Abstract
The discovery of interchromosomal interactions in higher eukaryotes points to a functional interplay between genome architecture and gene expression, challenging the view of transcription as a one-dimensional process. However, the extent of interchromosomal interactions and the underlying mechanisms are unknown. Here we present the first genome-wide analysis of transcriptional interactions using the mouse globin genes in erythroid tissues. Our results show that the active globin genes associate with hundreds of other transcribed genes, revealing extensive and preferential intra- and interchromosomal transcription interactomes. We show that the transcription factor Klf1 mediates preferential co-associations of Klf1-regulated genes at a limited number of specialized transcription factories. Our results establish a new gene expression paradigm, implying that active co-regulated genes and their regulatory factors cooperate to create specialized nuclear hot spots optimized for efficient and coordinated transcriptional control.
Increasing evidence suggests that long-range interactions between genomic regions contribute to the regulation of gene expression1. In higher eukaryotes individual chromosomes occupy discrete chromosome territories in the three-dimensional space of the nucleus2. However, chromosomal regions often loop out of their chromosome territories in association with activation3,4, and neighboring chromosomes can intermingle5, resulting in potentially functional contacts between genomic regions located on different chromosomes. Indeed, examples of interchromosomal interactions that regulate gene expression have been described6–8.
Genomic regions dynamically relocate to specialized subnuclear compartments that favor gene activation or silencing9–11. For example, RNA polymerase II (RNAPII) transcription occurs at transcription factories12–16, subnuclear compartments that are highly enriched in the active, hyperphosphorylated forms of RNAPII17. Transcription of most ‘active’ genes is not continuous but occurs in pulses of activity10,14,18–20. Gene activation is associated with relocation of genes to transcription factories13–15, indicating that transcriptional pulses occur by virtue of dynamic associations with transcription factories10. Previous studies on a limited number of transcriptionally active genes revealed long-range intra- and interchromosomal gene associations at shared transcription factories10,14. However, the extent and nature of these transcriptional associations is unclear, as is the question of whether they contribute to the control of gene expression.
Here we present a genome-wide analysis of gene co-associations in transcription factories with the mouse Hbb and Hba globin genes, using a new variant of the 3C technique21 combined with chromatin immunoprecipitation (ChIP). The results are corroborated by extensive FISH and immunofluorescence analyses of transcriptional co-associations at transcription factories. Collectively, our results show that co-regulated genes preferentially cluster at specialized transcription factories that seem to be optimized for their high-level transcription.
RESULTS
Preferential interchromosomal associations at factories
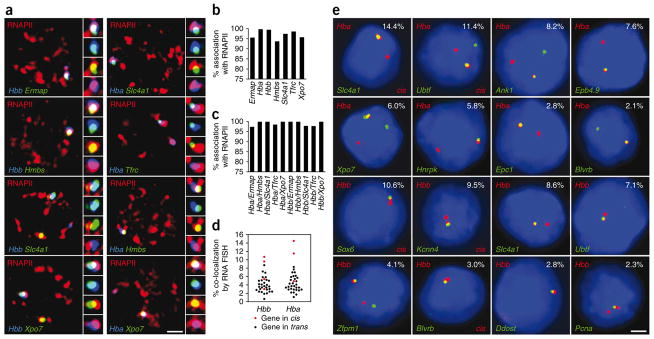
We used triple-label RNA immuno-FISH to examine interchromosomal associations between transcriptionally active genes at transcription factories. We chose five erythroid-expressed genes and analyzed their subnuclear location relative to transcription factories and the constitutively transcribed Hba and Hbb genes (Fig. 1a). In agreement with previous studies10,14,15, we found that the vast majority of RNA FISH signals (93–99%) overlapped with RNAPII foci (Fig. 1b). Furthermore, we found that essentially all colocalizing (overlapping) RNA FISH signals were localized to the same RNAPII focus (97–100%) (Fig. 1c). These data confirm earlier results10,12,15–17 indicating that virtually all gene transcription occurs at transcription factories. We conclude that overlapping RNA FISH signals are an excellent indicator of genes sharing the same transcription factory.
Figure 1.
Genes interchromosomally co-associate in transcription factories. (a) Triple-label RNA immuno-FISH of gene pairs and RNAPII factories. RNAPII-S5P staining is in red and RNA FISH signals are in green and blue, as indicated, for each gene pair. Side panels show enlarged images of the colocalizing FISH signals (top to bottom: triple label, blue and green, red and green, red and blue). Scale bar, 2 μm. (b,c) Bar charts showing percentages of RNA FISH signals that associate with an RNAPII factory (b) and percentages of colocalizing RNA FISH signals that co-associate within the same RNAPII focus (c). (d) Scatter plot showing distributions of RNA FISH colocalization frequencies for genes in cis (red) and trans (black) with Hba and Hbb. (e) Representative double-label RNA FISH in definitive erythroid cells for several erythroid-expressed genes (green) and Hba or Hbb (red), as indicated, with DAPI staining in blue. Colocalization frequencies are shown on each panel, and co-associations in cis are labeled. Scale bar, 2 μm.
Recent reports have noted that active genes up to 1 μm apart frequently associate with the same large nuclear Sc35 speckle domain, with the suggestion that Sc35 domains may spatially organize active genes22–24. We found that mouse erythroid cells lack large speckle domains (Supplementary Note and Supplementary Fig. 1a–c). Furthermore, we found that transcribed genes vary greatly in their frequency of association with Sc35 (Supplementary Fig. 1d), in agreement with previous reports22,23,25,26. These results argue strongly against a universal role for Sc35 domains in organizing active genes in eukaryotic nuclei.
To obtain a more comprehensive picture of transcriptional co- associations, we used double-label RNA FISH to quantify transcriptional associations between Hba, Hbb and 33 erythroid-expressed genes, located on 15 mouse chromosomes. We observed a considerable range of interchromosomal colocalization frequencies (up to 17-fold; Fig. 1d,e), indicating that genes nonrandomly co-associate with preferential transcription partners in factories (Supplementary Table 1). Consistent with previous results14, linked genes generally co-associated at higher frequencies than genes in trans. However, we found that co-association frequencies between some genes in trans can surpass those of linked genes (Fig. 1d and Supplementary Table 1). These findings demonstrate preferential interchromosomal co- associations between specific genes in shared transcription factories.
