L, lanes 13–15) together with GST, GST-AKT or GST-14-3-3ζ. Immunoblots showing expression of FoxO3 fragments, 18 T32-phosphorylated FoxO3, tubulin and the GST-fusion proteins in total cell extracts are presented.
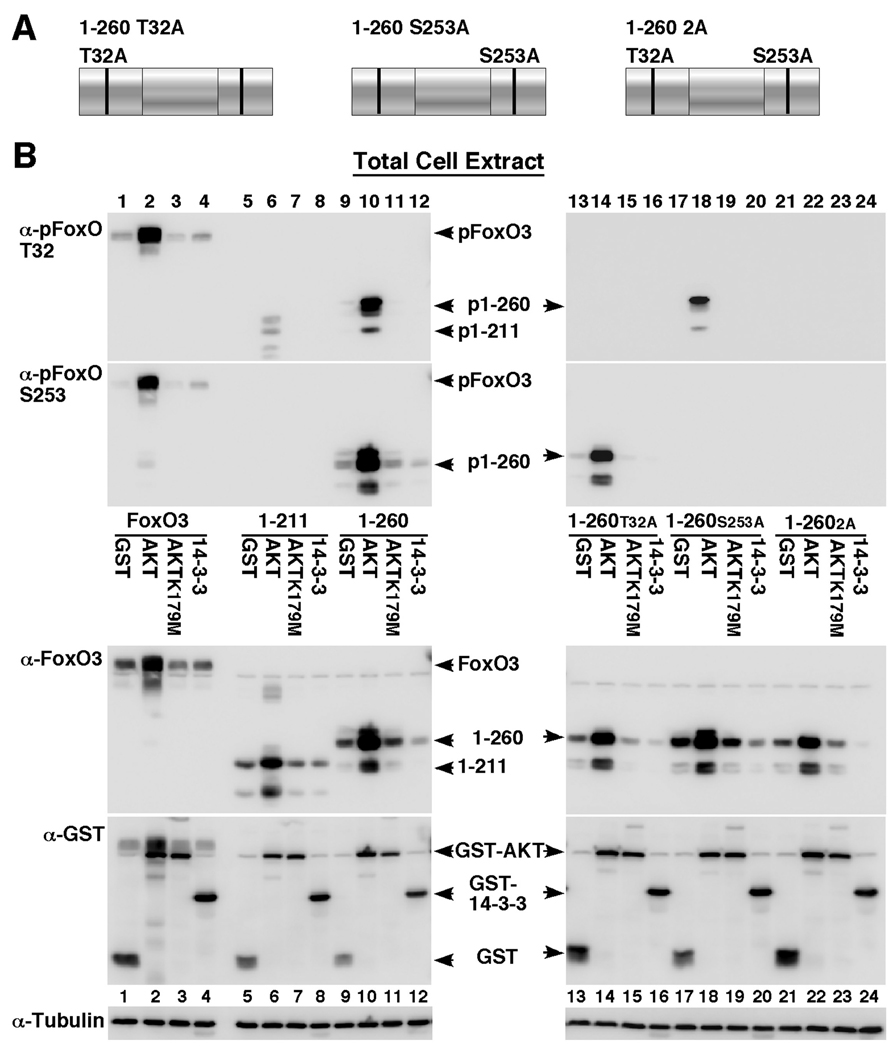
It has been previously reported that the second AKT phosphorylation residue on FoxO1, S256, is necessary for the subsequent phosphorylation of the remaining FoxO residues by AKT [43]. This study used a FoxO1 mutant containing an alanine substitution at the S256 site. However, there is a possibility that the FoxO1 S256A mutant limits the phosphorylation of the other sites by serving as a dominant negative form for AKT in trans rather than acting on FoxO1 itself in cis. To examine this question, we mutated the corresponding phosphorylation sites individually and in tandem on the 1–260 FoxO3 fragment (Fig. 5A) and assayed its phosphorylation on each of the sites (Fig. 5B). These results show that the 1–260 fragment is phosphorylated at both the T32 and S253 residues to the same extent as full-length FoxO3 (Fig. 5B, compare lanes 2 and 10), indicating that the C-terminus truncation that eliminates the third AKT phosphorylation site, S315, does not affect T32 and S253 phosphorylation. In addition, mutation of T32 did not have significant effect on S253 phosphorylation (Fig. 5B, compare lanes 10 and 14) and vice versa, mutation of S253 did not significantly affect T32 phosphorylation (Fig. 5B, compare lanes 10 and 18), suggesting that the phosphorylations at these sites are independent of each other in the context of the 1–260 fragment. Interestingly, an ATP-binding pocket AKT mutant, AKT K179M used as a control in these experiments, failed to increase the steady-state levels of FoxO3 in contrast to wildtype AKT (Fig. 5B, compare lanes 2, 6, 10, 14 18, 22 with lanes 3, 7, 11, 15, 19, 23), suggesting that an intact, kinase active AKT is needed for this function as further detailed below.
Fig. 5. FoxO3 T32 and S253 AKT phosphorylation sites are phosphorylated by AKT independent of each other.
(A) Depiction of FoxO3 variants used in this figure: 1–260 fragment containing alanine substitutions at the T32 and S253 AKT phosphorylation sites.
(B) HEK-293T cells were transfected for 24 hours with HA-FoxO3 (lanes 1–4), FoxO3 1–211 (lanes 5–8), FoxO3 1–260 (lanes 9–12), FoxO3 1–260 T32A (lanes 13–16), FoxO3 1–260 S253A (lanes 17–20) or FoxO3 1–260 2A fragment (T32A/S253A, lanes 21–24) together with GST, GST-AKT, GST-AKT K179M (an inactive, dominant negative AKT form) or GST-14-3-3ζ. Immunoblots showing expression of FoxO3, T32-phosphorylated FoxO3, S253-phosphorylated FoxO3, tubulin and the GST-fusion proteins in total cell extracts are presented.
