Abstract
Background
High Resolution Melting Analysis (HRMA) is becoming the preferred method for mutation detection. However, its accuracy in the individual clinical diagnostic setting is variable. To assess the diagnostic accuracy of HRMA for human mutations in comparison to DNA sequencing in different routine clinical settings, we have conducted a meta-analysis of published reports.
Methodology/Principal Findings
Out of 195 publications obtained from the initial search criteria, thirty-four studies assessing the accuracy of HRMA were included in the meta-analysis. We found that HRMA was a highly sensitive test for detecting disease-associated mutations in humans. Overall, the summary sensitivity was 97.5% (95% confidence interval (CI): 96.8–98.5; I2 = 27.0%). Subgroup analysis showed even higher sensitivity for non-HR-1 instruments (sensitivity 98.7% (95%CI: 97.7–99.3; I2 = 0.0%)) and an eligible sample size subgroup (sensitivity 99.3% (95%CI: 98.1–99.8; I2 = 0.0%)). HRMA specificity showed considerable heterogeneity between studies. Sensitivity of the techniques was influenced by sample size and instrument type but by not sample source or dye type.
Conclusions/Significance
These findings show that HRMA is a highly sensitive, simple and low-cost test to detect human disease-associated mutations, especially for samples with mutations of low incidence. The burden on DNA sequencing could be significantly reduced by the implementation of HRMA, but it should be recognized that its sensitivity varies according to the number of samples with/without mutations, and positive results require DNA sequencing for confirmation.
Introduction
Although DNA sequencing, including direct DNA sequencing and pyrosequencing [1], is considered as the “gold standard” for known/unknown mutation scanning, it still remains relatively expensive, laborious and time-consuming. Many other methods for mutation scanning have been developed to screen for differences between the two copies of DNA within an individual. These techniques include single-strand conformational polymorphism analysis (SSCP) [2], denaturing gradient gel electrophoresis (DGGE) [3], denaturing high performance liquid chromatography (DHPLC) [4], temperature gradient capillary electrophoresis (TGCE) [5] and mass spectroscopy [6]. All of these methods require separation of the sample on a gel or other matrix. Fluorescence monitoring of PCR product melting profiles is another alternative to DNA sequencing that permits the detection of DNA mutations in solution without the need for separation on a gel or other matrix [7]. Fluorescently labeled, probe-based methods, such dual hybridization [8], exonuclease (TaqMan) [9], or hairpin (Molecular Beacon) [10] probes, may be used for mutation detection, but only for the bases covered by the probe. Hence, these methods are not amenable to mutational scanning as mutational scanning requires methods that can detect mutations over larger regions. Furthermore, some of the above methods are not automated and are therefore labor intensive while others are complex, costly and require specialized instrumentation.
High resolution melting analysis (HRMA) is a simple, PCR-based method. In the presence of saturating concentrations of DNA binding dyes, the specific sequence of the amplicon determines the melting behavior as the temperature of the solution is increased. Fluorescence intensity decreases as the double stranded DNA becomes single stranded and the dye is released. The melting temperature (Tm) at which 50% of the DNA is in the double stranded state can be approximated by taking the derivative of the melting curve. The distinctive melting curve can used to detect DNA sequence variations in the amplicon without the need for any post-PCR processing. The method is easy to use, highly sensitive, specific, low cost and yields rapid sample turn-around [11]–[13], making HRMA an attractive choice for the detection of disease-associated mutational variants with applications in clinical diagnostic labs. Furthermore, HRMA is a nondestructive method. Therefore, subsequent analysis of the sample by other techniques, such as gel-electrophoresis or DNA sequencing, can still be performed after HRMA analysis. These characteristics make HRMA ideal for use in routine diagnostic settings. Due to its numerous advantages, HRMA has been widely applied in diagnostic laboratories for screening for disease-associated mutations. Since it was first introduced for genotyping in 2003 [14], HRMA has been used to detect mutations such as EGFR [15], [16], KRAS [13], [17], KIT [18], BRAF [19], [20], BRCA [21], TP53 [22]. In the setting of the EuroGenTest consortium, inter-laboratory evaluation and validation of HRMA, and generation of guidelines for implementing the method as a scanning technique for the discovery of new genes have been proposed [21]. One disadvantage of HRMA is that the sensitivity and specificity in an individual clinical diagnostic setting are variable [23]. According to the “OECD Guidelines for Quality Assurance in Molecular Genetic Testing” [24], there is an obligation for diagnostic laboratories to provide high quality results. Therefore, all methods implemented within a routine setting must be duly validated and achieve acceptable limits for sensitivity and specificity prior to their diagnostic use. Although reviews and reports on the use of HRMA for mutation scanning and genotyping have been published previously [23], [25]–[29], a systematic review of the application of the technique for diagnostic purposes has not been carried out. Therefore, the meta-analysis described in this study was performed to evaluate the diagnostic accuracy of HRMA and investigate the potential for implementation of HRMA in different routine clinical settings for the detection of human disease-associated mutations. The analysis includes a comparison to DNA sequencing. The purpose of the analysis is to provide clinicians and health managers with a more objective basis for decision-making regarding implementation of the technique and to assess areas where there is currently a lack of evidence regarding the technique [30].
Materials and Methods
Literature search strategy
A literature search was carried out between July and November 2010 using the following databases: Medline, Embase, Cochrane Library and the Medion databases. The following search words (all fields) were used: ‘high resolution melting analysis or high-resolution fluorescent melting curve analyses or High-resolution amplicon melting analysis’, ‘HRM or HRMA or HRMCA’, ‘mutation’, and ‘sequence or sequencing’. The CBMdisc databases were used for Chinese articles with the following keywords (in Chinese): ‘HRM or HRMA or HRMCA’ and ‘sequencing’. The results were limited to human species. The date of publication was limited to November 6, 2010. In addition, the following journals were screened manually: Human Mutation, Cancer Research, Human Molecular Genetics, Clinical Chemistry, Genetic Testing, Clinical Genetics, Nucleic Acid Research and the Journal of Medical Genetics and Human Genetics. Furthermore, the reference lists of the included studies were screened and additional search engines, including SUMsearch, TRIP database, Sciencedirect, Google, Database for Chinese Journals of Technology (Chinese) were used. The applicability of borderline publications was discussed by the authors until a consensus for inclusion or exclusion was reached. The Institutional Review Boards approved the conclusion that no ethical approval was required for this study.