Genome-wide detection of transcriptional co-associations
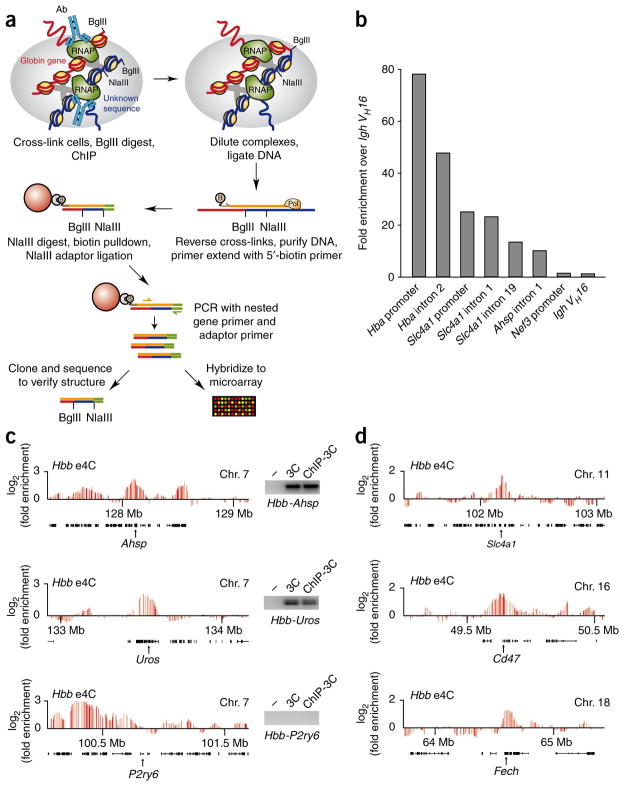
To obtain a global view of the repertoire of transcriptional interactions (the transcriptional interactome) for Hba and Hbb in erythroid nuclei, we developed e4C (enhanced ChIP-4C), a new genome-wide adaptation of the 4C assay, incorporating two major modifications (Fig. 2a). First, to focus on gene interactions within transcription factories, we included a ChIP step with an antibody recognizing the phosphorylated Ser5 residue of the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (RNAPII-S5P), the form implicated in transcriptional initiation and elongation27. Real-time PCR analysis of RNAPII-S5P ChIP DNA showed that active gene sequences were highly enriched (Fig. 2b). The immunoprecipitated chromatin was then diluted as for conventional 3C to favor ligation between DNA strands in the same cross-linked complex. Known long-range transcription factory associations between Hbb and distal active genes14 were detected by 3C assays on RNAPII-S5P ChIP material (Fig. 2c), demonstrating that the ChIP step maintains associations between genes in shared transcription factories. In contrast, no interaction of Hbb with the nearby inactive P2ry6 gene was detected14.
Figure 2.
e4C detects known and heretofore uncharacterized genomic co-associations with Hbb in cis and trans. (a) Overview of the e4C method. Nuclei are cross-linked, and the chromatin is digested with BglII as in conventional 3C, before immunoprecipitation with an antibody recognizing RNAPII-S5P. DNA ligation is performed on the immunoprecipitated chromatin under dilute conditions that favor ligation between DNA strands within cross-linked complexes. After reversal of the cross-links and DNA purification, a biotinylated primer (brown) specific to the bait gene (red) is used for primer extension into adjacent ligation products (blue). Biotinylated extension products are purified on streptavidin-coated magnetic beads (red sphere), digested with NlaIII and ligated to an adaptor (green). e4C products are amplified by PCR with a nested, bait-specific primer (yellow) and an adaptor-specific primer (green). e4C products are then analyzed by cloning and sequencing, or hybridization to a custom microarray. (b) Bar chart showing the enrichments of erythroid-expressed Hba, Slc4a1 and Ahsp sequences after ChIP using an antibody recognizing RNAPII-S5P. Enrichments of nonexpressed Nefm (formerly Nef3) and Igh VH16 sequences are shown as negative controls. Enrichments are shown relative to the VH16 control. (c) Hbb e4C microarray profiles for three ~2-Mb regions of genomic sequence in cis to Hbb, centered on Ahsp, Uros and P2ry6, showing the running mean enrichments of e4C signal over genomic signal for 100-kilobase (kb) windows. Black bars denote the positions of genes within these regions. Insets show PCR products for ligation products between Hbb and Ahsp, Uros or P2ry6 on water (−), 3C or RNAPII-S5P ChIP-3C templates. (d) Hbb e4C microarray profiles for three ~2 Mb regions of genomic sequence in trans to Hbb, centered on Slc4a1, Cd47 and Fech, showing the running mean enrichments of e4C signal over genomic signal for 100-kb windows.
Second, we incorporated a pre-enrichment step for bait-linked sequences. This provided a more than 100-fold increase in signal-to-noise ratio, affording the identification of distal cis- and trans- interacting loci with higher sensitivity and confidence (data not shown). In brief, the pre-enrichment step involves primer extension with a biotinylated bait-specific primer, and specific pulldown of bait-linked 3C products with streptavidin beads, before amplification with a bait-specific, nested primer. Cloning and sequencing of Hbb e4C products revealed that 95% (121 of 127) had the expected structure, indicating that e4C is highly efficient (Fig. 2a). We then hybridized the e4C DNA to custom microarrays covering most of the nonrepetitive mouse genome (see Online Methods).