3.4. AKT activation is required for FoxO3 binding and for increasing FoxO3 steady-state protein levels
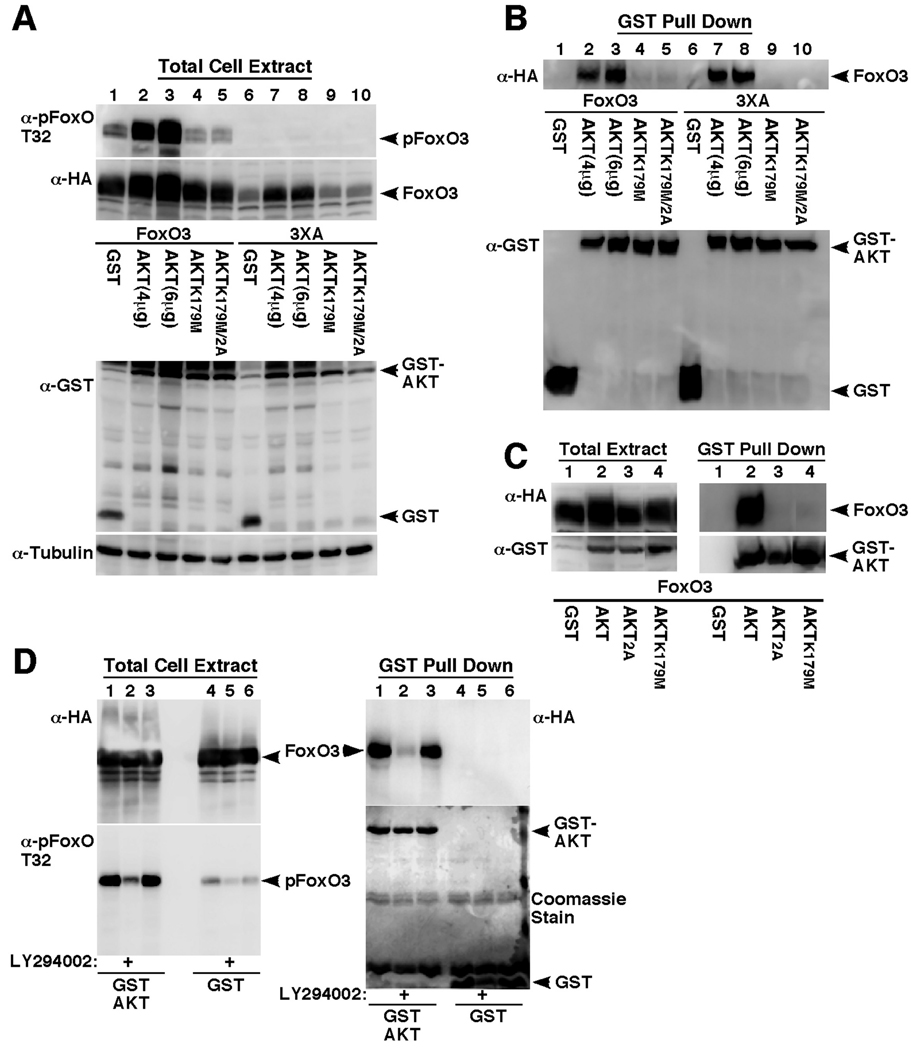
To further understand the mechanism of AKT-FoxO3 binding and the ability of AKT to increase FoxO3 protein levels, we examined the effects of inactivating AKT mutations on its binding to FoxO3 (Fig. 6). As seen in figure 6, an inactivating mutation in the ATP binding pocket (K179M), as well as mutations of the activating AKT phosphorylation sites (T308/S473) interfered with the ability of AKT to bind FoxO3 (Fig. 6B, compare lanes 2, 3 with 4 and 5 for wildtype FoxO3 and lanes 7, 8 with 9 and 10 for FoxO3-3XA mutant and Fig. 6C, compare lane 2 with 3 and 4). In addition, these mutations abolished the ability of AKT to increase FoxO3 steady-state protein levels (Fig. 6A, compare lanes 2, 3 with 4 and 5 and 7, 8 with 9 and 10). Notably, expression of the inactive AKT mutants resulted in lower FoxO3 phosphorylation levels, consistent with previous reports showing dominant negative effects of inactive AKT mutants on endogenous AKT [56]. Though these findings suggest that AKT needs to be active for FoxO3 binding, it is possible that the mutations result in conformational changes that indirectly affect the binding or that they result in altered AKT localization. To address this point, we examined the effect of the PI3K inhibitor LY294002 on AKT binding to FoxO3 (Fig. 6D). As seen in this figure, LY294002 treatment resulted in a marked decrease in FoxO3 phosphorylation, both in control GST-expressing cells and in the GST-AKT expressing cells, indicating on inhibition of AKT activity (Fig. 6D, left panel, compare lanes 2 and 5 with 1 and 4). Importantly, the AKT inhibition was accompanied by a significant dissociation of the AKT/FoxO3 complex (Fig. 6D, right panel, compare lanes 1 and 2), indicating that only active AKT binds to FoxO3, corroborating the results obtained with the inactivating AKT mutations. It still remains to be determined, however, whether the ability of AKT to increase FoxO3 steadystate protein levels depends on AKT binding to FoxO3 or strictly on AKT activity, since the AKT mutants we used and the LY294002 treatment have not distinguish between these two possibilities. Nonetheless, it is clear from the data that this effect is independent of the ability of AKT to phosphorylate FoxO3 itself as demonstrated by the effect of AKT on FoxO3 3XA and 3X-RxR mutants lacking the AKT phosphorylation sites.
Fig. 6. Intact activation sites and ATP binding pocket are required for AKT to bind FoxO3 and enhance its steady-state protein levels.
(A and B) HEK-293T cells were transfected for 24 hours with HA-FoxO3 (lanes 1–5) or HA-FoxO3 3XA (lanes 6–10) together with GST, GST-AKT (4 or 6 µg DNA), GST-AKT K179M or GST-AKT K179M/2A (K179M/T308A/S473A). Immunoblots showing expression of HA-FoxO3, T32-phosphorylated FoxO3, tubulin and the GST-fusion proteins in total cell extracts are presented in A and the recoveries of HA-FoxO3 and GST-fusion proteins following GST-pull-down are presented in B.
(C) HEK-293T cells were transfected for 24 hours with HA-FoxO3 together with GST (lane 1), GST-AKT (lane 2), GST-AKT 2A (T308A/S473A, lane 3) or GST-AKT K179M (lane 4). Immunoblots showing expression of HA-FoxO3 and the GST-fusion proteins in total cell extracts are presented in the left panel and the recoveries of HA-FoxO3 and GST-fusion proteins following GST-pull-down are presented in the right panel.
(D) HEK-293 cells were transfected with HA-FoxO3 together with GST control (lanes 4–6) or GST-AKT (lanes 1–3) and 24 hours after transfection the media was replaced with fresh complete media (lanes 1 and 4) or media lacking serum supplemented with vehicle (lanes 3 and 6 or with 20µM LY294002 (lanes 2 and 5). Immunoblots showing recovery of HA-FoxO3 following GST-pull-down are presented in the right panel. Also shown are GST-fusion protein recoveries and expression of HA-FoxO and pFoxO3 in total cell extracts (left two panels).