Inclusion and exclusion criteria
The inclusion criteria were as follows: (1) HRMA was applied to the study of disease-associated mutations in humans, (2) sequencing (including direct sequencing, dideoxy sequencing or sequencing of HRM products) was used as a reference standard, (3) only parts of mutated genes were investigated, (4) only some study data was compared to direct sequencing as a reference standard (only this data was included in the current study), (5) sensitivity and specificity were reported or could be calculated from the results reported, (6) the authors only reported that there were no false positive or false negative results, so that conclusions on sensitivity and specificity could be drawn without calculation of these parameters, (7) all fragments were included if one gene locus was amplified into multiple fragments and (8) the publication language was English or Chinese. The exclusion criteria were as follows: (1) studies were performed using only HRMA or comparing HRMA with non-sequencing techniques, (2) HRMA was combined with other detection methods, such as probes or qPCR, (3) studies used samples with artificially created sequences, (4) studies were aimed at detecting polymorphisms. Non-systematic/narrative reviews, letters, comments, and meeting abstracts were also excluded. Unpublished sources of data were not included. Publications identified as duplicates were excluded.
Assessment of Study Quality
The quality of the studies was assessed according to the “Quality Assessment of Diagnostic Accuracy Studies” (QUADAS) tool [31]. The modified tool was composed of 10 item questions summarized in Table 1, which were each answered “yes,” “unclear,” or “no.” Quality assessment of the studies was carried out independently by two reviewers (B.S. Li and F.L. Ma). If the quality assessment of the two reviewers were not in agreement, the discrepancies were resolved by consensus. The tool does not incorporate a global quality score. The main reason for this is that quality scores ignore the importance of individual items and potential biases related to individual items may vary according to context. Therefore, the application of quality scores may dilute or ignore potential associations [31].
Table 1. “Quality Assessment of Diagnostic Accuracy Studies” (QUADAS) Tool.
| Author | 1 | 2 | 3 | 6 | 8 | 9 | 10 | 11 | 13 | 14 |
| Bastien, R. (2008) | N | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Dagar, V. (2009) | N | Y | Y | Y | Y | U | U | U | Y | Y |
| Do, H. (2008) | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Doi, Y. (2009) | N | Y | Y | Y | Y | N | U | U | Y | Y |
| Fassina, A. (2009) | Y | Y | Y | Y | U | U | U | U | Y | Y |
| Franklin, W.A. (2010) | N | Y | Y | Y | Y | Y | U | U | Y | Y |
| Fukui, T. (2008) | N | Y | Y | N§ | U | U | U | U | Y | Y |
| Fuster, O. (2009) | N | Y | Y | Y | Y | Y | U | U | Y | Y |
| Gaucher, C. (2009) | N | Y | Y | Y | Y | U | U | U | Y | Y |
| Hung, C.C. (2008) | N | Y | Y | Y | Y | U | N | Y | Y | Y |
| Krenkova, P. (2009) | N | Y | Y | Y | Y | N | U | Y | Y | Y |
| Krypuy, M. (2006) | N | Y | Y | Y | Y | Y | U | U | Y | Y |
| Krypuy, M. FF (2007) | N | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Krypuy, M. FFPE (2007) | N | Y | Y | Y | Y | Y | U | U | Y | Y |
| Liyanage, K.E. (2008) | N | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Lopez-Villar, I. (2010) | N | Y | Y | Y | Y | Y | U | U | Y | Y |
| Ma, E.S. (2009) | N | Y | Y | Y | Y | Y | N | Y | Y | Y |
| Nomoto, K. (2006) | N | Y | Y | Y | Y | Y | U | Y | Y | Y |
| Olsen, R.K. (2010) | N | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Pichler, M. (2009) | N | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Polakova, K.M. (2008) | N | Y | Y | Y | Y | Y | U | U | Y | Y |
| Rapado, I. (2009) | Y | Y | Y | Y | Y | Y | U | U | Y | Y |
| Simi, L. (2008) | N | Y | Y | Y | Y | Y | U | U | Y | Y |
| Takano, T. (2007) | N | Y | Y | Y | U | U | Y | U | Y | Y |
| Tan, A.Y. (2008) | N | Y | Y | Y | Y | Y | U | U | Y | Y |
| van Eijk, R. (2010) | N | Y | Y | Y | Y | Y | U | U | Y | Y |
| Whitehall, V. (2009) | N | Y | Y | Y | Y | Y | U | U | Y | Y |
| Willmore, C. (2004) | N | Y | Y | Y | Y | N | U | U | Y | Y |
| Willmore-Payne, C. (2005) | N | Y | Y | Y | Y | Y | U | Y | Y | Y |
| Willmore-Payne, C. (2006 LC) | N | Y | Y | Y | Y | N | U | Y | Y | Y |
| Willmore-Payne, C. (2006) | N | Y | Y | Y | Y | N | U | Y | Y | Y |
| Xiao, J. (2009) | N | Y | Y | Y | Y | U | U | U | Y | Y |
| XinHui,Fu. (2009) | N | Y | Y | Y | Y | Y | U | U | Y | Y |
| YongPing,Lu. (2010) | N | Y | Y | Y | Y | U | U | U | Y | Y |
| ZhiHong,Chen. (2010) | N | Y | Y | Y | Y | Y | U | U | Y | Y |
Y = yes; N = no; U = unclear;
reference standard included sequencing and pyrosequencing; LC: lung cancer.
Items: 1) Was the spectrum of patients representative of the patients who will receive the test in practice? 2) Were the selection criteria clearly described? 3) Is the reference standard likely to classify the target condition correctly? 6) Did patients receive the same reference standard regardless of the index test result? 8) Was the execution of the index test described in sufficient detail to permit replication of the test? 9) Was the execution of the reference standard described in sufficient detail to permit replication? 10) Were the index test results interpreted without knowledge of the results of the reference standard result? 11) Were the reference standard results interpreted without knowledge of the results of the index test? 13) Were uninterpretable/intermediate test results reported? 14) Were withdrawals from the study explained?
Outcome parameters
The outcome parameters were sensitivity, specificity, positive predictive value and negative predictive value. Two ‘statistical’ units (per amplicon and per sample) and different definitions of ‘positive result’ were accepted as a basis for the calculation of these parameters. For example, ‘positive’ could mean any alteration, such as mutations, undetermined melting curves and polymorphisms.
Data extraction
The two reviewers (B.S. Li and F.L. Ma) independently extracted relevant data from each article using a standardized form (Table S1). The reviewers were not blinded with regard to information about the journal name, author names, author affiliations or year of publication since this has previously been shown to be unnecessary [32]. To resolve disagreement between reviewers, other authors assessed all discrepant items and the majority opinion was used for analysis.
Study characteristics
The QUADAS quality assessment tool was used to extract the relevant study design characteristics of each study (Table 1). In addition, other main study characteristics were recorded as follows: (1) year of publication, (2) disease type, (3) sample source, (4) prevalence of samples with mutation, (5) target fragment/mutation-type analyzed, (6) instrument used (7) dye used, (8) level of analysis (per amplicon and per sample) and (9) length of sequence (Table 2). The following features were also extracted: (1) sample size, (2) study site, (3) language and (4) design type.
