Abstract
Background
MicroRNA expression is severely disrupted in carcinogenesis, however limited evidence is available validating results from cell-line models in human clinical cancer specimens. MicroRNA-21 (mir-21) and microRNA-143 (mir-143) have previously been identified as significantly deregulated in a range of cancers including cervical cancer. Our goal was to investigate the expression patterns of several well-studied microRNA species in cervical samples and compare the results to cell line samples.
Methodology/Principal Findings
We measured the expression of mir-21 and mir-143 in 142 formalin-fixed, paraffin embedded (FFPE) cervical biopsy tissue blocks, collected from Dantec Oncology Clinic, Dakar, Senegal. MicroRNA expression analysis was performed using Taqman-based real-time PCR assays. Protein immunohistochemical staining was also performed to investigate target protein expression on 72 samples. We found that mir-21 expression increased with worsening clinical diagnosis but that mir-143 was not correlated with histology. These observations were in stark contrast to previous reports involving cervical cancer cell lines in which mir-143 was consistently down-regulated but mir-21 largely unaffected. We also identified, for the first time, that cytoplasmic expression of Programmed Cell Death Protein 4 PDCD4; a known target of mir-21) was significantly lower in women with invasive cervical carcinoma (ICC) in comparison to those with cervical intraepithelial neoplasia (2–3) or carcinoma in situ (CIN2-3/CIS), although there was no significant correlation between mir-21 and PDCD4 expression, despite previous studies identifying PDCD4 transcript as a known mir-21 target.
Conclusions
Whilst microRNA biomarkers have a number of promising features, more studies on expression levels in histologically defined clinical specimens are required to investigate clinical relevance of discovery-based studies. Mir-21 may be of some utility in predictive screening, given that we observed a significant correlation between mir-21 expression level and worsening histological diagnosis of cervical cancer.
Introduction
Early cancer detection strategies are based on the identification and validation of biomarkers which are highly indicative of disease progression from normal or precancerous tissue to early invasive cancers. MicroRNAs are a group of recently discovered short RNA species (∼21 nt) that are involved in the regulation of gene expression in a tissue-specific manner, affecting numerous cellular pathways including proliferation, differentiation and apoptosis [1]. It has been shown that microRNAs are aberrantly regulated in invasive cancer, and can act as tumor suppressors or enhancers in different tissues and environments [2]. Their recent discovery has led to a number of studies aimed at discovering novel cancer biomarkers (reviewed in [3]–[5]), however few have been validated in clinical specimens, especially those representative of pre-cancerous disease.
MicroRNAs are of increasing interest in cancer diagnostics due to the observation that a surprisingly small family of molecules can provide exquisite specificity in classifying tissue types, reflecting the developmental lineage and differentiation state. Lu et al. exploited this by using a microRNA panel of 217 species to classify poorly differentiated tumors with high concordance whilst a comparable mRNA panel containing ∼16,000 species failed [6]. Further studies have shown the potential for microRNAs to distinguish between cancer subtypes where histological diagnosis is complex or impossible, to diagnose tumors of unknown origin and in diagnosing cancer predisposition [4].
Investigations into the use of microRNAs as biomarkers for early cancer detection have identified surprising blood and tissue stability in contrast to mRNA [7], [8], and the development of highly sensitive and specific qPCR procedures is encouraging. However, in all cases to date, samples were extracted from patients who had already developed cancer [4], so the utility of microRNA as a marker of cancer progression from precancerous through to early invasive cancerous lesions remains unknown. Investigation of microRNA expression in samples spanning the entire range of histologically defined sample types, from normal to invasive cancer, is clearly required to properly determine the utility of microRNA expression levels as cancer biomarkers.
Cervical cancer is a disease for which stratification of histological types from normal through to invasive carcinoma is well characterized and supported by molecular techniques based on HPV genotyping. However, given the enormous success of cervical screening programs, only 35–65% CIN-3/CIS, 12–20% CIN-2 and <5% CIN-1 cases are expected to progress to more severe forms of dysplasia or invasive cancer – suggesting that markers with a higher predictive value for progression would be highly desirable [9], [10]. We therefore identified cervical cancer as an ideal test case for which to follow changes in specific microRNA expressions levels from normal through precancerous and cancerous tissues. Past studies have identified a range of aberrantly regulated microRNAs in cervical cancer cell lines, with mirs-127, 9, 203, 199a, 218, 21, 143, 205, 214,126, 15b, 16, 146a and 155 among the most common [3], [11]–[17]. However, only one of these studies included precancerous cervical specimens (CIN1-3), where high biological variability was noted in the microRNA expression levels, especially in normal samples (albeit with low sample sizes) [16].