Analysis of the microarray data shows that e4C detects known erythroid-specific associations between Hbb and the distal transcribed Ahsp (formerly Eraf) and Uros genes14,28 (Fig. 2c), whereas the nearby inactive P2ry6 gene is negative by e4C (Fig. 2c). However, in contrast to a previous 4C study28, the e4C results reveal extensive contacts between Hbb and selected genomic regions located on other chromosomes (examples shown in Fig. 2d). To identify the genomic regions that associate significantly with the globin genes, we used a sliding-window replicate Student’s t-test to determine clusters of array probes that were enriched in e4C products. Notably, 88% of the clusters identified were located in trans to Hbb (241 of 273), implying that the majority of potential Hbb contacts in transcription factories occur with sequences on other chromosomes (Supplementary Fig. 2 and Supplementary Table 2). We ruled out the possibility that random ligation between chromatin complexes could account for our results by mixing equal amounts of mouse and human chromatin before the RNAPII-S5P ChIP step (Fig. 2a) and then performing e4C with Hbb as bait (Supplementary Note). Sequencing the resultant e4C clones revealed that cross-ligation between mouse Hbb and human DNA was rare (1.3%; 1 of 77 clones) and could not account for the observed clustered-array hits.
We repeated the e4C assay with Hba as bait. The Hba e4C micro-array data indicate that the locus associates with genomic regions located both in cis and in trans (Supplementary Fig. 3). The majority of Hba e4C clusters (93%; 514 of 551) were located on different chromosomes (Supplementary Fig. 2 and Supplementary Table 3). Taken together, our results imply that both mouse globin genes undergo extensive intra- and interchromosomal associations in transcription factories in erythroid cells.
To verify that e4C detects interactions between transcribing genes, we profiled erythroid RNAPII-S5P ChIP DNA by sequencing of paired end tags (ChIP-PETs)29. We used a conservative threshold to identify erythroid-transcribed genes and genomic regions with high confidence (Supplementary Table 4). As expected, most e4C clusters were centered on active genes; 80% of the Hba e4C clusters and 90% of the Hbb e4C clusters contained one or more annotated genes enriched in RNAPII-S5P (Supplementary Fig. 4a). Another 7% and 2%, respectively, of the e4C clusters contained regions of enriched RNAPII-S5P binding devoid of known genes, which may represent unannotated expressed genes or intergenic transcribed regions. Overall, the Hbb e4C clusters contained 724 active genes, and the Hba e4C clusters contained 1,286 active genes (Supplementary Fig. 4b and Supplementary Note).
Preferential Hba and Hbb transcriptional networks
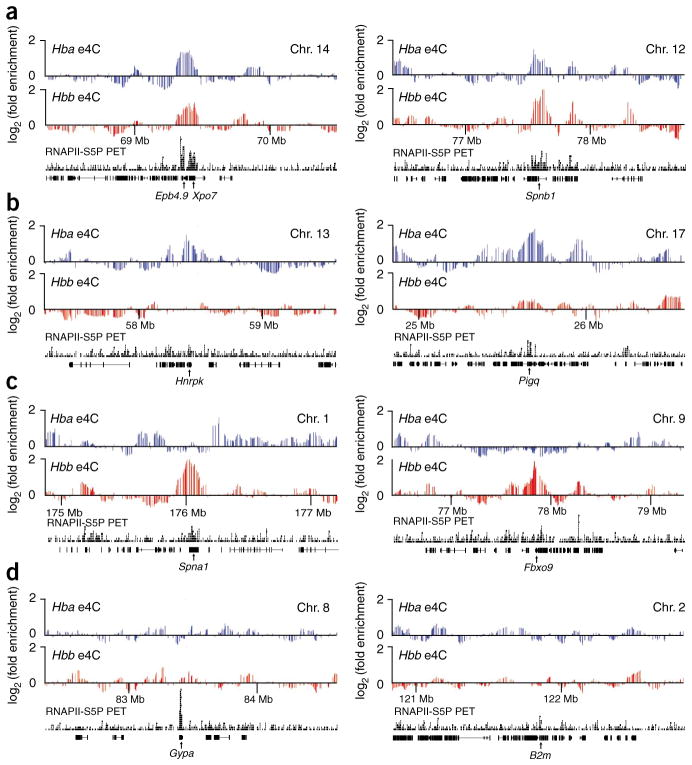
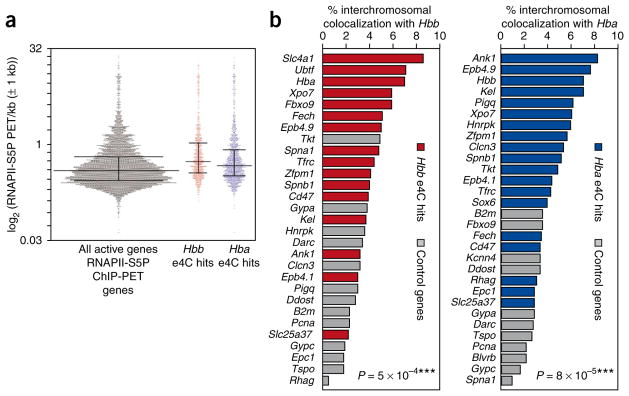
Because e4C includes an RNAPII-S5P ChIP enrichment step, one concern is that e4C simply detects associations between the globin genes and the most highly transcribed genes. Two lines of evidence argue against this possibility. First, e4C microarray profiles demonstrate that although some genes, such as Spnb1, Epb4.9 and Xpo7, associate with both globin genes (Fig. 3a), other genes with comparable RNAPII-S5P occupancy preferentially associate with only one globin gene (for example, Hnrpk, Pigq, Spna1 and Fbxo9; Fig. 3b,c) or with neither (for example, B2m and Gypa; Fig. 3d). These results indicate that co- associations detected by e4C are gene specific. In fact, 78% of the genes identified by e4C are unique to either Hba or Hbb, demonstrating that each locus associates preferentially with a specific subset or network of transcribed genes (Supplementary Fig. 4b). Second, although many highly transcribed genes associate with the globin genes, the majority of e4C hit genes are in the moderate to low range of RNAPII-S5P density (Fig. 4a). These results demonstrate that e4C is not solely identifying interactions between the most highly transcribed genes.
Figure 3.