Table 2. Result of the multivariable meta-regression model for the most important characteristics with backward regression analysis (Inverse Variance Weights).
| Variable | Coeff. | Std. Err. | p - value | RDOR | [95%CI] |
| Cte. | 1.838 | 0.9233 | 0.0516 | - | - |
| S | −0.629 | 0.1301 | 0.0000 | - | - |
| Instrument type | 0.440 | 0.2573 | 0.0930 | 1.55 | (0.93; 2.60) |
| Sample size | 2.577 | 0.5975 | 0.0001 | 13.15 | (3.97; 43.57) |
Examination results
2×2 tables were extracted on per sample or per amplicon basis, including the numbers of true-positive, true-negative, false-positive, and false-negative results in the detection of disease-associated mutations (Table S1).
Statistical analysis
Combined estimates of sensitivity, specificity, positive and negative likelihood ratios (LRs) and diagnostic odds ratio (DOR), together with their 95% confidence intervals (CI), were obtained from the available data reported in the selected studies (proportions of true positives, true negatives, false positives and false negatives). To handle studies with empty cells, 0.5 was added to all cells from all studies.
The heterogeneity of all indices was evaluated by graphical examination of forest plots, which are commonly used to detect heterogeneity in meta-analysis. As meta-analyses include small numbers of studies, the power of the usual Cochran's Q test is low. Therefore, they are poor at detecting true heterogeneity among studies as significant. An alternative approach to quantify the effect of heterogeneity is the I2 index that describes the percentage of total variation across studies that is due to heterogeneity rather than chance [33]. I2 is calculated and a value >50% indicates substantial heterogeneity [33]. Meta-analyses were performed by combining the sensitivities, specificities, LRs and DORs using the DerSimonian-Laird method, a random effects model [34], in order to incorporate variations among the studies. This approach was taken because including random effects has been previously reported as the more realistic and appropriate model for this type of meta-analysis [35], [36]. As a “threshold effect” was not detected by the Spearman test and the examination of sensitivity and specificity plots on a receiver operating characteristic (ROC) plane, summary receiver operating characteristic (SROC) curves were not constructed [37]. The analyses were carried out using Microsoft Excel 2003 (Microsoft, Seattle, WA, USA), SPSS 13.0 for Windows (SPSS, Chicago, IL, USA) and Meta-DiSc (Version 1.4) [38].
Meta-regression analysis
Meta-regression analysis was executed to determine whether diagnostic values were significantly affected by heterogeneity between the individual studies. First, single factor regression analysis was performed using variates including instrument type (HR-1 or other instrument (LightCycler4 80, Rotor-Gene 6000, LightScanner 96)), level of analysis (per amplicon or per sample), dye type (EvaGreen, LCGreen I, LCGreen plus, Resolight or Syto9), sample source (blood cell/bone marrow, fresh frozen (FF) tissue, formalin-fixed and paraffin-embedded (FFPE) tissue or cytologic slides), eligible/non-eligible sample size (eligible (>35 samples/amplicons with mutations and >35 samples/amplicons without mutation to yield 95% confidence intervals whose lower boundary is >90% sensitivity if the sensitivity is 100%) [39] and non-eligible (all other samples), disease type (tumorous or non-tumorous), study site (Europe, Asia, Oceania or North America), mutation type (TP53, EGFR, KRAS or others), study language (Chinese or English), design type (single-gate design or two-gate design) and answers from the 10 questions of the QUADAS quality assessment tool. Variates were considered as explanatory if their regression coefficients were statistically significant (P<0.05). Subsequently, we developed a multivariable regression model and using a backward stepwise algorithm, we identified the most important characteristics. Characteristics were retained in the regression model if P<0.05.
Subgroup analyses
Subgroup analyses were planned a priori depending on the following: (1) most important characteristics as selected via meta regression, (2) instrument type (HR-1 or other instrument), (3) dye type (EvaGreen, LCGreen I, LCGreen plus, Resolight or Syto9) and (4) sample source (blood/bone marrow cell, FF tissue, FFPE tissue or cytologic slides).
Results
Literature search outcome
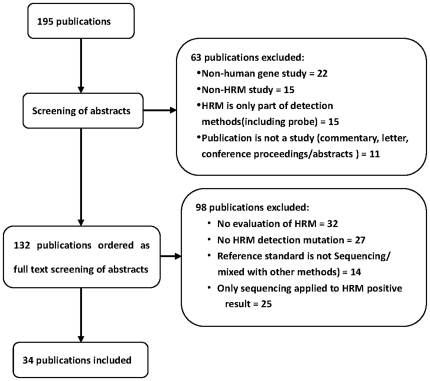
The results of the literature search and the stepwise exclusion process are illustrated in Figure 1. Out of 195 references found, only 34 articles met our inclusion criteria. These articles were divided into 58 ‘units’ for statistical analysis according to target fragment/mutation-type and sample source (Table S1). Of the 161 publications excluded, 22 were for non-human HRMA studies, such as viruses, bacteria, mosquitoes and other animals, 15 were non-HRMA studies applied to the human genome, 15 were studies where HRMA was used as part of other research methods, seven used multiple probes, eight combined HRMA with qPCR or other methods, 11 were not original research studies, one was a conference presentations, eight were reviews, two were letters, 32 were not for performance evaluation studies of HRMA, 27 were not of HRMA applied to mutation detection (20 SNPs, 4 methylation and 3 others), 14 did not exclusively use sequencing as the reference standard (8 dHPLC, 1 DGGE and 5 mixed methods) and 25 only applied sequencing to HRMA positive results. Amplicon size varied from 51–634 bp and the most common sample source was FFPE. The most frequently used dye was LCGreen I and the most commonly used instrument was the LC480.
Figure 1. Flowchart for the selection of articles for meta-analysis.
Study description
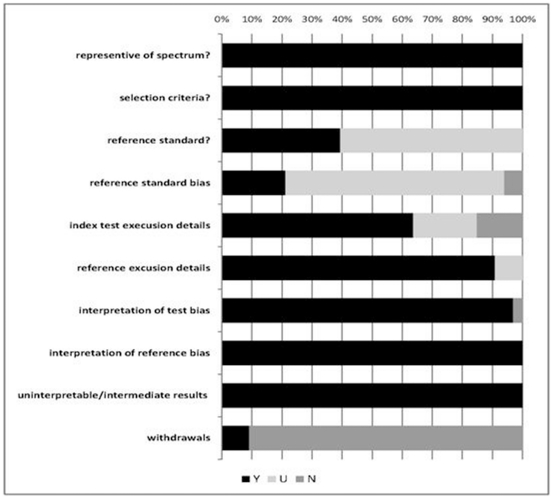
The 34 studies included in the meta-analysis included reports on the evaluation of the accuracy of HRMA for the detection of human disease-associated mutations (Table S1). Instrument types included HR-1 [15], [16], [19], [40]–[45] (n = 9), LightCycler480 [13], [17], [18], [20], [22], [46]–[55] (n = 14), RotorGene6000 [13], [56]–[62] (n = 8) and LightScanner96 [63]–[65] (n = 3). There were 27 single-gate designs and 7 two-gate designs. Disease types included 27 tumorous and seven non-tumorous diseases. The total answers to the QUADAS quality assessment tool included, ‘yes’ 251/350 (71.7%) and ‘unclear/no’99/350 (28.3%) (Figure 2). The study sites were distributed over 4 continents including Europe (10 total, 3 Spain, 1 Netherlands, 2 Italy, 2 Czech Republic, 1 Denmark, 1 France) Asia (9 total, 4 Japan, 5 China), Oceania (8 total, all Australia) and North America (7 total, all USA). Only three of the five studies carried out in China were Chinese language publications [55], [61], [63].