3.5. 14-3-3ζ binding protects FoxO3 from dephosphorylation
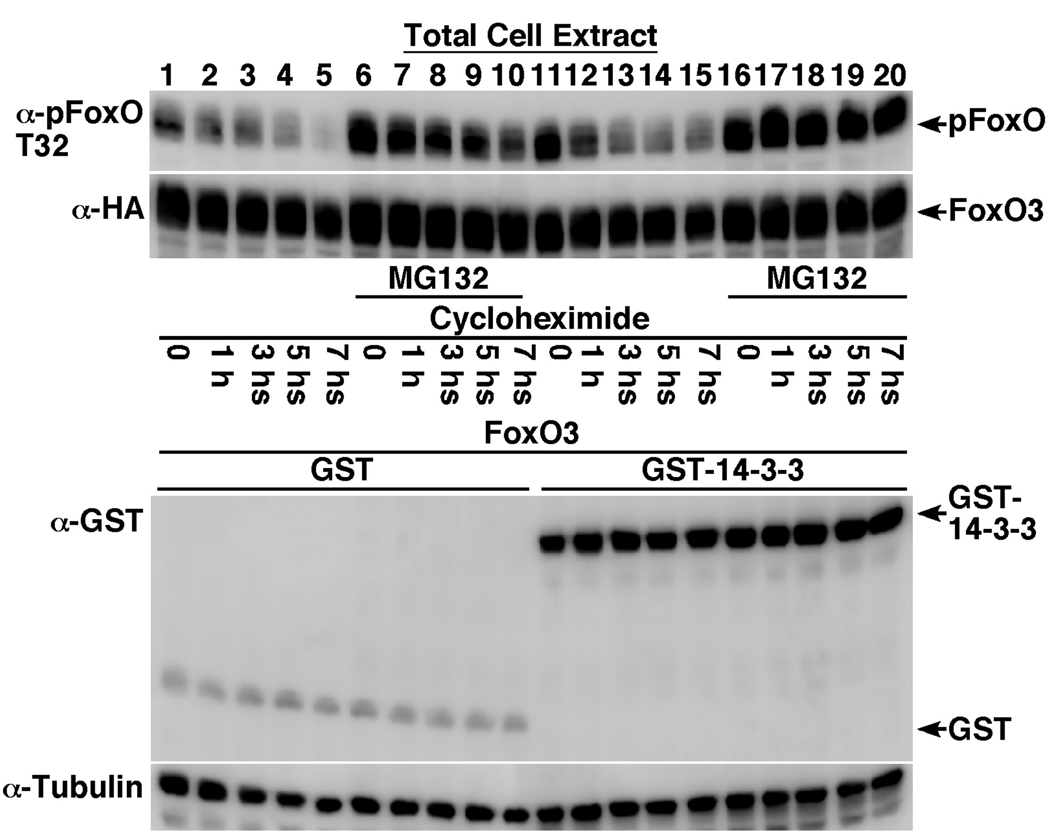
To elucidate the mechanism of 14-3-3ζ-induced increase in total and phosphorylated FoxO3 levels (Fig. 1), we examined the effect of 14-3-3ζ on FoxO3 protein half-life in the presence of the protein translation inhibitor, cycloheximide (Fig. 7). The proteasome inhibitor MG132 was used in these experiments to prevent proteasome-mediated degradation in order to determine whether the increase in phosphorylated-FoxO3 was due to protection from degradation or dephosphorylation. As seen in Fig. 7, basal phosphorylated-FoxO3 steady-state levels were higher in GST-14-3-3ζ expressing cells than in those expressing the GST control (Fig. 7, compare lane 1 and 11). Following 7-hour cycloheximide treatment, p-FoxO3 levels decreased in both, control and GST-14-3-3ζ expressing cells (Fig. 7, compare lane 1–5 with lanes 11–15). Treatment with MG132 resulted in increased p-FoxO3 levels in the control, GST expressing cells, indicating that phosphorylated FoxO3 is subject to degradation. More importantly, while p-FoxO3 levels gradually decreased following cycloheximide treatment in GST-expressing control cells, no decrease in p-FoxO3 levels was observed in GST-14-3-3ζ expressing cells treated with MG132 (Fig. 7, compare lanes 6–10 with lanes 16–20), suggesting that 14-3-3ζ primarily protects FoxO3 from dephosphorylation.
Fig. 7. 14-3-3ζ binding protects FoxO3 from dephosphorylation.
HEK-293T cells were transfected for 24 hours with HA-FoxO3 together with GST (lanes 1–10) or GST-14-3-3ζ (lanes 11–20). Cells were treated with vehicle (lanes 1–5 and 11–15) or 10µM MG132 (proteasome inhibitor, lanes 6–10 and 16–20) for 2 hours, followed by treatment with 25µg/ml cycloheximide (protein translation inhibitor) for the indicated periods. Cells were lysed and equal protein amounts were analyzed for total and T32-phosphorylated FoxO3 levels by immunoblotting. Also presented are GST and tubulin immunoblots showing equal protein loading.
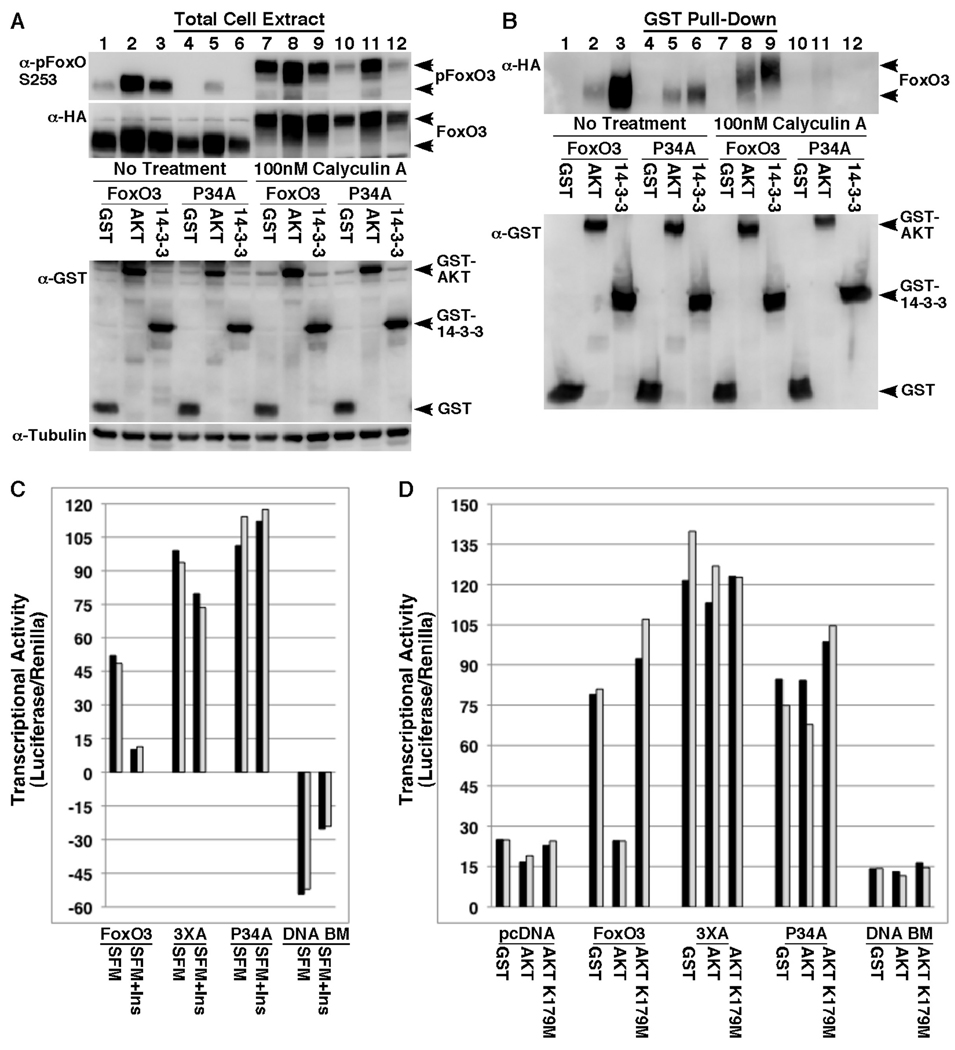
To further investigate this effect of 14-3-3ζ on phosphorylated-FoxO3 and to support the idea that 14-3-3ζ inhibits FoxO3 dephosphorylation, we generated a FoxO3 mutant, FoxO3 P34A. This mutant was designed to enable regulation by AKT (binding, phosphorylation and stabilization), but to interfere with 14-3-3 binding. This approach took advantage of the 14-3-3-binding requirements, which necessitates a proline at the +2 position of the phosphorylated residue; RSxpS/TxP [37, 57], P34 in FoxO3. Importantly, the +2 position has been shown to not have a significant effect on the ability of AKT to phosphorylate peptide substrates [36]. As seen in Fig. 8, the P34A FoxO3 mutant was indeed able to bind AKT to the same extent as wildtype FoxO3 (Fig. 8B, compare lanes 2 and 5), but showed a dramatic decrease in its ability to bind 14-3-3 (Fig. 8B, compare lanes 3 and 6), demonstrating the critical role of the proline at position 34 for 14-3-3 binding, while not affecting binding to AKT. In addition, AKT was able to increase the steady-state protein levels of both, the P34A mutant and wildtype FoxO3 (Fig. 8A, HA- blot, compare lane 1 with 2 and lane 4 with 5), while 14-3-3 affected wildtype FoxO3 but not the P34A mutant (Fig. 8, HA-blot, compare lane 1 with 3 and lane 4 with 6). This result was similar to the results obtained with the AKT phosphorylation site mutants that did not bind 14-3-3, i.e. the RxR and the 3XA FoxO3 mutants. Thus, these results established the P34A mutant as a useful tool for studying the role of 14-3-3 binding in the stabilization of phosphorylated-FoxO3, without seemingly affecting its regulation by AKT. To this effect, the FoxO3 P34A mutant exhibited diminished steady-state phosphorylation at the S253 site (Fig. 8A, p-FoxO blot, compare lanes 1–3 with 4–6). Importantly, addition of the phosphatase inhibitor calyculin A that inhibits PP2A and PP1A was able to restore phosphorylation, especially in the presence