In this study we investigated the expression profiles of two microRNAs (mir-21 and mir-143) and their previously validated target proteins in clinical samples from women with HPV infection without lesions, with histologically diagnosed pre-cancerous lesions, or with invasive cancer (ICC), as well as normal controls without HPV infection. The sequences were chosen based on previous findings from other groups identifying significant deregulation across a number of cervical cancer cell lines and some limited fresh frozen samples [12]. Our primary aim was to determine if there were significant correlations between microRNA expression and histological type which could point to a potential role for microRNA as an early marker of the oncogenic potential of pre-lesional HPV infection and/or of progression of precancer to cancer. Secondly, we compared microRNA expression in clinical samples and cancer cell lines to determine if cell lines should continue to be used as a basis for clinical investigations. We identified a significant association between mir-21 expression and histological diagnosis, but no association with mir-143. We also investigated the expression of known mir-21 target proteins (PDCD4, PTEN (phosphatase and tensin homolog), TPM1 (tropomyosin 1)) by immunohistochemistry analysis and identified a significant association between cytoplasmic PDCD4 expression and histological diagnosis. We identified discrepancies between the clinical samples and cervical cancer cell lines, suggesting that clinical samples must be investigated for validation purposes following discovery of candidate biomarkers in cell line discovery-focused studies.
Results
MicroRNA expression in cervical cancer cell lines
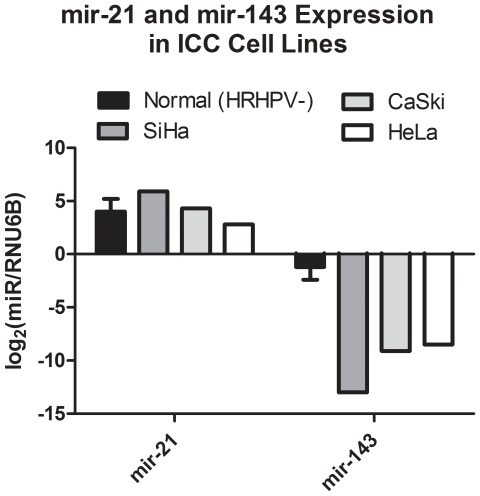
We investigated the expression of mir-21 and mir-143 in cervical cancer cell lines with respect to (Normal (HRHPV-) in order to confirm previous observations. Figure 1 shows that for all three cancer cell lines tested (SiHa, CaSki, HeLa), there was no consistent difference between mir-21 expression and that of normal samples. However, mir-143 expression of all three cancer cell lines was substantially decreased with respect to clinical samples However, we have not attached statistical analysis to these comparisons because of the different error distributions between clinical samples and cell lines, and because we would only have n = 3 in the case of the cell lines for comparison.
Figure 1. Expression profiling data for mir-21 and mir-143 in cervical cancer cell lines and normal specimens.
Mir-143 was significantly down-regulated in all three cervical cancer cell lines, in agreement with previous studies. However, mir-21 was not significantly different to the normal specimens, even though in previous studies mir-21 has been observed to be significantly up-regulated in the same cell lines.
MicroRNA expression in clinical samples
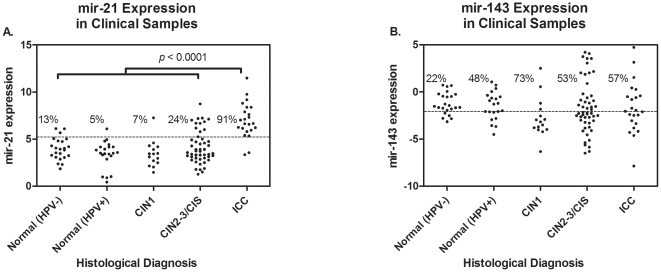
In contrast to cell line data, we observed a significant association between mir-21 expression and histologic type in clinical samples, but no association for mir-143 (Figure 2 and Table 1). Women with ICC showed significantly increased mir-21 expression levels compared to women with CIS/CIN2-3, CIN1, normal (HRHPV+) and normal (HRHPV-) (ANOVA, p<0.0001). Furthermore, women with CIS/CIN2-3 had marginally increased mir-21 expression levels compared to women with CIN1 (pu = 0.14 and pa = 0.24) and normal (HRHPV+) (pu = 0.07 and pa = 0.14). We found no association between age or HRHPV status and histological type (data not shown). A cutoff of 5.06 best distinguished ICC/Normal (HPV-) populations (determined from ROC curve) and was 91% sensitive and 87% specific. Figure 2 shows how that cutoff faired for the other two histology groups. Only 5% of the samples from women who are Normal (HPV+), 7% of CIN 1, and 24% of CIN 2-3/CIS samples had MIR-21 values greater than 5.06. Interestingly, 13% of samples from women with Normal (HPV-) histology showed mir-21 expression level greater than 5.06.
Figure 2. Expression profiling for (a) mir-21 and (b) mir-143 in 133 clinical samples stratified by histological diagnosis.
In contrast to the cell-line data presented in Figure 1, we observed a significant association between increased mir-21 expression an worsening histological diagnosis, however no significant associations for mir-143. There was a significant difference between all histological groups (ANOVA, p-value<0.0001) and furthermore, women with CIS/CIN2-3 had marginally higher mir-21 expression levels compared to those classified as Normal/HRHPV+ (pu = 0.0375 pa = 0.0563).