Figure 2. Assessment of quality items using modified QUADAS tool.
Y = yes; U = unclear; N = no.
Summary estimates of sensitivity, specificity, LR and DOR
The pooled sensitivity and specificity estimates were 97.5% (95% confidence interval (CI): 96.8–98.5; I2 = 27.0%) and 95.8% (95% CI: 95.3–96.3; I2 = 91.6%), respectively (Figure S1). The results from the analysis of all the pooled manuscripts are also shown in Table 3.
Table 3. Summary results of the pooled and subanalysis by meta disc 1.4.
| StatisticalIndex | Pooled | Sample Sizeϕ | Instrument TypeΦ | ||
| Eligiblea | Non-eligible | HR-1 | Other§ | ||
| Sensitivity (%) | 97.7 | 99.3 | 96.6 | 95.1 | 98.7 |
| (96.8, 98.5) | (98.1, 99.8) | (94.9–97.8) | (92.0, 97.2) | (97.7, 99.3) | |
| Cochran's | 0.034 | 0.551 | 0.119 | 0.008 | 0.682 |
| Q (P value) | |||||
| I2 | 27.0% | 0.0% | 19.9% | 53.1% | 0.0% |
| Specificity | 95.8 | 93.4 | 96.2 | 99.5 | 95.4 |
| (%) | (95.3, 96.3) | (91.7, 94.9) | (95.7, 96.7) | (98.6, 99.9) | (94.9, 95.9) |
| Cochran's | <0.0001 | <0.0001 | <0.0001 | 0.373 | <0.0001 |
| Q (P value) | |||||
| I2 | 91.6% | 92.2% | 91.6% | 7.1% | 93.2% |
| LR + | 34.72 | 28.51 | 37.82 | 32.06 | 32.24 |
| (22.37, 53.90) | (9.80, 82. 92) | (22.61, 63.24) | (17.61, 58.37) | (19.88, 52.26) | |
| Cochran's | <0.0001 | <0.0001 | <0.0001 | 0.539 | <0.0001 |
| Q (P value) | |||||
| I2 | 87.2% | 93.2% | 85.2% | 0.0% | 89.0% |
| LR − | 0.07 | 0.02 | 0.10 | 0.10 | 0.06 |
| (0.05, 0.09) | (0.01, 0.04) | (0.08, 0.13) | (0.06, 0.17) | (0.04, 0.08) | |
| Cochran's | 0.132 | 0.95 | 0.815 | 0.078 | 0.780 |
| Q (P value) | |||||
| I2 | 17.4% | 0.0% | 0.0% | 36.5% | 0.0% |
| DOR | 711.75 | 2198.5 | 522.16 | 634.96 | 816.23 |
| (427.18, | (735.39, | (304.06, | (262.66, | (431.39, | |
| 1185.9) | 6572.6) | 896.72) | 1535.0) | 1544.2) | |
| Cochran's | 0.031 | 0.31 | 0.102 | 0.829 | 0.004 |
| Q (P value) | |||||
| I2 | 27.2% | 14.4% | 21.2% | 0.0% | 39.5% |
: Other instruments included LightCycler480, Rotor-Gene6000, LightScanner96;
: the sensitivity of the eligible and other instruments groups were significantly higher than the non-eligible and HR-1 groups, while the opposite relationship was observed for specificity (P<0.0001).
: (>35 samples/amplicons with mutations and >35 samples/amplicons without mutations are needed to yield 95% confidence intervals whose lower bound is >90% sensitivity if the sensitivity is 100%).
Meta-regression analysis
After single factor regression analysis, two variables were found to be explanatory: sample size and instrument type. Therefore, we developed a multivariable regression model using a backward stepwise algorithm to evaluate sample size and instrument type as variables. From this regression model, sample size was determined to be the most important characteristic (Table 2).
Subgroup analysis
For the subgroup analysis of sample size and instrument type, the sensitivity of the eligible sample size subgroup was 99.3% (95% CI: 98.1, 99.8; I2 = 0.0%), non-eligible sample size subgroup was 96.6% (95% CI: 94.9, 97.8; I2 = 19.9%), other instruments subgroup was 98.7% (95% CI: 97.7, 99.3; I2 = 0.0%) and HR-1 instrument subgroup was 95.1% (95% CI: 92.0, 97.2; I2 = 53.1%) (Table 3). The sensitivity of the eligible sample size and other instruments subgroups were significantly higher than the non-eligible sample size and HR-1 instrument subgroups, respectively (P<0.0001, Figure S2 and Figure S3). The specificity of the eligible sample size subgroup was 93.4% (95% CI: 91.7, 94.9; I2 = 92.2%), non-eligible sample size subgroup was 96.2 (95% CI: 95.7, 96.7; I2 = 96.2%), other instruments subgroup was 95.4% (95% CI: 94.9, 95.9; I2 = 93.2%) and HR-1 instrument subgroup was 99.5% (95% CI: 98.6, 99.9; I2 = 7.1%). The specificity of the eligible sample size and other instruments subgroups were significantly lower than the non-eligible sample size and HR-1 instrument subgroups, respectively (P<0.0001, Figure S2 and Figure S3) but there were was no difference among other instruments within the other instruments subgroup (data not shown). No significant differences were detected in dye type and sample source subanalyses (data not shown).
The PRISMA 2009 checklist is provided as Checklist S1.
Discussion
In this systematic review, we obtained summary estimates for the diagnostic accuracy of HRMA in the detection of disease-associated mutations in humans. HRMA was found to be a high sensitive modality when compared with DNA sequencing.
It has been previously shown that studies of diagnostic performance of modalities with methodological shortcomings may lead to overestimates of the accuracy of the diagnostic test [66]. In this study, meta-regression analysis was used to evaluate the effect of different study characteristics, such as sample size, instrument type and dye type, on the diagnostic performance of HRMA. The advantage of the regression analysis performed here is that the model accounts not only for the heterogeneity between studies from different threshold settings but also for the error of estimation of the sensitivity and specificity values in each study. This random model also accounts for the residual heterogeneity that may remain even after adjusting for individual study characteristics and HRMA technical conditions [67]. The results of the meta-regression analysis indicated sample size was the most significant characteristic influencing diagnostic accuracy.