of AKT (Fig. 8A, compare lanes 7–9 with 10–12), suggesting that the mutant is impaired in its dephosphorylation rate rather than its accessibility to AKT. Calyculin A was selected for these experiments since PP2A was shown in previous studies to be the main phosphatase regulating FoxO dephosphorylation at the AKT phosphorylation sites [31, 58]. Similar results were obtained using antibodies for the T32 site (data not shown), however, since P34 could be part of the recognition site of the antibody, only the results obtained with the pS253 antibody offer conclusive evidence. Overall, our findings support the view that 14-3-3ζ binding protects FoxO3 from dephosphorylation, which in turn results in increased steady-state FoxO3 protein levels, thus offering a positive role of 14-3-3ζ in FoxO3 regulation in addition to its established negative role.
Fig. 8. Mutation of FoxO3 P34 diminishes binding to 14-3-3ζ and results in a constitutively active form.
(A and B) HEK-293T cells were transfected for 24 hours with HA-FoxO3 (lanes 1–3 and 7–9) or HA-FoxO3 P34A mutant (lanes 4–6 and 10–12) together with GST, GST-AKT or GST-14-3-3ζ. Cells were treated with vehicle (lanes 1–6) or with 100nM of the phosphatase inhibitor calyculin A (lanes 7–12) for one hour prior to harvesting. Cell lysates were analyzed directly (A) or following GST pull-down (B) for expression of FoxO3, S253-phosphorylated-FoxO3, tubulin and GST-fusion proteins by immunoblotting.
(C) HepG2 cells were transfected for 24h in duplicates with FHBE-Luciferase and RTK-Renilla control vector together with pcDNA control vector, HA-FoxO3, HA-FoxO3 3XA, HA-FoxO3 P34A or HA-FoxO3 DNA BM (R211A/S253E, DNA-binding mutant). Cells were incubated for additional 24h with serum-free media (SFM) or with serum-free media supplemented with 20µg/ml insulin (SFM+Ins) and analyzed for Luciferase activity using the Promega Dual-Glo Luciferase assay system. For calculating the activity of each FoxO variant, the Luciferase/Renilla ratio obtained in samples transfected with pcDNA control vector were subtracted from the values obtained with the FoxO variants. Each bar represented a single transfected sample.
(D) HepG2 cells were transfected for 48h in duplicates with FHBE-Luciferase and RTK-Renilla control vector together with pcDNA control vector, HA-FoxO3, HA-FoxO3 3XA, HA-FoxO3 P34A or HA-FoxO3 DNA BM (R211A/S253E, DNA-binding mutant) and together with GST control, GST-AKT or GST-AKT K179M (an inactive ATP-binding pocket mutant) as indicated. Cells were analyzed for Luciferase activity and presented are Luciferase/Renilla ratios. Each bar represented a single transfected sample.
3.6. Constitutive transcriptional activity of FoxO3 P34A mutant suggests that 14-3-3 binding rather than AKT phosphorylation is the key factor in negative FoxO regulation
To determine the functional significance of 14-3-3 binding to FoxO3, we compared the transcriptional activity of FoxO3 P34A mutant with that of wildtype FoxO3 and the 3XA FoxO3 mutant lacking the three AKT phosphorylation sites (Fig. 8C and D). These experiments show that wildtype FoxO3 activity can be suppressed by insulin treatment in HepG2 cells, while the 3XA mutant exhibits constitutive activity, as previously reported. The P34A FoxO3 mutant exhibited as high activity as the 3XA mutant, and was not affected by insulin treatment, providing evidence that the P34A mutation results in constitutive activation of FoxO3 (Fig. 8C). In addition, even co-expression with AKT, which results in a strong inhibition of wildtype FoxO3, failed to inhibit the transcriptional activity of the P34A mutant (Fig. 8D), demonstrating its usefulness in study models that have high AKT activity. Indeed, we obtained similar results in PC3 prostate cancer cells that carry high AKT activity due to inactivation of the PTEN lipid phosphatase (data not shown). These results suggest that 14-3-3 binding rather than AKT phosphorylation per se is the key factor in the negative regulation of FoxO3 transcriptional activity.