Table 1. MicroRNA expression in FFPE cervical specimens.
| Normal (HPV-) (n = 23) | Normal (HPV+) (n = 21) | CIN1 (n = 15) | CIN2-3/CIS (n = 51) | ICC (n = 23) | p-Value* | |
| Age | 42.6±5.6 | 47.2±8.7 | 40.4±5.5 | 43.7±8.7 | 45.4±9.5 | 0.11 |
| Mir-21 | 4.0±1.2 | 3.4±1.4 | 3.4±1.4 | 4.1±1.7 | 7.0±1.9 | <0.0001 |
| Mir-143 | −1.2±1.2 | −1.5±1.5 | −2.7±2.1 | −1.6±2.8 | −1.7±2.7 | 0.38 |
*ANOVA.
PDCD4, PTEN and TPM1 expression
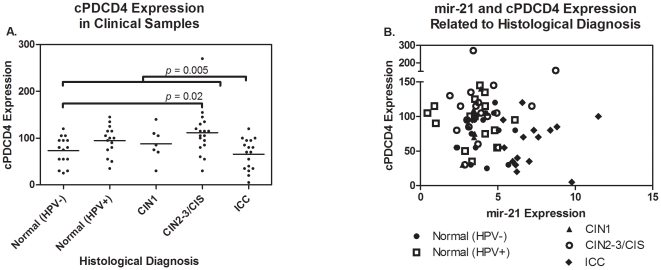
In order to investigate the effect of mir-21 deregulation on tissue-specific target protein expression in clinical samples, we measured protein expression levels of PDCD4 and PTEN by immunohistochemistry (IHC) in 72 of the samples tested for mir-21 expression. TPM1 expression was evaluated on a subset of samples, however the level of TPM1 expression in cervical epithelium was found to be below the threshold of detection by immunohistochemistry. We observed a significant association between cytoplasmic PDCD4 (cPDCD4) expression and sample histology (Table 2, Figure 3 and Figure 4). Samples from women with ICC had significantly lower cPDCD4 expression compared to those with CIN2-3/CIS (pu = 0.0005 or pa = 0.003) and marginally lower expression compared to (HRHPV+) (pu = 0.03 or pa = 0.11). Women in the normal (HRHPV-) group had significantly lower cPDCD4 expression compared to CIS/CIN2-3 (pu = 0.005 or pa = 0.02). There also appeared to be a marginal decrease in nuclear PDCD4 (nPDCD4) in women with ICC in comparison to those with CIN2-3/CIS. There was no significant correlation between PDCD4 expression (nuclear or cytoplasmic) and mir-21 expression in these samples, however we observed a general grouping of ICC with high mir-21 but not PDCD4, and CIN-2,3/CIS with higher PDCD4 but non-elevated mir-21 (Figure 4). There was also no association between PTEN protein expression and sample histology or microRNA expression. We considered analyzing IHC by both the frequency of the maximum intensity score and the frequency of the predominant intensity score. However, we observed similar results compared to the composite score analysis and determined there is no advantage for using the frequency of intensity scores over the composite scores.
Table 2. Protein expression results for 72 clinical samples.
| Normal (HPV-) (n = 15) | Normal (HPV+) (n = 15) | CIN1 (n = 7) | CIN2-3/CIS (n = 18) | ICC (n = 17) | p-value* | |
| Age | 42.1±4.9 | 46.6±9.3 | 42.3±4.8 | 45.5±10.6 | 46.4±10.7 | 0.55 |
| nPDCD4 | 122.0±47.9 | 143.0±49.3 | 124.3±51.8 | 152.8±61.4 | 110.9±40.7 | 0.13 |
| cPDCD4 | 73.3±30.2 | 94.7±31.4 | 87.9±35.0 | 111.9±50.8 | 65.6±33.1 | 0.006 |
| nPTEN | 70.3±40.8 | 81.7±35.6 | 44.3±37.2 | 68.1±28.5 | 64.4±46.9 | 0.32 |
| cPTEN | 64.0±31.8 | 69.0±29.3 | 47.9±43.1 | 68.3±28.2 | 69.7±36.0 | 0.62 |
*ANOVA.
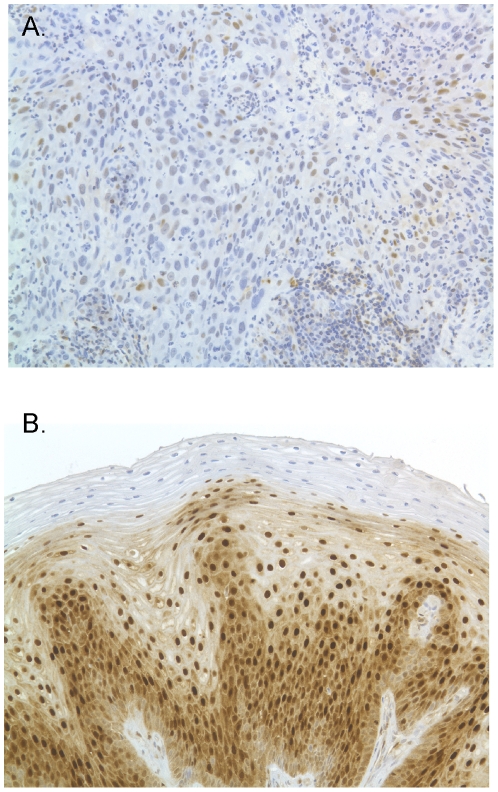
Figure 3. Micrographs showing PDCD4 staining of (a) Normal (HRHVP-) specimen and (b) ICC specimen.