Specificity of interchromosomal transcriptional networks. Hba (blue) and Hbb (red) e4C microarray profiles for ~2 Mb regions of genomic sequence in trans to both globin genes, showing the running mean enrichments of e4C signal over genomic signal for 100-kb windows. (a–d) Profiles are centered on genes identified by e4C as interacting with both Hba and Hbb (Epb4.9, Xpo7 and Spnb1) (a), interacting with Hba (Hnrpk and Pigq) (b), interacting with Hbb (Spna1 and Fbxo9) (c) and interacting with neither globin gene (Gypa and B2m) (d). Positions of genes and RNAPII-S5P ChIP-PET profiles (black) are shown below the e4C microarray profiles.
Figure 4.
Validation of the e4C assay. (a) Scatter plot showing distributions of all erythroid-expressed genes (gray dots), Hbb e4C hits (red) and Hba e4C hits (blue), ranked by RNAPII-S5P PET density. Horizontal lines represent the median, 25th and 75th percentiles. (b) Validation of e4C by interchromosomal RNA FISH colocalization frequencies with Hba and Hbb. Genes identified by e4C as associating with Hba are highlighted in blue, Hbb e4C–associating genes are in red and non-hit genes in gray. P values are for differences in colocalization frequencies between e4C hit and non-hit genes.
Finally, to validate the e4C results, we used the double-label RNA FISH data set described above (Fig. 1d,e). We reasoned that co-associating gene pairs detected by e4C should share transcription factories at higher frequencies. Indeed, we found that genes identified as Hba or Hbb interacting partners by e4C colocalized with the globin genes at significantly higher frequencies (P = 8 × 10−5 for Hba, P = 5 × 10−4 for Hbb) than other genes (Fig. 4b). These results authenticate the e4C assay as a reliable indicator of genes that preferentially engage the same transcription factory. We conclude that the mouse globin genes preferentially associate with distinct but overlapping networks of transcribed genes.
Co-association of specific gene triplets at transcription factories
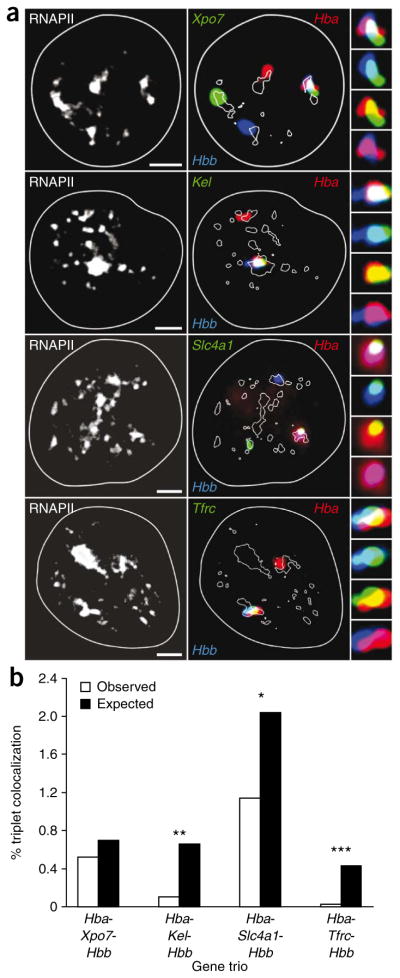
Our e4C results indicate that each globin gene associates with hundreds of other genomic regions from nearly all mouse chromosomes (Supplementary Fig. 2). Previous estimates have suggested that a single transcription factory may be occupied by, on average, eight transcription units30. To address the question whether more than two genes could occupy the same factory, we performed quadruple-label RNA immuno-FISH to simultaneously visualize three active genes and RNAPII transcription sites. We assayed Hba, Hbb and four other erythroid-expressed genes (Kel, Slc4a1, Tfrc, Xpo7) identified by e4C as interacting partners for both Hba and Hbb. We found multiple examples of three different gene transcription signals associated with the same RNAPII focus (Fig. 5a). Thus, a single transcription factory can indeed accommodate the simultaneous transcription of at least three genes, located on three different chromosomes.
Figure 5.
Co-association of three active genes within the same transcription factory. (a) Quadruple-label RNA immuno-FISH of three transcribed genes and RNAPII factories. RNAPII-S5P staining is shown in the panels on the left; RNA FISH signals in the same cells are shown in red (Hba), blue (Hbb) and green (top to bottom: Xpo7, Kel, Slc4a1, Tfrc) in the panels on the right. Side panels show enlarged images of the colocalizing FISH signals (top to bottom: triple label, blue and green, red and green, red and blue). Scale bar, 2 μm. (b) Bar chart showing the observed versus expected frequencies of gene triplet associations at a shared transcription factory. *P < 0.05; **P < 0.005; ***P < 0.001.
We next scored the frequencies of simultaneous overlap between the three genes at the same RNAPII focus. Using the pairwise colocalization frequencies (Supplementary Table 1), we calculated expected triple-interaction frequencies, assuming independence between interacting gene pairs (Supplementary Note). For Hba-Xpo7-Hbb, we found that the observed frequency of triplet associations mirrored the expected frequencies (Fig. 5b). Xpo7 seems to associate preferentially with Hba and Hbb independent of whether they are together or separate. In contrast, the Hba-Kel-Hbb, Hba-Slc4a1-Hbb and Hba-Tfrc-Hbb triplets occur at significantly lower-than-expected frequencies. These results suggest that Kel, Slc4a1 and Trfc preferentially colocalize with Hbb when it is not associated with Hba, and vice versa. These findings reveal a potential complex hierarchy of preferential associations within the transcription networks, which may represent mutually exclusive associations or physical constraints within the nucleus.
Globin transcription networks are enriched in Klf1-regulated genes
To understand the molecular basis for preferential associations in transcription factories, we searched the promoter sequences of e4C Hba- and Hbb-interacting genes for known transcription factor binding motifs. We found an enrichment of CACC motifs—potential binding sites for the erythroid-specific transcription factor Klf131 (Kruppel-like factor 1). Klf1 binds the Hbb-b1 promoter32, the Hbb locus control region (LCR)32 and the Hba promoter and upstream enhancer region in erythroid cells32,33. Hbb-b1 transcription is reduced by >80% in Klf1-deficient erythroid cells, whereas Hba transcription is less markedly affected34,35. We compiled a list of Klf1-regulated genes based on published expression profiles36–38 (Supplementary Table 5). We found that genes upregulated by Klf1 were overrepre-sented among the Hbb e4C interacting genes (P = 3 × 10−8). Hba e4C interacting genes were also significantly overrepresented in genes upregulated by Klf1, but to a lesser extent (P = 3 × 10−4), perhaps reflecting Hba’s partial dependency on Klf1. In contrast, genes down-regulated by Klf1 were not enriched (P = 0.8 for Hba, P = 0.9 for Hbb). These data suggest that genes upregulated by Klf1 preferentially share transcription factories and that shared regulatory factors may be the basis for transcription network specificity.