4. Discussion
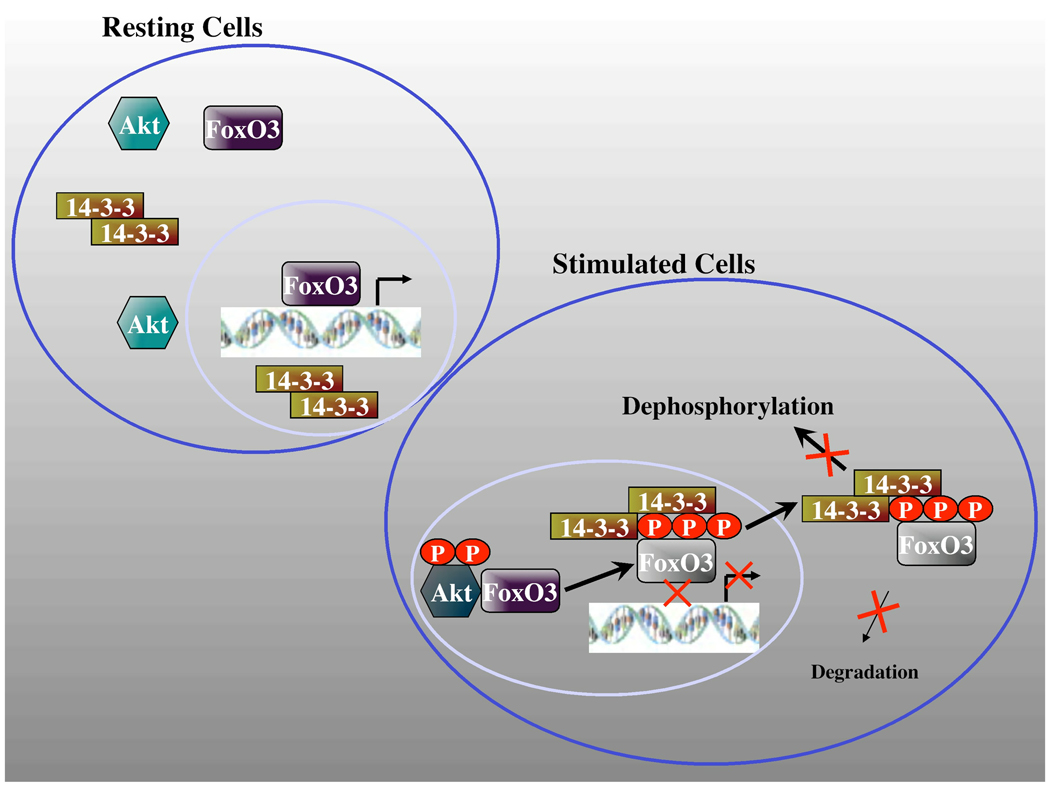
The results presented here provide several new findings that extend our understanding of the complex regulation of FoxO3 by AKT and 14-3-3 and offer a new model of FoxO regulation were both 14-3-3 and AKT have positive roles in FoxO3 regulation besides their established negative roles (Fig. 9). In this model, we propose that active AKT binds FoxO3 through a docking point distinct from the RxRxxS consensus AKT-phosphorylation recognition motif and increases the steady-state levels of FoxO3 protein, either directly or indirectly through the activation of other downstream targets. Since we have not observed binding of phosphorylated-FoxO3 with AKT, we propose that following FoxO phosphorylation, AKT-FoxO complexes dissociate, possibly as a result of competition with 14-3-3. The phosphorylation by AKT does not promote by-itself the inhibition of FoxO3 transcriptional activity, rather, 14-3-3 binding serves as the key point in this inhibition, presumably via blocking FoxO3-DNA binding and promoting its nuclear exclusion. At the same time, however, 14-3-3 protects FoxO3 from dephosphorylation and degradation, thus, maintaining a cytoplasmic pool of FoxO3 that can be recycled. Hence, the availability of free 14-3-3 in the cell could dictate the rate of FoxO3 inactivation and also the fate of phosphorylated-FoxO3; either toward degradation or recycling.
Fig. 9. A Model for FoxO3 regulation by AKT and 14-3-3.
AKT activation by growth factors that activate the PI3K pathway induces AKT binding to FoxO3, resulting in increased steady-state FoxO3 levels. Phosphorylated FoxO3 dissociates from AKT, possibly as a consequence of competition with 14-3-3. 14-3-3 binding to phosphorylated FoxO3 blocks its DNA binding and transcriptional activity and also promotes its nuclear exclusion. In the cytoplasm, 14-3-3 binding inhibits FoxO3 dephosphorylation and degradation, thus maintaining a pool that can be recycled. See text for more details.
The proposed model is supported by results demonstrating that AKT binding to FoxO3 is not dependent on the AKT phosphorylation recognition motif RxRxxS/T, as far as neither mutating the phosphorylation site or the arginines has affected FoxO3-AKT interaction. This finding suggests that AKT has a docking point on FoxO3, which is different from the established phosphorylation recognition motif. This is a significant finding that could be relevant also to other AKT substrates that were reported to stably associate with AKT. Currently our understanding of AKT binding to its substrates is very limited and the domains in AKT or the substrate that mediate the binding have not been defined [46, 51, 52]. Crystallography analyses of AKT kinase domain co-crystallized with a substrate peptide pinpointed residues in the AKT kinase pocket that interact with the arginines in the substrate peptide, however, their role in AKT-target binding has not been examined [53, 59, 60]. Also, it has not been determined whether these interactions are of high enough affinity to mediate the observed stable interaction of AKT with its substrates. Our results clearly show that mutation of the phosphorylation recognition motif does not affect the binding of AKT to FoxO3, thus at least in the case of FoxO3, there appears to be a different domain on FoxO3 that mediates the binding. Surprisingly, however, although the phosphorylation recognition motif was not required for AKT binding, AKT inhibition and mutations that impair its activation or catalytic function blocked the binding to FoxO3. This finding suggests that AKT activation state and the examined residues, directly or indirectly contribute to the interaction. One possible explanation for the observed results is that AKT autophosphorylation plays a role in attaining or maintaining the right conformation for FoxO3 binding [61]. In this regard, AKT binding to several of its substrates has been shown to be impaired by the K179 or the T308/S473 mutations (e.g., SETDB1, B23/NPM and Merlin) [48, 62, 63], however, in the case of several other substrates, no effect of these mutations was seen with regards to substrate binding (e.g., Bad and PEA-15) [64, 65]. Detailed structural studies will be probably required to resolve these questions and to reveal the mechanistic aspects of AKT interactions with FoxO and its other targets.