Figure 4. Protein expression data measured by immunohistochemistry for cytoplasmic PDCD4 stratified by histological diagnosis.
Here we observed a significant association between cytoplasmic PDCD4 expression and sample histology.
Discussion
While microRNA expression profiling has been shown to be highly specific for cancer typing, tumor diagnosis and identifying tumors of unknown origin, it is not yet clear if microRNAs can perform as markers of disease progression – a crucial requirement for identifying those conditions which require treatment. Ideally, such markers of progression would show systematic changes in expression in precancerous tissues, which correlate with disease progression to invasive cancer. Clearly, this requires access to large numbers of clinical samples which cover the range of histologically defined tissue types involved in disease progression. In this study we investigated the expression levels of two key MicroRNAs identified in the literature (and their validated protein targets) in cervical tissue stratified by histological diagnosis in order to determine their utility as biomarkers for early cancer diagnosis. We found that mir-21 expression was significantly increased only in ICC with respect to other clinical samples and that mir-143 expression did not correlate with histology at all. Cervical cancer cell line results, while consistent with previous studies on the same cell lines, appeared to give discordant microRNA expression results in comparison to clinical samples. Interestingly, PDCD4 protein expression increased in precancerous lesions then decreased significantly in ICC, possibly as an effect of mir-21-induced silencing. These results highlight the need for further studies focusing on tissue profiling in preference to cell lines, where expression levels of candidate MicroRNA biomarkers can be assessed in pre-cancerous disease, where their utility in cancer diagnostics would prove most beneficial. We identified key inconsistencies in microRNA expression profiles in our clinical specimens as compared to cervical cancer cell lines. However, recent investigations show significant differences in microRNA expression based on cell culture conditions suggesting that cell line experiments may not be particularly informative when searching for clinically relevant microRNAs [12]. Cheng et al. 2005 [18] showed significant increase in HeLa cell growth and proliferation in response to inhibition of mir-21, whereas Yao et al 2009 [14] showed significant decrease in cell growth and proliferation for the same treatment. Wang et al 2008 [12] further showed that changing culture conditions from a monoculture to a suspension raft culture led to a completely different set of significantly deregulated microRNAs. Similar discordant results have been seen in other studies investigating the relevance of cell culture models in comparison to in vivo tissue of spheroidal cultures [19], [20]. Also, other gene/protein expression differences have recently been identified, including the presence of WNT5A expression in gastric cancers (not expressed in cell lines) [21] and differences in DNA methylation [22] and gene expression [23] signatures between colorectal carcinomas and colon cancer cell lines. Clearly, a key problem with the cell line approach is that potential biomarkers are identified in cells immortalized from invasive cancers – it is often completely unknown if these biomarkers can be detected in pre-cancerous disease, which requires analysis of clinical samples. A further complication in the current study may be the racial background (West African) in comparison to cancer cell lines which are Caucasian – however little information is known about such variation.
Increased mir-21 expression has been identified in a large number of tumor types [24]–[29] and in this study has shown significant association with the development of histologically-defined ICC. Furthermore, our results show that mir-21 may be of some utility in predicting the progression of pre-cancerous lesions into ICC, as women with high mir-21 values may be expected to progress. However, further studies involving larger sample sizes and patient follow-up are required to investigate this aspect, as there are clearly insufficient data for proper testing of this hypothesis by splitting into training and testing sets. Furthermore, while the mir-21 expression levels of women with normal pathology with and without HRHPV infection are not statistically different, the HRHPV- group showed approximately double the number of samples above the optimal cut-off compared to the HRHPV+ group. The significance of this result is again difficult to quantify due to the relatively low sample sizes involved, but it may indicate that a panel of biomarkers may be necessary to help reduce false positives.
A number of studies have identified mir-143 as significantly underexpressed in cancer, including breast and cervical cancers [12], [17], [30]. Whilst we also observed significant underexpression in cervical cancer cell lines with respect to normal (HRHPV-) samples, we did not find any statistically significant associations with respect to clinical samples from women across a wide spectrum of neoplastic disease. Previous studies investigating microRNA expression in cervical samples again show conflicting results. Lee et al 2008 [11] did not include either mir-21 or mir-143 in their initial screening experiments and as such did not identify these species as being involved in cancer development; this highlights a clear challenge in identifying potential biomarkers, in cases where leading candidates are ignored. However, Wang et al 2008 [12] suggested that reduction of mir-143 was required for tumor development whilst Lui et al 2007 [17] showed mixed results for mir-143, mainly due to the low level of expression detected in either normal or cancerous samples. Inconsistencies may be due to the relatively low level mir-143 expression in cervical tissues, which may require a more precise extraction method (e.g. laser microdissection) for tumor extraction – none of the studies listed here used such methods.