Data from the subgroup analysis indicated differences for sample size. After studies were divided into eligible sample size and non-eligible sample size subgroups, the heterogeneity was significantly decreased. The eligible sample size subgroup of studies had significantly higher sensitivity and was less heterogeneous that the non-eligible sample size subgroup. These improvements may result from differences in the prevalence of samples/amplicons with mutations, as the number of mutations in the eligible sample size subgroup was significantly higher (550/1543, 35.6%), than the non-eligible sample size subgroup (697/6274, 11.1%). Therefore, the results showed that the number of samples with/without mutations in a study has an important influence on diagnostic accuracy [39].
Although the multivariable regression analysis presented here showed that the instrument type was not a significant characteristic, previous studies have shown that instrument type does affect the sensitivity and specificity of HRMA [11], [68]–[71]. The subgroup analysis of instrument type indicated some differences. For example, other instruments were more sensitive than the HR-1 instrument. This may be because the other instruments were some of the latest real-time thermal cyclers modified to incorporate HRMA, and yield high-resolution data quality by melting 18-times slower than the HR-1 instrument [25].
We found that HRMA was a highly sensitive method for mutation detection that yielded low negative LR without substantial heterogeneity. The sensitivity of all publications in the study, the eligible/non-eligible sample size subgroup and other instruments subgroups were 97.5%, 99.3%/96.6% and 98.7%, respectively and the negative LR were 0.07, 0.02/0.10 and 0.05, respectively. These results compare well with a recent compilation of 19 studies for constitutional variants that found an overall sensitivity of 99.3% (n = 5839) [72]. The high sensitivity of HRMA means that the technique can be considered as SnNOut (high sensitivity, negative, rules out) [73]–[75]. In this scenario, a negative HRMA test result rules out mutations. Therefore, when implemented correctly, the need for subsequent sequencing disappears for the pooled group (sensitivity 81.0%, 6322/7817, 32 false negatives), and the other instruments (sensitivity 82.2%, 5726/6967, 12 false negatives) and non-eligible sample size subgroups (sensitivity 85.6%, 5373/6274, 25 false negatives). These results are consistent with Provaznikova et al. [76] who reported avoiding unnecessary sequencing of more than 85% of the MYH9 gene. HRMA takes only a few minutes and costs only 11% of the cost of sequencing one exon [28], significantly reducing costs and saving time. However, in the eligible sample size subgroup, the reduction of sequencing is less (61.5%, 949/1543, 7 false negative) due to the greater number of mutations. Thus, the results showed HRMA is more suitable for screening for lower incidence mutations.
In general, as the sensitivity of diagnostic tests improves, the specificity decreases. Therefore, the specificity of the eligible sample size and other instruments subgroups was significantly lower than the non-eligible sample size and HR-1 subgroups. Specificity was homogeneous in the HR-1 instrument subgroup. This may be due to the fact that most of the samples were from only two research institutions (635/933 units of statistical analysis). However, the overall specificity of HRMA showed considerable heterogeneity between studies. This may be related to additional factors, such as the sequence length, GC content and sequence, that are properties of the individual sequences under study [77]–[79]. Other factors that are independent of the sequence, such as the presence of substances such as DMSO or betaine [80], [81], may also affect specificity. It is difficult to quantitatively analyse these factors.
In addition, we found that the sample source and dye used had no impact on HRMA accuracy. This is contrast to previous studies that found that the sample source and dye used affected HRMA accuracy [15], [20], [22], [41], [72]. The discrepancies between studies may result from differences in sample size, the focus of the researchers and/or the methods of statistical analysis. The continent of origin, design type and diagnostic accuracy also showed no significant effect in the meta-regression analysis.
In this study, amplicon length had some impact on the sensitivity and specificity of HRMA, as in previous reports [23], [82]. For example, for PCR products of less than 400 bp, sensitivity and specificity were 100%. While for PCR products 400–1000 bp long, the sensitivity was reduced to 96.1% and specificity to 99.4%. In this study, the majority of amplicon lengths were in the recommended amplicon length range (less than 300 bp) [23]. Therefore, the impact of amplicon length was not investigated further here. Many factors, including sequence-dependent and non-sequence dependent factors, affected HRMA accuracy. Therefore, standardization of DNA preparation, PCR and HRMA operating procedures are essential. Also, it remains necessary to subsequently sequence positive results from HRMA for confirmation. The current meta-analysis has some limitations in that the studies were heterogeneous and most studies used small sample sizes. In addition, the effect of language selection bias and literature type cannot be ignored as we only chose published articles in Chinese and English. In order to avoid this bias, the search should not be language limited and all literature types should be searched. However, most of the articles found on HRMA were published in English so the language bias was minimized. The selection bias was further minimized by maximizing the sensitivity of the search words, performing the search over a long search time and using a variety of databases/search engines including Medline, Embase, Cochrane Library, Medion, CBMdisc, Sciencedirect, SUMsearch, Google, Database for Chinese Journals of Technology (Chinese) and selected Journal Special Issues. In addition, the reference lists from articles obtained from the automated searches were checked manually.
Publication bias is a potential limitation of any systematic review. Smaller studies are associated with a greater diagnostic accuracy [83]. However, studies about publication bias focus mostly on randomized trials, and these types of studies are registered. The registration of studies for diagnostic studies is either limited or difficult to achieve. Due to the smaller sample sizes used for diagnostic studies, fewer studies were identified by the searches were for inclusion in this review. We examined publication bias by assessing whether the sample size of studies was associated with diagnostic accuracy, and found an association between sample size and HRMA diagnostic performance in the subgroup analysis. Therefore, assessment of the effect of sample size on HRMA accuracy was not ignored in further studies. In addition, there was no consistent relationship between language restriction and publication bias [83].
In conclusion, the sensitivity, simplicity, and low cost of HRMA make it the method of choice to screen patients for disease-associated variants, especially those diseases with lower incidence mutations. HRMA sensitivity is higher in the eligible sample size subgroup and is affected by instrument type but not by sample source or dye type. The DNA sequencing burden can be significantly reduced by the implementation of HRMA, but positive results still require sequencing for diagnostic confirmation. Further clinical studies of HRMA need to pay attention to the impact of sample size on diagnostic accuracy. However, as HRMA is still a relatively new technology, increases in accuracy can be expected as the diagnostic technology improves with time.
Supporting Information
Meta-analysis of studies evaluating HRMA sensitivity and specificity from pooled estimates.
(TIF)
Meta-analysis of studies evaluating HRMA sensitivity and specificity from the subanalysis of sample size.
(TIF)
Meta-analysis of studies evaluating HRMA sensitivity and specificity from the subanalysis of instrument type.