An interesting characteristic of AKT-FoxO3 interaction was our inability to detect association of phosphorylated FoxO3 with AKT, suggesting that the complex dissociates following phosphorylation, either through conformational change in FoxO3 or as a result of displacement by 14-3-3.
Another novel aspect of this study is the demonstration that co-expressing AKT with FoxO3 increases the steady-state levels of FoxO3 protein. This effect of AKT was not dependent on the FoxO3 RxRxxS/T motifs and a fragment containing the first 1–260 amino acids exhibited same increase as the full-length protein. It was, however, dependent on having an intact AKT, since mutations of AKT activation sites or the ATP binding pocket, which also abrogated the binding of AKT to FoxO3, abolished the enhancement. Thus, since FoxO3 phosphorylation itself was not needed for this enhancement and our AKT mutants were unable to distinguish between the binding requirements and catalytic activity, the possibilities for this effect are that AKT increases the protein levels through downstream signaling events or through direct binding to FoxO3. Initial experiments excluded the involvement of the mTORC1 complex, as rapamycin treatment did not block this effect (data not shown). Elucidation of the FoxO3 interaction point with AKT and vice versa, may help in addressing this question. AKT binding has been shown to decrease the degradation of several of its targets, including PEA-15 and B23/NPM [63, 65], raising the 11 possibility that also in the case of FoxO3, AKT may be increasing the protein levels through direct binding rather than downstream signaling events or a feedback mechanism. Nonetheless, the observed increase in steady-state FoxO3 protein levels suggests a potential positive regulatory role of AKT in FoxO3 regulation besides the established negative role. The correlation between AKT activation and FoxO protein expression has been observed in T cells: it has been seen in two separate studies that AKT activation by PTEN deletion correlated with increased FoxO protein levels [66], while inhibition of AKT associated with decreased FoxO protein levels [67].
The regulation of FoxO proteins by 14-3-3 has been studied extensively, establishing a negative regulatory role through blocking their DNA binding and sequestering them in the cytoplasm [5, 26–28, 43, 68]. The results presented here show that 14-3-3 proteins can also serve a positive regulatory role by increasing the steady-state protein levels of phosphorylated and total FoxO3. This effect of 14-3-3 was dependent on the ability of 14-3-3 to bind FoxO3, since various FoxO3 mutants impaired in 14-3-3 binding were not affected. The stabilization involved primarily protection of phosphorylated FoxO3 from dephosphorylation at the AKT phosphorylation sites. Interestingly, a positive role of 14-3-3 in protecting their targets from proteolytic cleavage was observed in Arabidopsis [69] and 14-3-3 was shown to protect FoxO1 degradation in vitro by Arabidopsis-derived proteases. Our findings are also in agreement with the current literature showing that significant phosphorylated-FoxO protein levels can be found in the cytoplasm, indicating that they are not being immediately degraded. This observation suggests that there may be a time frame in which phosphorylated-FoxO can be salvaged through dephosphorylation and shuttled back into the nucleus [58, 70]. Therefore, the availability of unbound 14-3-3 in the cell, which can be modulated by various factors and cell conditions [57, 71], could affect the fate of phosphorylated FoxO3 to either stabilization and recycling or degradation.
An important finding in this study was the observation that the FoxO3 P34A mutant, which has a decreased capacity for 14-3-3 binding, but can be regulated by AKT in a similar fashion to the wildtype, exhibited a constitutive transcriptional activity that could not be inhibited even by the overexpression of AKT (Fig. 8D). This finding suggests that 14-3-3 binding rather than AKT phosphorylation itself serves as the key point in the negative regulation of FoxO3. This view is in agreement with the results of Obsil et al. [27], showing that FoxO4-14-3-3 binding motifs are necessary for the inhibition of FoxO4-DNA binding. The significance of this finding is that it suggests that under stress conditions such as oxidative stress, were 14-3-3 proteins have been shown to undergo inactivating phosphorylation by stress kinases such as JNK [72, 73], FoxO proteins may be less prone to negative regulation by AKT, since phosphorylated FoxO will still be functional without having 14-3-3 exerting the negative regulation. It remains to be determined whether under these conditions FoxO proteins undergo rapid dephosphorylation and recycling, without having 14-3-3 proteins protecting their dephosphorylation, or whether they are subjected to accelerated degradation.
Overall, our data provide a more comprehensive view of FoxO3 regulation by AKT and 14-3-3 and suggest positive roles for both 14-3-3 and AKT in FoxO3 regulation besides the previously described negative regulatory roles.
Research Highlights.
AKT binding to FoxO3 is independent of the RxRxxS/T AKT phosphorylation recognition motif
AKT activation is required for FoxO3 binding
AKT expression increases FoxO3 steady-state protein levels
14-3-3 binding stabilizes phosphorylated FoxO3
FoxO3 P34A mutation impairs 14-3-3 binding, resulting in a constitutively active form
Acknowledgments
This work was supported in part by the National Institute of Health Grant R01 GM 067134 (G. T.) and NCI T32-CA009531 Training Grant (M.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 2.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, Medema RH. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 4.Tang TT, Dowbenko D, Jackson A, Toney L, Lewin DA, Dent AL, Lasky LA. The forkhead transcription factor AFX activates apoptosis by induction of the BCL-6 transcriptional repressor. J Biol Chem. 2002;277:14255–14265. doi: 10.1074/jbc.M110901200. [DOI] [PubMed] [Google Scholar]