HBB transgenes relocate to the Hbb transcriptional interactome
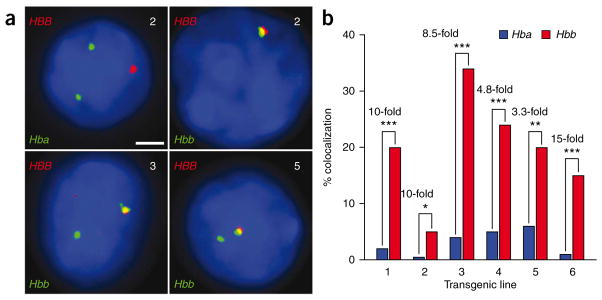
As a functional test of the theory that Klf1-regulated genes are preferentially organized in three-dimensional space to share factories, we investigated the subnuclear localization of Klf1-regulated transgenes relative to the endogenous Hbb and Hba genes. We chose the human β-globin (HBB) locus transgene, which is strongly dependent on Klf1 for transcription39,40. Two HBB transgenic lines contain large YACs encompassing the entire HBB locus, whereas four other lines contain a reporter gene driven by a fully functional microlocus LCR and HBB promoter41. We performed double-label RNA FISH to test whether the HBB transgenes could share transcription factories with the endogenous Hbb or Hba globin genes (Fig. 6a). Notably, the HBB transgene showed a strong preference for localizing to a factory occupied by Hbb as compared with Hba (Fig. 6b). This result cannot be explained by physical linkage of the transgene to the Hbb locus, in that DNA FISH on metaphase spreads confirmed that none of the transgenes were integrated on chromosome 7 (data not shown). Colocalization frequencies between the transgene and endogenous Hbb locus were remarkably high (Fig. 6b), demonstrating that randomly integrated HBB transgenes have a strong preference for interchromosomal association with their co-regulated, endogenous counterparts in transcription factories. These data imply that the presence of a transcriptionally active transgene locus can spatially reorganize an ectopic chromosomal site to be near a co-regulated gene locus.
Figure 6.
Ectopic genes enter endogenous transcriptional networks. (a) Representative double-label RNA FISH for HBB transgenes (red) and endogenous Hba or Hbb genes (green) in definitive erythroid cells from transgenic lines (as indicated at top right of each panel), DAPI staining in blue. Scale bar, 2 μm. (b) Interchromosomal colocalization frequencies between HBB and Hba (blue bars) and HBB and Hbb (red bars) from six transgenic lines. Fold differences in transgene colocalization with Hbb versus Hba are shown along with the corresponding indicators of P values *P < 0.05; **P < 0.005; ***P < 0.001.
Specialized transcription factories
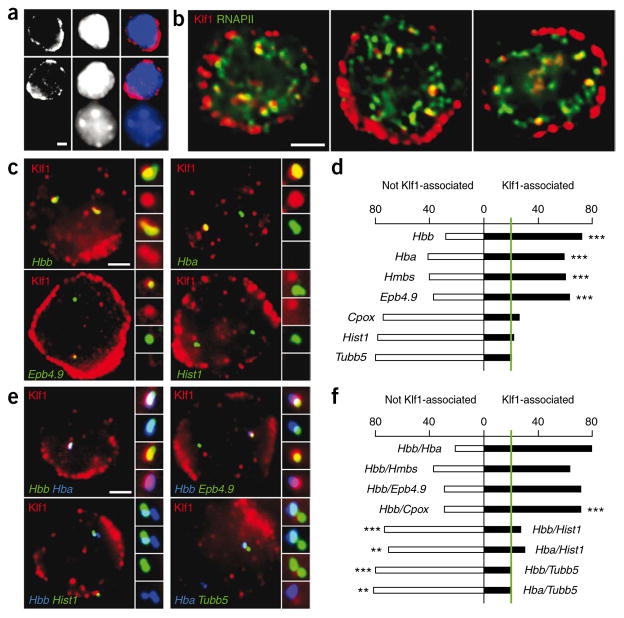
The above results imply that a subset of transcription factories may be specialized to preferentially transcribe a specific network of genes, such as Klf1-regulated genes. We therefore assessed the spatial distribution of Klf1 relative to RNAPII factories by immunofluorescence in mouse erythroid cells. In agreement with a recent study42, we found that most Klf1 is located in the cytoplasm of mouse erythroid cells, whereas nuclear Klf1 is present in discrete sites (on average 40 foci per nucleus) (Fig. 7a). Nearly all Klf1 foci overlapped with RNAPII-S5P foci, indicating that 10–20% of transcription factories contain high levels of Klf1 (Fig. 7b).
Figure 7.