Previous studies revealed that PDCD4, PTEN and TPM1 are all transcripts targeted by mir-21 in various tissues [14], [17], [24], [26], [31], [32] and that PTEN is progressively underexpressed from normal tissue through to cervical cancer [33]–[35]. One recent study has also reported reduced PDCD4 expression in cervical cancer cell lines [14] which is in agreement with our findings in the clinical samples examined in this study. In fact recent studies have demonstrated not only a direct relation between PDCD4 expression and tumor progression (Wei et al, 2009 [36]) but also the key role of PDCD4 and mir-21 in apoptotic inflammatory response (Sheedy et al, 2009 [37]). Moreover, recent cell model studies support suggest a role for loss of PDCD4 expression in increased sensitivity of cells to agents that cause DNA damage (Singh et al, 2009 [38]), all of which would be expected to provide favourable conditions for tumor growth. Mice expressing high levels of PDCD4 are also more resistant to tumor growth in comparison to normal controls. We hypothesise that cPDCD4 expression (and, to a lesser extent, nPDCD4 expression) is increased in pre-cancerous specimens due to increased apoptosis, followed by a sharp decrease in expression for ICC specimens, where apoptotic mechanisms are subverted. Similar trends have been observed for other tumor suppressor genes in relation to oncogenesis and apoptosis [39], [40]. Interestingly, we did not identify any correlation between PTEN expression and histological diagnosis; loss of expression has been noted previously, and we have observed lower expression in a breast cancer study using the same methods (data not shown).
In this study we have found that the expression level of mir-21 is significantly correlated with histological diagnosis of ICC. A known target of mir-21, PDCD4 was also downregulated but was not significantly correlated with mir-21 expression. Expression of mir-143 was not correlated with histological diagnosis of clinical samples, however it was significantly down-regulated in cervical cancer cell lines. These findings are important in the quest to gauge the utility of MicroRNA biomarkers for diagnostic applications, a key component of which is to measure expression levels in precancerous tissues as well as normal and invasive cancers. Clearly, future work must focus on clinically relevant samples, in which predictive markers expressed in pre-cancerous tissue can be identified for further validation.
Materials and Methods
Selection of microRNAs
MiRs investigated in this study were chosen based on previous observations in the literature (Table 3). We specified that each microRNA should also have an experimentally verified mRNA target or set of target genes and at least one of the predicted targets had to be reported as involved in oncogenesis and progression in cervical or other neoplasms (based on sequences in the miRBase database [41]–[43]). MiR-21 is reported to be overexpressed in a number of cancers [29], [44] and targets transcripts of the PTEN [24], TPM1 [26] and PDCD4 [14], [31], [32] genes, whilst miR-143 is underexpressed in many cancers and targets ERK5 transcripts [17], [29], [30]. Both mir-21 overexpression and mir-143 underexpression have been observed in cervical cancer cell lines and clinical specimens [12], [17].
Table 3. Cancer associations for MicroRNAs used in this study.
| MicroRNA | Cancer Association | Validated Target | Reference |
| Mir-21 | Cervical | PDCD4 | [14] |
| Colon | PDCD4 | [31] | |
| Prostate | MARCKS | [50] | |
| Stomach | PTEN | [51] | |
| Breast | PDCD4, TPM1 | [26], [32] | |
| Liver | PTEN | [24] | |
| Lung, pancreas | – | [17], [29], [44] | |
| Mir-143 | Breast | ERK5 | [30] |
| B-cell tumors | ERK5 | [52] | |
| Cervical, prostate, colon | – | [17], [29], [53], [54] |
Samples
Clinical specimens were collected from the Dantec Oncology Clinic, Dakar, Senegal from studies of cervical cancer [45]; all study procedures were approved by the institutional review boards of the University of Washington (Seattle, WA) and University of Dakar (Dakar, Senegal) and informed written consent was provided by study participants. Histological diagnoses were produced at the University of Washington, Department of Pathology, Harborview Medical Center, Seattle, WA by a pathologist (NBK). Samples collected included 142 cervical biopsies preserved as FFPE tissue blocks. HPV genotyping was performed as described previously [46], with High-Risk HPV (HRHPV) subtypes HPV16/18/26/31/33/35/39/45/51/52/53/56/58/59/66/67/68/73/82 as defined by Munoz et. al. 2006 [47]. Clinical specimens were grouped according to histological diagnosis, with groups defined by treatment regimens per current guidelines [48]. Normal (HRHPV-) identified those women with a normal histology and a negative test result for HRHPV (n = 23). Normal (HRHPV+) indicated those women with a normal histology and a positive test for HRHPV (n = 21). Mildly dysplastic CIN1 lesions (n = 15) typically spontaneously regress and women with these lesions are usually followed with repeat Pap smear. Women with precancerous CIN2-3/CIS lesions (n = 51) would all be referred for treatment to prevent development of invasive disease ICC (n = 23). These groupings allowed us to determine if the microRNA biomarkers under investigation could identify various stages of neoplastic disease which require different clinical management. The average age of the 133 subjects from who clinical specimens were used was 44 ranging from 29 to 72 years. All four histologic groups were frequency matched on age in order to achieve similar age distributions and means, which ranged between 43 and 45 years. Cell lines and culture media were purchased from ATCC and Gibco (Invitrogen, Carlsbad, CA).