- 5.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 6.Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ, Jr, DiStefano PS, Chiang LW, Greenberg ME. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 7.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 8.van den Heuvel AP, Schulze A, Burgering BM. Direct control of caveolin-1 expression by FOXO transcription factors. Biochem J. 2005;385:795–802. doi: 10.1042/BJ20041449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan WQ, Wang K, Lv DY, Li PF. Foxo3a inhibits cardiomyocyte hypertrophy through transactivating catalase. J Biol Chem. 2008;283:29730–29739. doi: 10.1074/jbc.M805514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carnero A, Blanco-Aparicio C, Renner O, Link W, Leal JF. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets. 2008;8:187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 11.Dansen TB, Burgering BM. Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 2008;18:421–429. doi: 10.1016/j.tcb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 13.Hosaka T, Biggs WH, Tieu D, Boyer AD, Varki NM, Cavenee WK, Arden KC. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. P Natl Acad Sci USA. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tieu DD, Hosaka T, Biggs WH, Cavenee WK, Arden KC. FKHR1 (FOXO1) is essential for mouse embryonic angtogenesis and vasculogenesis. J Invest Med. 2003;51:S177–S177. [Google Scholar]
- 15.Biggs WH, 3rd, Cavenee WK, Arden KC. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm Genome. 2001;12:416–425. doi: 10.1007/s003350020002. [DOI] [PubMed] [Google Scholar]
- 16.Barthel A, Schmoll D, Unterman TG. FoxO proteins in insulin action and metabolism. Trends Endocrin Met. 2005;16:183–189. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, Kobayashi R, Hung MC. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 18.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 19.Vogt PK, Jiang H, Aoki M. Triple layer control: phosphorylation, acetylation and ubiquitination of FOXO proteins. Cell Cycle. 2005;4:908–913. doi: 10.4161/cc.4.7.1796. [DOI] [PubMed] [Google Scholar]
- 20.Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 21.Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, Bos JL, Burgering BM. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biggs WH, 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci U S A. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 24.Rena G, Guo S, Cichy SC, Unterman TG, Cohen P. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J Biol Chem. 1999;274:17179–17183. doi: 10.1074/jbc.274.24.17179. [DOI] [PubMed] [Google Scholar]
- 25.Woods YL, Rena G. Effect of multiple phosphorylation events on the transcription factors FKHR, FKHRL1 and AFX. Biochem Soc Trans. 2002;30:391–397. doi: 10.1042/bst0300391. [DOI] [PubMed] [Google Scholar]
- 26.Cahill CM, Tzivion G, Nasrin N, Ogg S, Dore J, Ruvkun G, Alexander-Bridges M. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J Biol Chem. 2001;276:13402–13410. doi: 10.1074/jbc.M010042200. [DOI] [PubMed] [Google Scholar]
- 27.Obsil T, Ghirlando R, Anderson DE, Hickman AB, Dyda F. Two 14-3-3 binding motifs are required for stable association of Forkhead transcription factor FOXO4 with 14-3-3 proteins and inhibition of DNA binding. Biochemistry. 2003;42:15264–15272. doi: 10.1021/bi0352724. [DOI] [PubMed] [Google Scholar]
- 28.Silhan J, Vacha P, Strnadova P, Vecer J, Herman P, Sulc M, Teisinger J, Obsilova V, Obsil T. 14-3-3 protein masks the DNA binding interface of forkhead transcription factor FOXO4. J Biol Chem. 2009;284:19349–19360. doi: 10.1074/jbc.M109.002725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aoki M, Jiang H, Vogt PK. Proteasomal degradation of the FoxO1 transcriptional regulator in cells transformed by the P3k and Akt oncoproteins. Proc Natl Acad Sci U S A. 2004;101:13613–13617. doi: 10.1073/pnas.0405454101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, Tindall DJ. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan L, Lavin VA, Moser LR, Cui Q, Kanies C, Yang E. PP2A regulates the pro-apoptotic activity of FOXO1. J Biol Chem. 2008;283:7411–7420. doi: 10.1074/jbc.M708083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC, Huang H, Kuo HP, Lee DF, Li LY, Lien HC, Cheng X, Chang KJ, Hsiao CD, Tsai FJ, Tsai CH, Sahin AA, Muller WJ, Mills GB, Yu D, Hortobagyi GN, Hung MC. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–148. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W, Dolloff NG, El-Deiry WS. ERK and MDM2 prey on FOXO3a. Nat Cell Biol. 2008;10:125–126. doi: 10.1038/ncb0208-125. [DOI] [PubMed] [Google Scholar]
- 34.Fu W, Ma Q, Chen L, Li P, Zhang M, Ramamoorthy S, Nawaz Z, Shimojima T, Wang H, Yang Y, Shen Z, Zhang Y, Zhang X, Nicosia SV, Pledger JW, Chen J, Bai W. MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J Biol Chem. 2009;284:13987–14000. doi: 10.1074/jbc.M901758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 36.Obata T, Yaffe MB, Leparc GG, Piro ET, Maegawa H, Kashiwagi A, Kikkawa R, Cantley LC. Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J Biol Chem. 2000;275:36108–36115. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- 37.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 38.Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 39.Kikuchi S, Nagai T, Kunitama M, Kirito K, Ozawa K, Komatsu N. Active FKHRL1 overcomes imatinib resistance in chronic myelogenous leukemia-derived cell lines via the production of tumor necrosis factor-related apoptosis-inducing ligand. Cancer Sci. 2007;98:1949–1958. doi: 10.1111/j.1349-7006.2007.00623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H, Zhao R, Yang HY, Lee MH. Constitutively active FOXO4 inhibits Akt activity, regulates p27 Kip1 stability, and suppresses HER2-mediated tumorigenicity. Oncogene. 2005;24:1924–1935. doi: 10.1038/sj.onc.1208352. [DOI] [PubMed] [Google Scholar]
- 41.Park KW, Kim DH, You HJ, Sir JJ, Jeon SI, Youn SW, Yang HM, Skurk C, Park YB, Walsh K, Kim HS. Activated forkhead transcription factor inhibits neointimal hyperplasia after angioplasty through induction of p27. Arterioscler Thromb Vasc Biol. 2005;25:742–747. doi: 10.1161/01.ATV.0000156288.70849.26. [DOI] [PubMed] [Google Scholar]
- 42.Nakae J, Barr V, Accili D. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J. 2000;19:989–996. doi: 10.1093/emboj/19.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rena G, Prescott AR, Guo S, Cohen P, Unterman TG. Roles of the forkhead in rhabdomyosarcoma (FKHR) phosphorylation sites in regulating 14-3-3 binding, transactivation and nuclear targetting. Biochem J. 2001;354:605–612. doi: 10.1042/0264-6021:3540605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du K, Tsichlis PN. Regulation of the Akt kinase by interacting proteins. Oncogene. 2005;24:7401–7409. doi: 10.1038/sj.onc.1209099. [DOI] [PubMed] [Google Scholar]