Co-regulated genes cluster in specialized transcription factories. (a) Immunofluorescence staining for Klf1 in definitive erythroid cells. Klf1 is shown in red, DAPI in blue. Note nonerythroid, Klf1-negative cell (bottom) demonstrating antibody specificity. Scale bar, 2 μm. (b) Immunofluorescence detection of Klf1 (red) and RNAPII-S5P (green) in definitive erythroid cells. Scale bar, 2 μm. (c) Double-label RNA immuno-FISH of nascent transcripts (green) and Klf1 foci (red). Side panels show enlarged images of selected nascent transcript signals with corresponding red channel (Klf1). Scale bar, 2 μm. (d) Graph showing proportions of RNA FISH signals found associated with Klf1 foci (black), or not associated with Klf1 foci (white). Green line shows ‘background’ association level of 20%, based on proportions of transcription factories containing high levels of Klf1. Indicators of P values are shown for Klf1 association frequencies higher than the background level (***P < 0.001). (e) Triple-label RNA immuno-FISH for pairs of nascent transcripts (blue and green, as indicated) and Klf1 foci (red). Side panels show enlarged images of the colocalizing FISH signals (top to bottom: triple label, blue and green, red and green, red and blue). Scale bar, 2 μm. (f) Graph showing proportions of colocalized pairs of RNA FISH signals found associated with Klf1 foci (black) or not associated with Klf1 foci (white). Green line shows ‘background’ association level of 20%, based on proportions of transcription factories containing high levels of Klf1. Indicators of P values are shown for increase in Hbb-Cpox Klf1 association frequency compared with Cpox Klf1 association frequency, and for decreases in globin-Hist1h3f Klf1 and globin-Tubb5 Klf1 association frequencies compared to globin Klf1 association frequencies (**P < 0.005; ***P < 0.001).
We next assessed the position of several transcriptionally active, Klf1-regulated genes relative to Klf1 foci by RNA immuno-FISH (Fig. 7c,d). We found that the majority (59–72%) of actively transcribed alleles of Hbb, Hba, Hmbs and Epb4.9 were preferentially associated with ‘Klf1 transcription factories’. In contrast, Cpox, which has only slightly decreased expression in the Klf1 knockout36, associated with Klf1 factories at only marginally higher frequencies (26%) than expected by a purely random distribution. Actively transcribed alleles of the Klf1-independent Tubb5 and H2A/H2B histone genes (within the Hist1h3f (formerly Hist1) cluster on chromosome 13) showed no preferential localization to Klf1-containing factories (21% and 22%, respectively).
To test if Klf1 is involved in clustering Klf1-regulated genes, we assessed colocalizing pairs of genes relative to Klf1 foci (Fig. 7e). Using triple-label RNA immuno-FISH, we found that colocalizing pairs of Klf1-regulated genes are associated with Klf1 transcription factories at very high frequencies (63–79%; Fig. 7f). Notably, 71% of the Cpox-Hbb colocalizing pairs are associated with Klf1 foci, much higher than the frequency for Cpox alone (P = 2 × 10−4). This result clearly shows that Cpox alleles associate with Hbb preferentially in Klf1 factories. In contrast, colocalizing Tubb5-globin and histone-globin gene pairs show no preferential association with Klf1 transcription factories. This is significantly different from the percentage of globin alleles associating with Klf1 alone (Hba, P = 1 × 10−3 for Tubb5 and P = 2 × 10−3 for histones; Hbb, P = 1 × 10−5 for Tubb5 and P = 4 × 10−7 for histones) and shows that the vast majority of Hbb and Hba alleles that colocalize with Tubb5 or Hist1h3f are not associated with Klf1 factories. Collectively, these data clearly show that transcribed, Klf1-regulated genes are preferentially located in transcription factories containing high levels of Klf1, and strongly suggest that these factories are shared by many other Klf1-regulated genes. Thus, there are specialized transcription factories where active alleles of Klf1-regulated genes are more often found and are preferentially clustered with other Klf1-regulated genes.
Klf1 is required for clustering of Klf1-regulated genes
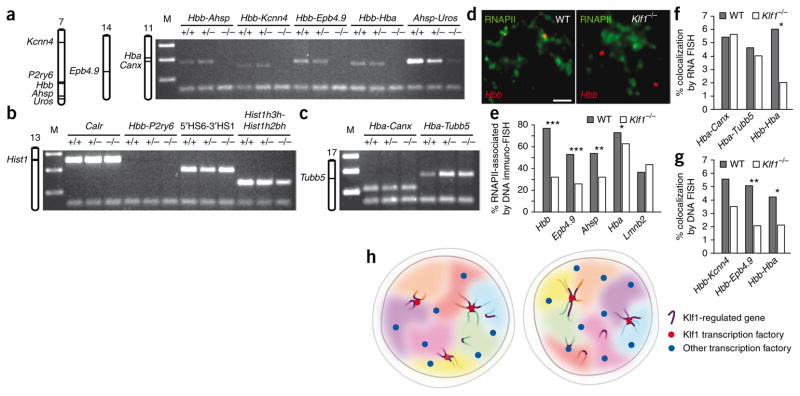
To test whether Klf1 is required for clustering of Klf1-regulated genes, we assessed intra- and interchromosomal associations in erythroid nuclei from wild-type and Klf1-knockout mice via 3C. Associations between Hbb and the Klf1-regulated genes Ahsp and Kcnn4 were specifically disrupted in Klf1−/− erythroid cells but intact in Klf1+/− and wild type (Fig. 8a). Furthermore, interchromosomal associations between Hbb and Epb4.9, and between Hbb and Hba, were also disrupted specifically in the knockout. This effect is not limited to the Hbb locus, in that long-range associations between Ahsp and Uros were also decreased in Klf1−/− mice. In contrast, a known Klf1-independent interaction within the Hbb locus (LCR 5′HS6 and 3′HS1; ref. 43) was maintained in Klf1−/− mice (Fig. 8b), as was a long-range association between Klf1-independent histone genes (Hist1h3h and Hist1h2bh, separated by 1.8 megabases (Mb) on chromosome 13). Intra- and interchromosomal associations between Hba and the Klf1-independent Canx and Tubb5 genes were also maintained in the knockout mice (Fig. 8c). These results show that interactions between Klf1-regulated genes are specifically disrupted in the absence of Klf1.
Figure 8.