MicroRNA qPCR expression analysis
Extraction of microRNA from blocks was performed using the RecoverAll Total Nucleic Acid Isolation Kit (Applied BioSystems, Foster City, CA), according to manufacturer's instructions. The only alteration was that for cell line extraction, the deparaffination steps were omitted on the advice of Applied Biosystems staff. Reverse transcription (RT) reactions and quantitative polymerase chain reactions (qPCR) were performed using the MicroRNA TaqMan Reverse Transcription Kit and the TaqMan MicroRNA Assays (Applied BioSystems) for mir-21, mir-143 and RNU6B control shRNA. RT and qPCR for the RNU6B control shRNA analysis was performed side-by-side with the mir-21 and mir-143 reactions for each sample. A standard curve was generated by extracting total RNA from VK2 cells and performing RT reactions on 10-fold dilutions, subsequently including each serial dilution in each qPCR run, throughout the whole study.
Immunohistochemistry
PDCD4 rabbit polyclonal antibody and TPM1 mouse monoclonal antibody was purchased from Abcam (Abcam Inc., Cambridge MA, ab51495, ab17784 respectively), PTEN mouse monoclonal antibody was purchased from Dako (Dako Corporation; Carpinteria, CA, clone 6H2.1) Four-micron sections of formalin-fixed paraffin-embedded tissue were cut and placed on Superfrost Plus microscope slides. The tissue sections were deparaffinized and rehydrated through a series of graded alcohols. Antigen retrieval was carried out with Tris/EDTA pH 9.0 buffer in a tabletop autoclave. Endogenous peroxidase activity was blocked with 3% H2O2. Primary antibodies were varied to determine the optimal antibody concentration resulting in the most intense staining with no background. Antibodies for PDCD4 were diluted 1∶2000 and PTEN antibody was diluted to 1∶500 and applied to each slide. TPM1 antibody was diluted to 1∶100.The slides were washed in ImmPRESS (Vector Laboratories; Burlingame, CA), then an anti-mouse (for PTEN and TPM1) and an anti-rabbit (for PDCD4) IgG polymerized reporter enzyme was applied to each tissue. Color development was accomplished by incubation in diaminobenzidine (Dako). The slides were counterstained in hematoxylin (Dako), dehydrated through graded alcohols, cleared in xylene, and coverslipped with permanent mounting media. A negative tissue control was run on each tissue, where antibody diluent buffer was substituted for the primary antibody. In addition, positive control tissues known to express the antigen in question were run in conjunction with each batch of slides. PDCD4 and PTEN staining was evaluated semi-quantitatively using the H-score scoring method [49]. This method of quantification takes into account the staining intensity and the percentage of cells staining at a particular intensity. Staining intensity was scored as 0 (no staining), 1 (weak staining), 2 (moderate staining), 3 (intense staining). Each intensity score was multiplied by the percentage of cells staining at that level. The resulting values were summed to yield a composite score ranging from 0 to 300. PDCD4 and PTEN expression was evaluated in both the nuclear and cytoplasmic compartments.
Statistical Methods
Taqman qPCR reactions were performed in triplicate redundancy and the Ct values converted to a ratio with respect to the RNU6B control. Non-cancer controls were frequency age-matched to cases with invasive cancer. Values of both microRNAs were log10-transformed in order to normalize the distribution of these measurements for all statistical analyses. ANOVA was used to compare all histologic groups with respect to the continuous factors of interest. P-values are reported as unadjusted (pu) and adjusted for multiple comparisons using false discovery rates (pa). Receiver Operating characteristic (ROC) curves were used to assess the predictability of cervical disease progression. Optimal cutoffs were determined to best distinguish Normal (HPV-) women from women with ICC. A two-sided 0.05 test level determined statistical significance for all analyses. All analyses were conducted using SAS version 9.2 (SAS Institute Inc., Cary, NC).