(TIF)
Characteristics of the 34 studies included in the meta-analysis.
(DOC)
PRISMA 2009 checklist.
(DOC)
Acknowledgments
We thank Medjaden Bioscience Limited for assisting in the preparation of this manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by a Natural Science Foundation of Guangdong Province Grant (No. 10151600301), and a Medical Science and Technology of Guangdong Province Grant (A2010692). The funders had no role in study design,data collection and analysis, decision to publish, or preparation of manuscript.
References
- 1.Ahmadian A, Gharizadeh B, Gustafsson AC, Sterky F, Nyren P, et al. S ingle-nucleotide polymorphism analysis by pyrosequencing. Anal Biochem. 2000;280(1):103–110. doi: 10.1006/abio.2000.4493. [DOI] [PubMed] [Google Scholar]
- 2.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989;86(8):2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lerman LS, Silverstein K. Computational simulation of DNA melting and its application to denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:482–501. doi: 10.1016/0076-6879(87)55032-7. [DOI] [PubMed] [Google Scholar]
- 4.Xiao W, Oefner PJ. Denaturing high-performance liquid chromatography: A review. Hum Mutat. 2001;17(6):439–474. doi: 10.1002/humu.1130. [DOI] [PubMed] [Google Scholar]
- 5.Li Q, Liu Z, Monroe H, Culiat CT. Integrated platform for detection of DNA sequence variants using capillary array electrophoresis. Electrophoresis. 2002;23(10):1499–1511. doi: 10.1002/1522-2683(200205)23:10<1499::AID-ELPS1499>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 6.Bocker S. Simulating multiplexed SNP discovery rates using base-specific cleavage and mass spectrometry. Bioinformatics. 2007;23(2):e5–12. doi: 10.1093/bioinformatics/btl291. [DOI] [PubMed] [Google Scholar]
- 7.Wittwer CT, Herrmann MG, Gundry CN, Elenitoba-Johnson KS. Real-time multiplex PCR assays. Methods. 2001;25(4):430–442. doi: 10.1006/meth.2001.1265. [DOI] [PubMed] [Google Scholar]
- 8.Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997;22(1):130–131, 134–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
- 9.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6(10):986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 10.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14(3):303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 11.De Leeneer K, Coene I, Poppe B, De Paepe A, Claes K. Rapid and sensitive detection of BRCA1/2 mutations in a diagnostic setting: comparison of two high-resolution melting platforms. Clin Chem. 2008;54(6):982–989. doi: 10.1373/clinchem.2007.098764. [DOI] [PubMed] [Google Scholar]
- 12.Kramer D, Thunnissen FB, Gallegos-Ruiz MI, Smit EF, Postmus PE, et al. A fast, sensitive and accurate high resolution melting (HRM) technology-based assay to screen for common K-ras mutations. Cell Oncol. 2009;31(3):161–167. doi: 10.3233/CLO-2009-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitehall V, Tran K, Umapathy A, Grieu F, Hewitt C, et al. A Multicenter Blinded Study to Evaluate KRAS Mutation Testing Methodologies in the Clinical Setting. J Mol Diagn. 2009;11(6):543–552. doi: 10.2353/jmoldx.2009.090057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49(6):853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 15.Takano T, Ohe Y, Tsuta K, Fukui T, Sakamoto H, et al. Epidermal growth factor receptor mutation detection using high-resolution melting analysis predicts outcomes in patients with advanced non small cell lung cancer treated with gefitinib. Clin Cancer Res. 2007;13(18):5385–5390. doi: 10.1158/1078-0432.CCR-07-0627. [DOI] [PubMed] [Google Scholar]
- 16.Fukui T, Ohe Y, Tsuta K, Furuta K, Sakamoto H, et al. Prospective study of the accuracy of EGFR mutational analysis by high-resolution melting analysis in small samples obtained from patients with non-small cell lung cancer. Clin Cancer Res. 2008;14(15):4751–4757. doi: 10.1158/1078-0432.CCR-07-5207. [DOI] [PubMed] [Google Scholar]
- 17.Franklin WA, Haney J, Sugita M, Bemis L, Jimeno A, et al. KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer . J Mol Diagn. 2010;12(1):43–50. doi: 10.2353/jmoldx.2010.080131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuster O, Barragan E, Bolufer P, Cervera J, Larrayoz MJ, et al. Rapid detection of KIT mutations in core-binding factor acute myeloid leukemia using high-resolution melting analysis. J Mol Diagn. 2009;11(5):458–463. doi: 10.2353/jmoldx.2009.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willmore-Payne C, Holden JA, Tripp S, Layfield LJ. Human malignant melanoma: detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis. Hum Pathol. 2005;36(5):486–493. doi: 10.1016/j.humpath.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Pichler M, Balic M, Stadelmeyer E, Ausch C, Wild M, et al. Evaluation of high-resolution melting analysis as a diagnostic tool to detect the BRAF V600E mutation in colorectal tumors. J Mol Diagn. 2009;11(2):140–147. doi: 10.2353/jmoldx.2009.080100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Stoep N, van Paridon CD, Janssens T, Krenkova P, Stambergova A. Diagnostic guidelines for high-resolution melting curve (HRM) analysis: an interlaboratory validation of BRCA1 mutation scanning using the 96-well LightScanner. Hum Mutat. 2009;30(6):899–909. doi: 10.1002/humu.21004. [DOI] [PubMed] [Google Scholar]
- 22.Bastien R, Lewis TB, Hawkes JE, Quackenbush JF, Robbins TC, et al. High-throughput amplicon scanning of the TP53 gene in breast cancer using high-resolution fluorescent melting curve analyses and automatic mutation calling. Hum mutat. 2008;29(5):757–764. doi: 10.1002/humu.20726. [DOI] [PubMed] [Google Scholar]
- 23.Montgomery JL, Sanford LN, Wittwer CT. High-resolution DNA melting analysis in clinical research and diagnostics. Expert Rev Mol Diagn. 2010;10(2):219–240. doi: 10.1586/erm.09.84. [DOI] [PubMed] [Google Scholar]
- 24.OECD website. Available: http://www.oecd.org/dataoecd/43/6/3883978.pdf. Accessed 2011 May 10.