Klf1 mediates specific intra- and interchromosomal co-associations. (a–c) 3C analyses of genomic associations in definitive erythroid cells derived from wild-type (+/+), heterozygous (+/−) and Klf1-null (−/−) mice, including karyoview showing positions of the genes tested. The lower band in all lanes represents the loading control. Shown are 3C products between Klf1-regulated gene pairs as indicated (a); between Klf1-independent controls (ligation between two adjacent fragments in the Calr gene was used as a positive control, and association between Hbb and P2ry6 served as a negative control; see Fig. 2c) (b); and between Hba and Klf1-independent genes. M, DNA size marker (c). (d) Double-label DNA immuno-FISH of Hbb (red) and RNAPII-S5P (green). WT, wild type. Scale bar, 2 μm. (e) Graph showing proportions of DNA FISH signals found associated with transcription factories in WT and Klf1−/− fetal liver cells, with the corresponding indicators of P values (*P < 0.05; **P < 0.005; ***P < 0.001). (f) Bar chart showing RNA FISH colocalization frequencies of the indicated gene pairs in WT and Klf1−/− cells. (g) Bar chart showing DNA FISH colocalization frequencies of the indicated gene pairs in WT and Klf1−/− cells. (h) Model of dynamic associations between genes in specialized transcription factories. Schematic representation of two cells that could represent the same cell at different times, or two cells with differing genome conformations within a population. Transcription factories are depicted as blue dots; Klf1-containing transcription factories are shown as red dots. Chromatin loops containing Klf1-regulated genes (aubergine segments) from the same or different chromosomes territories (colored areas) preferentially co-transcribe in the limited number of specialized Klf1-containing transcription factories. Temporarily nontranscribed alleles are positioned away from transcription factories. We propose that interactions between transcription network members are dynamic and may change over time, which may influence chromosomal conformations and chromosome positioning.
Because our data support the concept that associations between active genes occur in transcription factories, we asked whether key Klf1-regulated genes show reduced factory occupancy in Klf1−/− erythroid cells. Indeed, Hbb, Epb4.9 and Ahsp all show markedly reduced association with transcription factories in Klf1−/− erythroid cells, consistent with their high dependence on Klf1 (Fig. 8d,e). In contrast, the percentage of Hba alleles associated with factories was only mildly decreased, possibly reflecting Hba’s partial dependence on Klf1. Factory occupancy of the Klf1-independent Lmnb2 gene was unchanged. Thus, Klf1-regulated genes show reduced association with transcription factories in the absence of Klf1, which may in part explain their reduced expression36–38.
We confirmed by RNA FISH that the number of active Hbb alleles was markedly reduced in Klf1−/− erythroid cells (to 5% that in wild-type cells). Notably, those Hbb alleles that were still active in Klf1−/− erythroid cells showed markedly reduced association with Hba (Fig. 8f). In contrast, colocalization frequencies between Hba, Canx and Tubb5 pairs were unchanged in the knockout (Fig. 8f), in agreement with the 3C results (Fig. 8c). Thus, the partially Klf1-dependent Hba maintains associations with Klf1-independent genes, whereas its association with the highly Klf1-dependent Hbb gene is specifically disrupted. We noted that RNA FISH signals for Hbb were considerably weaker in the knockout, suggesting a reduced rate of transcriptional firing or elongation. This precluded RNA FISH analyses of other Klf1-regulated genes with considerably lower transcription rates than Hbb. We therefore used DNA FISH to determine changes in colocalization frequencies between Hbb, Epb4.9 and Kcnn4 pairs. We found that intrachromosomal (Hbb-Kcnn4) and interchromosomal (Hbb-Hba; Hbb-Epb4.9) interaction frequencies were reduced in Klf1−/− cells (Fig. 8g). Collectively, these results clearly demonstrate that Klf1 is required for preferential colocalization of Klf1-regulated genes at shared transcription factories.
DISCUSSION
Using a combination of techniques (e4C, 3C, RNA FISH, DNA FISH and immuno-FISH), we have shown that the mouse globin genes preferentially associate with hundreds of other transcribed genomic loci in transcription factories. The globin-interacting genes are distributed over nearly all mouse chromosomes, uncovering extensive intra- and interchromosomal transcriptional interaction networks in erythroid nuclei. It is highly unlikely that all of these interactions occur simultaneously in the same cell. Instead, our data on gene triplet associations at factories imply that genome organization is inherently plastic. We propose that multiple different genome conformations exist and that each brings the globin genes into close proximity with a varied subset of their preferred transcriptional partners (Fig. 8h). Although gene associations at factories seem to be dynamic44, it is presently not known whether these whole-genome conformations are dynamically interchangeable within one nucleus or whether individual conformations are relatively stable in a subset of nuclei.
Within the globin transcriptional networks, Klf1-regulated genes are preferentially transcribed at a limited number of specialized transcription factories containing large amounts of Klf1. Our data also show that Klf1-regulated genes share Klf1-containing factories and that Klf1 is required for clustering of these co-regulated genes. It is important to point out that the majority of Klf1-regulated genes are not dependent on Klf1 for expression but instead require Klf1 for increased expression in the definitive erythroid lineage. For example, many of the genes encoding proteins involved in iron uptake and heme synthesis are ubiquitously expressed in all cell types but are highly upregulated by Klf1 during erythropoiesis36,37. We propose that specialized transcription factories boost the expression of clustered, co-regulated genes by concentrating specific transcription factors required for their coordinate or increased transcription (Fig. 8h). This may occur through a self-organization process whereby locally elevated concentrations of transcription factors and their cognate binding sites increase the probability of gene re- initiation, thus increasing occupancy time and transcriptional output at a shared factory. Such a model does not propose that specific pairwise interactions of genes are essential for their expression, but rather that individual genes may indirectly benefit from cooperative associations in these specialized microenvironments. Consequently, removal of a member of the network would not be expected to affect transcription of other network members to any great extent. In agreement with this, transcription and expression of several genes that associate with Hbb did not change in Hbb LCR knockout mice (data not shown), in which Hbb factory association and expression are markedly reduced15.
The concept of transcription factor–mediated spatial genome organization is supported by the observation that ectopic Klf1-regulated transgenes enter the same transcription sites as a co-regulated endogenous gene. This finding implies that transcriptional associations may considerably influence genome organization. This is supported indirectly by observed alterations in chromosomal co-associations when transcription is inhibited5,13. Thus, we propose that preferential associations in transcription factories substantially affect higher order chromosomal conformations and are a major driving force in tissue-specific chromosome positioning45.