Acknowledgments
The authors would like to acknowledge Pap Toure, Papa Salif Sow, and Amadou Dembele for their clinical work, recruitment and management of study patients, and collection of specimens in Senegal. In addition, we would like to thank Donna Kenney and Limin Hu for their technical assistance in storing, preparing and sectioning specimen blocks.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors acknowledge funding contributions from the National Institutes of Health/National Cancer Institutes (NIH/NCI) (R01 CA75920 and R01 CA115713; Dr. Kiviat) and the American Australian Association (Merck Company Foundation Postdoctoral Fellowship; Dr. Corrie). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cowland JB, Hother C, Gronbaek K. MicroRNAs and cancer. APMIS. 2007;115:1090–1106. doi: 10.1111/j.1600-0463.2007.apm_775.xml.x. [DOI] [PubMed] [Google Scholar]
- 2.Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84:55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- 3.Reshmi G, Pillai MR. Beyond HPV: Oncomirs as new players in cervical cancer. Febs Letters. 2008;582:4113–4116. doi: 10.1016/j.febslet.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Paranjape T, Slack FJ, Weidhaas JB. MicroRNAs: tools for cancer diagnostics. Gut. 2009;58:1546–1554. doi: 10.1136/gut.2009.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Lee CGL. MicroRNA and cancer - focus on apoptosis. Journal of Cellular and Molecular Medicine. 2009;13:12–23. doi: 10.1111/j.1582-4934.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.Kai-Hung W, Hwan-Wun L, Shih-Rung L, Da-Ching D, Tang-Yuan C. Field methylation silencing of the protocadherin 10 gene in cervical carcinogenesis as a potential specific diagnostic test from cervical scrapings. Cancer Science. 2009;9999 doi: 10.1111/j.1349-7006.2009.01285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, et al. Circulating microRNAs as Novel Minimally Invasive Biomarkers for Breast Cancer. Annals of Surgery. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 9.Holowaty P, Miller AB, Rohan T, To T. Natural history of dysplasia of the uterine cervix. Journal of the National Cancer Institute. 1999;91:252–258. doi: 10.1093/jnci/91.3.252. [DOI] [PubMed] [Google Scholar]
- 10.Hawes SE, Kiviat NB. Screening for cervical cancer. In: Holmes K, Sparling P, Stamm W, Piot P, Wasserheit J, et al., editors. Sexually transmitted diseases. 3rd ed. New York: McGraw-Hill; 1999. pp. 1399–1408. [Google Scholar]
- 11.Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ, et al. Altered MicroRNA expression in cervical carcinomas. Clinical Cancer Research. 2008;14:2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Tang S, Le S-Y, Lu R, Rader JS, et al. Aberrant Expression of Oncogenic and Tumor-Suppressive MicroRNAs in Cervical Cancer Is Required for Cancer Cell Growth. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002557. Article No.: e2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, et al. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575–2582. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao Q, Xu H, Zhang Q-Q, Zhou H, Qu L-H. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochemical and Biophysical Research Communications. 2009;388:539–542. doi: 10.1016/j.bbrc.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 15.Yang ZZ, Chen S, Luau XJ, Li YX, Liu M, et al. MicroRNA-214 is Aberrantly Expressed in Cervical Cancers and Inhibits the Growth of HeLa Cells. IUBMB Life. 2009;61:1075–1082. doi: 10.1002/iub.252. [DOI] [PubMed] [Google Scholar]
- 16.Pereira PM, Marques JP, Soares AR, Carreto L, Santos MAS. MicroRNA expression variability in human cervical tissues. PLoS One. 2010;5:e11780. doi: 10.1371/journal.pone.0011780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lui W-O, Pourmand N, Patterson BK, Fire A. Patterns of Known and Novel Small RNAs in Human Cervical Cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 18.Lai JC, Cheng YW, Chiou HL, Wu MF, Chen CY, et al. Gender difference in estrogen receptor alpha promoter hypermethylation and its prognostic value in non-small cell lung cancer. Int J Cancer. 2005;117:974–980. doi: 10.1002/ijc.21278. [DOI] [PubMed] [Google Scholar]
- 19.Grill J, Lamfers MLM, van Beusechem VW, Dirven CM, Pherai DS, et al. The organotypic multicellular spheroid is a relevant three-dimensional model to study adenovirus replication and penetration in human tumors in vitro. Molecular Therapy. 2002;6:609–614. [PubMed] [Google Scholar]
- 20.Kaaijk P, Troost D, Dast PK, Leenstra S, Bosch DA. Long-term culture of organotypic multicellular glioma spheroids – a good culture model for studying. Neuropathology and Applied Neurobiology. 1995;21:386–391. doi: 10.1111/j.1365-2990.1995.tb01075.x. [DOI] [PubMed] [Google Scholar]
- 21.Saitoh T, Mine T, Katoh M. Frequent up-regulation of WNT5A mRNA in primary gastric cancer. International Journal of Molecular Medicine. 2002;9:515–519. [PubMed] [Google Scholar]