- 25.Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;89(6):597–608. doi: 10.2217/14622416.8.6.597. [DOI] [PubMed] [Google Scholar]
- 26.Erali M, Voelkerding KV, Wittwer CT. High resolution melting applications for clinical laboratory medicine. Exp Mol Pathol. 2008;85(1):50–58. doi: 10.1016/j.yexmp.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor CF. Mutation scanning using high-resolution melting. Biochem Soc Trans. 2009;37(2):433–437. doi: 10.1042/BST0370433. [DOI] [PubMed] [Google Scholar]
- 28.Vossen RH, Aten E, Roos A, den Dunnen JT. High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum mutat. 2009;30(6):860–866. doi: 10.1002/humu.21019. [DOI] [PubMed] [Google Scholar]
- 29.Wittwer CT. High-resolution DNA melting analysis: advancements and limitations. Hum mutat. 2009;30(6):857–869. doi: 10.1002/humu.20951. [DOI] [PubMed] [Google Scholar]
- 30.Erali M, Voelkerding KV, Wittwer CT. High resolution melting applications for clinical laboratory medicine. Exp Mol Pathol. 2008;85(1):50–58. doi: 10.1016/j.yexmp.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whiting P, Rutjes AW, Reitsma JB, Bossuyt PM, Kleijnen J. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol. 2003;3(10):25. doi: 10.1186/1471-2288-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berlin JA. Does blinding of readers affect the results of meta-analyses? Lancet. 1997;350(9072):185–186. doi: 10.1016/s0140-6736(05)62352-5. [DOI] [PubMed] [Google Scholar]
- 33.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 35.Lijmer JG, Bossuyt PM, Heisterkamp SH. Exploring sources of heterogeneity in systematic reviews of diagnostic tests. Stat Med. 2002;21(11):1525–1537. doi: 10.1002/sim.1185. [DOI] [PubMed] [Google Scholar]
- 36.Macaskill P. Empirical Bayes estimates generated in a hierarchical summary ROC analysis agreed closely with those of a full Bayesian analysis. J Clin Epidemiol. 2004;57(9):925–932. doi: 10.1016/j.jclinepi.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 37.Deeks JJ. Systematic reviews in health care: Systematic reviews of evaluations of diagnostic and screening tests. BMJ. 2001;323(7305):157–162. doi: 10.1136/bmj.323.7305.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6(12):31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Assessing Diagnostic Technologies. Technology Assessment Program Report No. 1. Boston: US Dept of Veterans Affairs; 1996. [Google Scholar]
- 40.Willmore C, Holden JA, Zhou L, Tripp S, Wittwer CT, et al. Detection of c-kit-activating mutations in gastrointestinal stromal tumors by high-resolution amplicon melting analysis. Am J Clin Pathol. 2004;122(2):206–216. doi: 10.1309/4E6U-YBY6-2N2F-CA6N. [DOI] [PubMed] [Google Scholar]
- 41.Nomoto K, Tsuta K, Takano T, Fukui T, Fukui T, et al. Detection of EGFR mutations in archived cytologic specimens of non-small cell lung cancer using high-resolution melting analysis. Am J Clin Pathol. 2006;126(4):608–615. doi: 10.1309/N5PQNGW2QKMX09X7. [DOI] [PubMed] [Google Scholar]
- 42.Willmore-Payne C, Holden JA, Layfield LJ. Detection of epidermal growth factor receptor and human epidermal growth factor receptor 2 activating mutations in lung adenocarcinoma by high-resolution melting amplicon analysis: correlation with gene copy number, protein expression, and hormone receptor expression. Hum Pathol. 2006;37(6):755–763. doi: 10.1016/j.humpath.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Willmore-Payne C, Holden JA, Layfield LJ. Detection of EGFR- and HER2-activating mutations in squamous cell carcinoma involving the head and neck. Mod Pathol. 2006;19(5):634–640. doi: 10.1038/modpathol.3800552. [DOI] [PubMed] [Google Scholar]
- 44.Hung CC, Lee CN, Chang CH, Jong YJ, Chen CP, et al. Genotyping of the G1138A mutation of the FGFR3 gene in patients with achondroplasia using high-resolution melting analysis. Clin Biochem. 2008;41(3):162–166. doi: 10.1016/j.clinbiochem.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 45.Liyanage KE, Hooper AJ, Defesche JC, Burnett JR, van Bockxmeer FM. High-resolution melting analysis for detection of familial ligand-defective apolipoprotein B-100 mutations. Ann Clin Biochem. 2008;45(2):170–176. doi: 10.1258/acb.2007.007077. [DOI] [PubMed] [Google Scholar]
- 46.Polakova KM, Lopotova T, Klamova H, Moravcova J. High-resolution melt curve analysis: initial screening for mutations in BCR-ABL kinase domain. Leuk Res. 2008;32(8):1236–1243. doi: 10.1016/j.leukres.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 47.Tan AY, Westerman DA, Carney DA, Seymour JF, Juneja S, et al. Detection of NPM1 exon 12 mutations and FLT3 - internal tandem duplications by high resolution melting analysis in normal karyotype acute myeloid leukemia. J Hematol Oncol. 2008;1(29):10. doi: 10.1186/1756-8722-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doi Y, Sasaki D, Terada C, Mori S, Tsuruda K, et al. High-resolution melting analysis for a reliable and two-step scanning of mutations in the tyrosine kinase domain of the chimerical bcr-abl gene. Int J Hematol. 2009;90(1):37–43. doi: 10.1007/s12185-009-0337-y. [DOI] [PubMed] [Google Scholar]
- 49.Fassina A, Gazziero A, Zardo D, Corradin M, Aldighieri E, et al. Detection of EGFR and KRAS mutations on trans-thoracic needle aspiration of lung nodules by high resolution melting analysis. J Clin Pathol. 2009;62(12):1096–1102. doi: 10.1136/jcp.2009.067587. [DOI] [PubMed] [Google Scholar]
- 50.Gaucher C, Walrant-Debray O, Nguyen TM, Esterle L, Garabedian M, et al. PHEX analysis in 118 pedigrees reveals new genetic clues in hypophosphatemic rickets. Hum Genet. 2009;125(4):401–411. doi: 10.1007/s00439-009-0631-z. [DOI] [PubMed] [Google Scholar]
- 51.Ma ES, Wong CL, Law FB, Chan WK, Siu D. Detection of KRAS mutations in colorectal cancer by high-resolution melting analysis. J Clin Pathol. 2009;62(10):886–891. doi: 10.1136/jcp.2008.063677. [DOI] [PubMed] [Google Scholar]
- 52.Rapado I, Grande S, Albizua E, Ayala R, Hernandez JA, et al. High resolution melting analysis for JAK2 Exon 14 and Exon 12 mutations: a diagnostic tool for myeloproliferative neoplasms. J Mol Diagn. 2009;11(2):155–161. doi: 10.2353/jmoldx.2009.080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao J, Bastian RW, Perlmutter JS, Racette BA, Tabbal SD, et al. High-throughput mutational analysis of TOR1A in primary dystonia. BMC Med Genet. 2009;10(11):24. doi: 10.1186/1471-2350-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopez-Villar I, Ayala R, Wesselink J, Morillas JD, Lopez E, et al. Simplifying the detection of MUTYH mutations by high resolution melting analysis. BMC cancer. 2010;10(5):408. doi: 10.1186/1471-2407-10-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhi-hong C, Ai-lin G, She-juan AN, You-wei Z, Dong MA, et al. High resolution melting analysis for the rapid and sensitive of KRAS cond 12 and 13 mutations in coloretal cancer. Chin J Lab Med. 2010;33(3):209–212. [Google Scholar]