Our results contrast with previous subgenomic 4C studies that detected few interchromosomal contacts for Hbb and concluded that active Hbb loci interact primarily in cis28, regardless of transcription46. We propose that the reasons for this discrepancy are probably the increased sensitivity and specificity of e4C. The biotin pre-enrichment step removes the thousands of copies of genomic DNA in the PCR reaction, which contribute to the probe in other 4C assays, potentially causing background noise upon microarray hybridization. In addition, the RNAPII-S5P ChIP step specifically enriches for transcriptionally active alleles, thus focusing on interactions at transcription factories and excluding cells not transcribing the bait gene. Conventional 3C and 4C approaches inherently average the interactions of all alleles in a population, active and temporarily nontranscribed; thus, preferential associations among the transcribed subpopulation of alleles might escape detection with these techniques.
Previous studies have focused on small numbers of genes or genetic elements that functionally cluster in three dimensions, such as nucleolar rDNA repeats47, tRNA genes in yeast48, silenced Hox genes in Drosophila49, virally induced associations between the interferon-β gene and NF-κB–bound sites6, between EZH2-bound sites50, and clustering of transiently transfected reporter constructs51. Here we used a genome-wide e4C screen to identify extensive spatial networks of active genes, demonstrating that interchromosomal clustering of genes is a widespread principle of nuclear organization.
Our results imply that transcriptional regulation should be considered in the context of the three-dimensional organization of the genome rather than as a mechanism that acts on single genes in isolation. Co-regulated genes cluster in nuclear space and potentially collaborate to create transcription sites that are optimized for their regulated expression. Thus, the concept of regulation of cell-type gene expression patterns by combinatorial transcription factor control is played out in the nucleus as combinatorial associations between multiple genes at specialized transcription sites, creating functional overlapping transcription networks.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Supplementary Material
Acknowledgments
We thank all members of the Laboratory of Chromatin and Gene Expression for their help and advice, and also thank C. Osborne and P. Schoenfelder for critical reading of the manuscript, A. Segonds-Pichon for help with the statistical analyses, and L. Mercer and M. Anderton for handling mouse strains. We are indebted to J. Cunningham, T. Abramova and V. Jansen for providing tissue from Klf1−/− mice. D.U. is recipient of an European Molecular Biology Organisation long-term fellowship. P.F. is a Senior Fellow of the Medical Research Council. This work was supported by the Medical Research Council and the Biotechnology and Biological Sciences Research Council, UK.
Footnotes
Accession codes. GEO: RNAPII-S5P ChIP-PET data, GSE18873. ArrayExpress: e4C microarray data, E-TABM-822.
Note: Supplementary information is available on the Nature Genetics website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
AUTHOR CONTRIBUTIONS
T.S. and S.S. carried out e4C experiments, L.C. and S.S. performed RNA FISH and RNA immuno-FISH experiments, and T.S. conducted the microarray analysis. N.F.C. performed 3C experiments, A.H. contributed transgene data, S.A. performed computational analysis and S.K. helped to establish 3C and e4C assays. J.A.M. and D.U. carried out ChIP experiments, C.H.E. contributed electron microscopy data, D.S.D. and N.F.C. conducted DNA FISH experiments. Y.L., C.-L.W. and Y.R. provided sequencing and mapping of ChIP material. J.J.B. provided the Klf1 antibody and advice on Klf1 experiments. S.S., T.S., L.C. and P.F. designed the study and wrote the manuscript. P.F. supervised this research and obtained funding for the study.
References
- 1.Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 2.Bolzer A, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volpi EV, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113:1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- 5.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apostolou E, Thanos D. Virus infection induces NF-κB-dependent interchromosomal associations mediating monoallelic IFN-β gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 7.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 9.Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- 10.Osborne CS, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat Struct Mol Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 12.Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osborne CS, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 15.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wansink DG, et al. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 18.Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 20.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 22.Brown JM, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown JM, et al. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Q, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. Processing of endogenous pre-mRNAs in association with SC35 domains is gene specific. J Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xing Y, Johnson CV, Moen PT, Jr, McNeil JA, Lawrence J. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC35 domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 28.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 29.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 30.Pombo A, et al. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shyu YC, et al. Chromatin-binding in vivo of the erythroid Kruppel-like factor, EKLF, in the murine globin loci. Cell Res. 2006;16:347–355. doi: 10.1038/sj.cr.7310045. [DOI] [PubMed] [Google Scholar]
- 33.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nuez B, Michaolovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 35.Perkins AC, Sharpe AH, Orkin SH. Lethal β-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 36.Drissen R, et al. The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Mol Cell Biol. 2005;25:5205–5214. doi: 10.1128/MCB.25.12.5205-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodge D, et al. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107:3359–3370. doi: 10.1182/blood-2005-07-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilson DG, Sabatino DE, Bodine DM, Gallagher PG. Major erythrocyte membrane protein genes in EKLF-deficient mice. Exp Hematol. 2006;34:705–712. doi: 10.1016/j.exphem.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 39.Perkins AC, Gaensler KM, Orkin SH. Silencing of human fetal globin expression is impaired in the absence of the adult β-globin gene activator protein EKLF. Proc Natl Acad Sci USA. 1996;93:12267–12271. doi: 10.1073/pnas.93.22.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wijgerde M, et al. The role of EKLF in human β-globin gene competition. Genes Dev. 1996;10:2894–2902. doi: 10.1101/gad.10.22.2894. [DOI] [PubMed] [Google Scholar]
- 41.Talbot D, et al. A dominant control region from the human β-globin gene locus conferring integration site-independent gene expression. Nature. 1989;338:352–355. doi: 10.1038/338352a0. [DOI] [PubMed] [Google Scholar]
- 42.Quadrini KJ, Gruzglin E, Bieker JJ. Non-random subcellular distribution of variant EKLF in erythroid cells. Exp Cell Res. 2008;314:1595–1604. doi: 10.1016/j.yexcr.2008.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drissen R, et al. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 45.Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;5:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palstra RJ, et al. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS One. 2008;3:e1661. doi: 10.1371/journal.pone.0001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- 48.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimaud C, et al. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 50.Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 2008;18:1171–1179. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J Cell Biol. 2008;181:615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.