- 22.Lind GE, Thorstensen L, Lovig T, Meling GI, Hamelin R, et al. A CpG island hypermethylation profile of primary colorectal carcinomas and colon cancer cell lines. Molecular cancer. 2004;3:28. doi: 10.1186/1476-4598-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez A, Smith F, Wang QD, Wang XF, Evers BM. Assessment of differential gene expression patterns in human colon cancers. Annals of Surgery. 2000;232:576–584. doi: 10.1097/00000658-200010000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, et al. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, et al. Altered Expression of miR-21, miR-31, miR-143 and miR-145 Is Related to Clinicopathologic Features of Colorectal Cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 26.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 27.Si M-L, Zhu S, Wu H, Lu Z, Wu F, et al. miR-21-mediated tumor growth. 2006;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 28.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, et al. Altered MicroRNA Expression Confined to Specific Epithelial Cell Subpopulations in Breast Cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 29.Iorio MV, Ferracin M, Liu C-G, Veronese A, Spizzo R, et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 30.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, et al. MicroRNA-143 Regulates Adipocyte Differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 31.Asangani IA, Rasheed SAK, Nikolova DA, Leupold JH, Colburn NH, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. 2007;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 32.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, et al. Programmed Cell Death 4 (PDCD4) Is an Important Functional Target of the MicroRNA miR-21 in Breast Cancer Cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Joh K, Yatsuki H, Zhao W, Soejima H, et al. Retinoic acid receptor beta2 is epigenetically silenced either by DNA methylation or repressive histone modifications at the promoter in cervical cancer cells. Cancer Lett. 2007;247:318–327. doi: 10.1016/j.canlet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 34.El-Mansi MT, Williams AR. Evaluation of PTEN expression in cervical adenocarcinoma by tissue microarray. Int J Gynecol Cancer. 2006;16:1254–1260. doi: 10.1111/j.1525-1438.2006.00569.x. [DOI] [PubMed] [Google Scholar]
- 35.Lee JS, Choi YD, Lee JH, Nam JH, Choi C, et al. Expression of PTEN in the progression of cervical neoplasia and its relation to tumor behavior and angiogenesis in invasive squamous cell carcinoma. J Surg Oncol. 2006;93:233–240. doi: 10.1002/jso.20493. [DOI] [PubMed] [Google Scholar]
- 36.Wei NA, Liu SS, Leung TH, Tam KF, Liao XY, et al. Loss of Programmed cell death 4 (Pdcd4) associates with the progression of ovarian cancer. Mol Cancer. 2009;8:70. doi: 10.1186/1476-4598-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 38.Singh P, Marikkannu R, Bitomsky N, Klempnauer KH. Disruption of the Pdcd4 tumor suppressor gene in chicken DT40 cells reveals its role in the DNA-damage response. Oncogene. 2009;28:3758–3764. doi: 10.1038/onc.2009.239. [DOI] [PubMed] [Google Scholar]
- 39.Xia HHX, Talley NJ. Apoptosis in gastric epithelium induced by Helicobacter pylori infection: Implications in gastric carcinogenesis. American Journal of Gastroenterology. 2001;96:16–26. doi: 10.1111/j.1572-0241.2001.03447.x. [DOI] [PubMed] [Google Scholar]
- 40.Birchall MA, Winterford CM, Allan DJ, Harmon BV. Apoptosis in normal epithelium, premalignant and malignant lesions of the oropharynx and oral cavity: a preliminary study. European journal of cancer Part B, Oral oncology. 1995;31B:380–383. doi: 10.1016/0964-1955(95)00048-8. [DOI] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucl Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucl Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffiths-Jones S. The microRNA Registry. Nucl Acids Res. 2004;32:D109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Q, Balasubramanian A, Hawes SE, Toure P, Sow PS, et al. Detection of Hypermethylated Genes in Women With and Without Cervical Neoplasia. Journal of the National Cancer Institute. 2005;97:273–282. doi: 10.1093/jnci/dji041. [DOI] [PubMed] [Google Scholar]
- 46.Xi LF, Toure P, Critchlow CW, Hawes SE, Dembele A, et al. Prevalence of specific types of human papillomavirus and cervical squamous intraepithelial lesions in consecutive, previously unscreened, West-African women over 35 years of age. International Journal of Cancer. 2003;103:803–809. doi: 10.1002/ijc.10876. [DOI] [PubMed] [Google Scholar]
- 47.Munoz N, Castellsague X, de Gonzalez AB, Gissmann L. HPV in the etiology of human cancer. Vaccine. 2006;24:1–10. doi: 10.1016/j.vaccine.2006.05.115. [DOI] [PubMed] [Google Scholar]
- 48.American College of Obstetricians and Gynecologists. ACOG Practice Bulletin No. 99: management of abnormal cervical cytology and histology. Obstet Gynecol. 2008;112:1419–1444. doi: 10.1097/AOG.0b013e318192497c. [DOI] [PubMed] [Google Scholar]
- 49.Brockenbrough JS, Souquet T, Morihara JK, Stern JE, Hawes SE, et al. Tumor 39-Deoxy-39-18F-Fluorothymidine (18F-FLT) Uptake by PET Correlates with Thymidine Kinase 1 Expression: Static and Kinetic Analysis of 18F-FLT PET Studies in Lung Tumors. Journal of Nuclear Medicine. 2011;52:1181–1188. doi: 10.2967/jnumed.111.089482. [DOI] [PubMed] [Google Scholar]
- 50.Li T, Li D, Sha JJ, Sun P, Huang YR. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochemical and Biophysical Research Communications. 2009;383:280–285. doi: 10.1016/j.bbrc.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 51.Guo JM, Miao Y, Xiao BX, Huan R, Jiang Z, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. Journal of Gastroenterology and Hepatology. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 52.Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and-145 in B-cell malignancies. Cancer Science. 2007;98:1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clape C, Fritz V, Henriquet C, Apparailly F, Fernandez P, et al. miR-143 Interferes with ERK5 Signaling, and Abrogates Prostate Cancer Progression in Mice. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C, Zhou Z, Wang L, Yang L, Zhou B, et al. Clinicopathological significance of microRNA-31,-143 and-145 expression in colorectal cancer. Disease Markers. 2009;26:27–34. doi: 10.3233/DMA-2009-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]