- 45.Zheng WH, Kar S, Quirion R. Insulin-like growth factor-1-induced phosphorylation of the forkhead family transcription factor FKHRL1 is mediated by Akt kinase in PC12 cells. J Biol Chem. 2000;275:39152–39158. doi: 10.1074/jbc.M002417200. [DOI] [PubMed] [Google Scholar]
- 46.Brazil DP, Park J, Hemmings BA. PKB binding proteins. Getting in on the Akt. Cell. 2002;111:293–303. doi: 10.1016/s0092-8674(02)01083-8. [DOI] [PubMed] [Google Scholar]
- 47.Goswami A, Burikhanov R, de Thonel A, Fujita N, Goswami M, Zhao Y, Eriksson JE, Tsuruo T, Rangnekar VM. Binding and phosphorylation of par-4 by akt is essential for cancer cell survival. Mol Cell. 2005;20:33–44. doi: 10.1016/j.molcel.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 48.Tang X, Jang SW, Wang X, Liu Z, Bahr SM, Sun SY, Brat D, Gutmann DH, Ye K. Akt phosphorylation regulates the tumour-suppressor merlin through ubiquitination and degradation. Nat Cell Biol. 2007;9:1199–1207. doi: 10.1038/ncb1641. [DOI] [PubMed] [Google Scholar]
- 49.Klein JB, Barati MT, Wu R, Gozal D, Sachleben LR, Jr, Kausar H, Trent JO, Gozal E, Rane MJ. Akt-mediated valosin-containing protein 97 phosphorylation regulates its association with ubiquitinated proteins. J Biol Chem. 2005;280:31870–31881. doi: 10.1074/jbc.M501802200. [DOI] [PubMed] [Google Scholar]
- 50.Vandermoere F, El Yazidi-Belkoura I, Demont Y, Slomianny C, Antol J, Lemoine J, Hondermarck H. Proteomics exploration reveals that actin is a signaling target of the kinase Akt. Mol Cell Proteomics. 2007;6:114–124. doi: 10.1074/mcp.M600335-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 53.Yang J, Cron P, Good VM, Thompson V, Hemmings BA, Barford D. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat Struct Biol. 2002;9:940–944. doi: 10.1038/nsb870. [DOI] [PubMed] [Google Scholar]
- 54.Anderson MJ, Viars CS, Czekay S, Cavenee WK, Arden KC. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics. 1998;47:187–199. doi: 10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]
- 55.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 56.Cichy SB, Uddin S, Danilkovich A, Guo S, Klippel A, Unterman TG. Protein kinase B/Akt mediates effects of insulin on hepatic insulin-like growth factor-binding protein-1 gene expression through a conserved insulin response sequence. J Biol Chem. 1998;273:6482–6487. doi: 10.1074/jbc.273.11.6482. [DOI] [PubMed] [Google Scholar]
- 57.Tzivion G, Shen YH, Zhu J. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene. 2001;20:6331–6338. doi: 10.1038/sj.onc.1204777. [DOI] [PubMed] [Google Scholar]
- 58.Singh A, Ye M, Bucur O, Zhu S, Tanya Santos M, Rabinovitz I, Wei W, Gao D, Hahn WC, Khosravi-Far R. Protein phosphatase 2A reactivates FOXO3a through a dynamic interplay with 14-3-3 and AKT. Mol Biol Cell. 2010;21:1140–1152. doi: 10.1091/mbc.E09-09-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cameron AJ, De Rycker M, Calleja V, Alcor D, Kjaer S, Kostelecky B, Saurin A, Faisal A, Laguerre M, Hemmings BA, McDonald N, Larijani B, Parker PJ. Protein kinases, from B to C. Biochem Soc Trans. 2007;35:1013–1017. doi: 10.1042/BST0351013. [DOI] [PubMed] [Google Scholar]
- 60.Calleja V, Laguerre M, Larijani B. 3-D structure and dynamics of protein kinase B-new mechanism for the allosteric regulation of an AGC kinase. J Chem Biol. 2009;2:11–25. doi: 10.1007/s12154-009-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li X, Lu Y, Jin W, Liang K, Mills GB, Fan Z. Autophosphorylation of Akt at threonine 72 and serine 246. A potential mechanism of regulation of Akt kinase activity. J Biol Chem. 2006;281:13837–13843. doi: 10.1074/jbc.M602060200. [DOI] [PubMed] [Google Scholar]
- 62.Gao H, Yu Z, Bi D, Jiang L, Cui Y, Sun J, Ma R. Akt/PKB interacts with the histone H3 methyltransferase SETDB1 and coordinates to silence gene expression. Mol Cell Biochem. 2007;305:35–44. doi: 10.1007/s11010-007-9525-3. [DOI] [PubMed] [Google Scholar]
- 63.Lee SB, Xuan Nguyen TL, Choi JW, Lee KH, Cho SW, Liu Z, Ye K, Bae SS, Ahn JY. Nuclear Akt interacts with B23/NPM and protects it from proteolytic cleavage, enhancing cell survival. Proc Natl Acad Sci U S A. 2008;105:16584–16589. doi: 10.1073/pnas.0807668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 65.Trencia A, Perfetti A, Cassese A, Vigliotta G, Miele C, Oriente F, Santopietro S, Giacco F, Condorelli G, Formisano P, Beguinot F. Protein kinase B/Akt binds and phosphorylates PED/PEA-15, stabilizing its antiapoptotic action. Mol Cell Biol. 2003;23:4511–4521. doi: 10.1128/MCB.23.13.4511-4521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Finlay DK, Sinclair LV, Feijoo C, Waugh CM, Hagenbeek TJ, Spits H, Cantrell DA. Phosphoinositide-dependent kinase 1 controls migration and malignant transformation but not cell growth and proliferation in PTEN-null lymphocytes. J Exp Med. 2009;206:2441–2454. doi: 10.1084/jem.20090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waugh C, Sinclair L, Finlay D, Bayascas JR, Cantrell D. Phosphoinositide (3,4,5)-triphosphate binding to phosphoinositide-dependent kinase 1 regulates a protein kinase B/Akt signaling threshold that dictates T-cell migration, not proliferation. Mol Cell Biol. 2009;29:5952–5962. doi: 10.1128/MCB.00585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brunet A, Kanai F, Stehn J, Xu J, Sarbassova D, Frangioni JV, Dalal SN, DeCaprio JA, Greenberg ME, Yaffe MB. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol. 2002;156:817–828. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cotelle V, Meek SE, Provan F, Milne FC, Morrice N, MacKintosh C. 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J. 2000;19:2869–2876. doi: 10.1093/emboj/19.12.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Birkenkamp KU, Coffer PJ. Regulation of cell survival and proliferation by the FOXO (Forkhead box, class O) subfamily of Forkhead transcription factors. Biochem Soc Trans. 2003;31:292–297. doi: 10.1042/bst0310292. [DOI] [PubMed] [Google Scholar]
- 71.Tzivion G, Luo ZJ, Avruch J. Calyculin A-induced vimentin phosphorylation sequesters 14-3-3 and displaces other 14-3-3 partners in vivo. J Biol Chem. 2000;275:29772–29778. doi: 10.1074/jbc.M001207200. [DOI] [PubMed] [Google Scholar]
- 72.Yoshida K, Yamaguchi T, Natsume T, Kufe D, Miki Y. JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat Cell Biol. 2005;7:278–285. doi: 10.1038/ncb1228. [DOI] [PubMed] [Google Scholar]
- 73.Tsuruta F, Sunayama J, Mori Y, Hattori S, Shimizu S, Tsujimoto Y, Yoshioka K, Masuyama N, Gotoh Y. JNK promotes Bax translocation to mitochondria through phosphorylation of 14-3-3 proteins. EMBO J. 2004;23:1889–1899. doi: 10.1038/sj.emboj.7600194. [DOI] [PMC free article] [PubMed] [Google Scholar]