- 56.Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC cancer. 2006;6(21):295. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krypuy M, Ahmed AA, Etemadmoghadam D, Hyland SJ Group AOCS. High resolution melting for mutation scanning of TP53 exons 5–8. BMC cancer. 2007;7(31):168. doi: 10.1186/1471-2407-7-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Do H, Krypuy M, Mitchell PL, Fox SB, Dobrovic A. High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies. BMC cancer. 2008;8(21):142. doi: 10.1186/1471-2407-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simi L, Pratesi N, Vignoli M, Sestini R, Cianchi F, et al. High-resolution melting analysis for rapid detection of gene mutations in colorectal cancer. Am J Clin Pathol. 2008;130(2):247–253. doi: 10.1309/LWDY1AXHXUULNVHQ. [DOI] [PubMed] [Google Scholar]
- 60.Dagar V, Chow CW, Ashley DM, Algar EM. Rapid detection of SMARCB1 sequence variation using high resolution melting. BMC cancer. 2009;9(13):437. doi: 10.1186/1471-2407-9-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fu X, Deng Y, Fan X, Luo Y, Chen D, et al. Detecting KRAS mutations in colorectal carcinoma patients with high resolution melting curve analysis. Chin Arch Gen Surg (Electronic Edition) 2009;3(5):376–378. [Google Scholar]
- 62.Krenkova P, Norambuena P, Stambergova A, Macek M., Jr Evaluation of high-resolution melting (HRM) for mutation scanning of selected exons of the CFTR gene. Folia Biol (Praha) 2009;55(6):238–242. [PubMed] [Google Scholar]
- 63.Lu Y, Li Z, Liu Q, Feng W, Wei XU, et al. Detecting the TGFA Gene Taq I Mutation with High Resolution Melting Method. Journal of China Medical University. 2010;39(4):281–282, 285. [Google Scholar]
- 64.Olsen RK, Dobrowolski SF, Kjeldsen M, Hougaard D, Simonsen H, et al. High-resolution melting analysis, a simple and effective method for reliable mutation scanning and frequency studies in the ACADVL gene. J Inherit Metab Dis. 2010;33(3):247–260. doi: 10.1007/s10545-010-9101-y. [DOI] [PubMed] [Google Scholar]
- 65.van ER, van PM, Chhatta AR, Gupta N, Vossen RH, et al. Sensitive and specific KRAS somatic mutation analysis on whole-genome amplified DNA from archival tissues. J Mol Diagn. 2010;12(1):27–34. doi: 10.2353/jmoldx.2010.090028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lijmer JG, Mol BW, Heisterkamp S, Bonsel GJ, Prins MH, et al. Empirical evidence of design-related bias in studies of diagnostic tests. JAMA. 1999;282(11):1061–1066. doi: 10.1001/jama.282.11.1061. [DOI] [PubMed] [Google Scholar]
- 67.Rutter CM, Gatsonis CA. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Stat Med. 2001;20(19):2865–2884. doi: 10.1002/sim.942. [DOI] [PubMed] [Google Scholar]
- 68.Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT, Voelkerding KV. Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin Chem. 2006;52(3):494–503. doi: 10.1373/clinchem.2005.063438. [DOI] [PubMed] [Google Scholar]
- 69.Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT, Voelkerding KV. Instrument comparison for heterozygote scanning of single and double heterozygotes: a correction and extension of Herrmann et al., Clin Chem 2006;52:494–503. Clin Chem. 2007;53(1):150–152. doi: 10.1373/clinchem.2006.081240. [DOI] [PubMed] [Google Scholar]
- 70.Herrmann MG, Durtschi JD, Wittwer CT, Voelkerding KV. Expanded instrument comparison of amplicon DNA melting analysis for mutation scanning and genotyping. Clin Chem. 2007;53(8):1544–1548. doi: 10.1373/clinchem.2007.088120. [DOI] [PubMed] [Google Scholar]
- 71.Ugo V, Tondeur S, Menot ML, Bonnin N, Le GG, et al. Interlaboratory development and validation of a HRM method applied to the detection of JAK2 exon 12 mutations in polycythemia vera patients. PLoS One. 2010;5(1):e88–93. doi: 10.1371/journal.pone.0008893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Farrar JS, Reed GH, Wittwer CT, editors. High resolution melting curve analysis for molecular diagnositics. Burlington: Elsevier; 2009. [Google Scholar]
- 73.Sackett DL, Haynes RB, Guyatt GH, Tugwell P. Clinical epidemiology, a basic science for clinical medicine. Boston: Little Brown; 1992. [Google Scholar]
- 74.Obuchowski NA, Graham RJ, Baker ME, Powell KA. Ten criteria for effective screening: their application to multislice CT screening for pulmonary and colorectal cancers. AJR Am J Roentgenol. 2001;176(6):1357–1362. doi: 10.2214/ajr.176.6.1761357. [DOI] [PubMed] [Google Scholar]
- 75.Straus SE, Richardson WS, Glasziou P, Haynes RB, editors. Evidence-based medicine: how to practice and teach EBM. New York: Elsevier/Churchill Livingstone; 2005. [Google Scholar]
- 76.Provaznikova D. High-resolution melting analysis for detection of MYH9 mutations. Platelets. 2008;19(6):471–475. doi: 10.1080/09537100802140013. [DOI] [PubMed] [Google Scholar]
- 77.Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245(2):154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 78.Liew M, Pryor R, Palais R, Meadows C, Erali M, et al. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50(7):1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- 79.Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004;50(10):1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 80.Rees WA, Yager TD, Korte J, von HPH. Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry. 1993;32(1):137–144. doi: 10.1021/bi00052a019. [DOI] [PubMed] [Google Scholar]
- 81.von AN, Oellerich M, Schutz E. Limitations of genotyping based on amplicon melting temperature. Clin Chem. 2001;47(7):1331–1332. [PubMed] [Google Scholar]
- 82.Reed GH. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clinical Chemistry. 2004;50(10):1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- 83.Song F, Khan KS, Dinnes J, Sutton AJ. Asymmetric funnel plots and publication bias in meta-analyses of diagnostic accuracy. Clinical Chemistry. 2002;31(1):88–95. doi: 10.1093/ije/31.1.88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Meta-analysis of studies evaluating HRMA sensitivity and specificity from pooled estimates.
(TIF)
Meta-analysis of studies evaluating HRMA sensitivity and specificity from the subanalysis of sample size.
(TIF)
Meta-analysis of studies evaluating HRMA sensitivity and specificity from the subanalysis of instrument type.
(TIF)
Characteristics of the 34 studies included in the meta-analysis.
(DOC)
PRISMA 2009 checklist.
(DOC)