Degradation of mutant Ndc10 is mediated by the E3 ligase Doa10 at the endoplasmic reticulum/nuclear envelope membrane. An autonomous degradation motif was localized to the C-terminal region of Ndc10. The motif is composed of two indispensable elements: a hydrophobic surface of an amphipathic helix and a loosely structured, hydrophobic C-terminal tail.
Abstract
Proper functioning of the protein-folding quality control network depends on the network's ability to discern diverse structural perturbations to the native states of its protein substrates. Despite the centrality of the detection of misfolded states to cell homeostasis, very little is known about the exact sequence and structural features that mark a protein as being misfolded. To investigate these features, we studied the requirements for the degradation of the yeast kinetochore protein Ndc10p. Mutant Ndc10p is a substrate of a protein-folding quality control pathway mediated by the E3 ubiquitin (Ub) ligase Doa10p at the endoplasmic reticulum (ER)/nuclear envelope membrane. Analysis of Ndc10p mutant derivatives, employing a reverse genetics approach, identified an autonomous quality control–associated degradation motif near the C-terminus of the protein. This motif is composed of two indispensable hydrophobic elements: a hydrophobic surface of an amphipathic helix and a loosely structured hydrophobic C-terminal tail. Site-specific point mutations expose these elements, triggering ubiquitin-mediated and HSP70 chaperone–dependent degradation of Ndc10p. These findings substantiate the ability of the ER quality control system to recognize subtle perturbation(s) in the native structure of a nuclear protein.
INTRODUCTION
Accumulation of aberrant proteins within the cell can often have deleterious consequences. To circumvent these consequences, prokaryotic and eukaryotic cells employ various co- and posttranslational quality control systems. These systems include molecular chaperones and proteases designed to recognize damaged proteins and subsequently repair or eliminate them. Under different conditions—when these quality control pathways demonstrate low performance or when they are otherwise overwhelmed—misfolded proteins accumulate and often form insoluble aggregates that can lead to cellular dysfunction (Kubota, 2009).
Protein degradation by the ubiquitin (Ub)-proteasome system is an essential mechanism that plays an imperative role in protein quality control surveillance (Buchberger et al., 2010). The ability of this system to respond correctly to varied physiological conditions requires diverse and somewhat conflicting features from its substrates (Ravid and Hochstrasser, 2008). On the one hand, there is a requirement for increased substrate specificity for degrading targets involved in regulatory cellular processes, such as transcription or cell cycle progression. On the other hand, plasticity of the recognition process is essential for rapidly detecting proteins undergoing nonnative conformational changes. Plasticity of substrate recognition is particularly important for quality control–associated degradation pathways that operate to reduce the load of futile proteins by targeting unfolded, misfolded, and misassembled proteins for proteolysis by the 26S proteasome (Buchberger et al., 2010).
A principal site for cellular protein quality surveillance in all eukaryotes is the endoplasmic reticulum (ER) membrane, which in yeast is continuous with the outer layer of the nuclear envelope. The ER quality control machinery recognizes a diverse array of substrates, all of which reach temporal or permanent improperly folded states. These include: ER-membrane proteins, such as a mutant form of Ste6p (Loayza and Michaelis, 1998), Ubc6p (Walter et al., 2001), and 3-hydroxy-3-methylglutaryl CoA (HMG CoA) reductase (Hampton and Rine, 1994; Ravid et al., 2000); plasma membrane proteins, such as the cystic fibrosis transmembrane conductance regulator (CFTR; Jensen et al., 1995; Ward et al., 1995) and the T-cell receptor α chain (Yu et al., 1997); mutant secretory proteins that get stuck in the ER lumen, such as a mutant form of carboxypeptidase Y (CPY; Hiller et al., 1996) and ribophorin I (de Virgilio et al., 1999); synthetic substrates expressed in the cytosol (Gilon et al., 1998; Metzger et al., 2008); and nuclear proteins, such as the yeast transcription repressor Matα2 (Hochstrasser and Varshavsky, 1990). How can the ER quality control system recognize such a diverse collection of substrates? In general, misfolded substrates display degradation signals, or degrons (Varshavsky, 1991), recognized by a small number of specific Ub-ligation complexes. For example, in the baker's yeast Saccharomyces cerevisiae, two ubiquitylation complexes are embedded in the ER membrane (Carvalho et al., 2006; Denic et al., 2006). These complexes are named according to the E3 ligases involved, Hrd1p/Der3p and Doa10p (Hampton et al., 1996; Bordallo et al., 1998; Swanson et al., 2001), and their orthologues are present in all higher eukaryotes. In yeast, each of the Ub-ligation complexes recognizes a distinct set of substrates, with minor overlaps (Vashist and Ng, 2004). The Hrd1p complex recognizes ER transmembrane substrates and substrates with lesions facing the ER lumen, while Doa10p is responsible for ubiquitylating substrates with cytosolic and nuclear lesions. Unlike Hrd1p, Doa10p is localized to both the cytosolic leaflet of the ER and the inner nuclear membrane, and it is engaged in ubiquitylating substrates from both compartments (Deng and Hochstrasser, 2006).
The triage decision for Hrd1p substrates is largely based on their folding or maturation state in the ER lumen, whereas the features that characterize degrons of Doa10p diverse substrates are largely unknown. To date, no substantial structural data are available for proteins in their misfolded state, including Ub-system degradation substrates. However, several studies suggest that exposure of hydrophobic regions, such as the ones usually located inside globular proteins or inserted into membranes, are required for Doa10p-dependent degradation (Johnson et al., 1998; Gilon et al., 2000; Arteaga et al., 2006).
Our study focuses on the degradation signals that trigger quality control substrate surveillance by the Doa10p ubiquitylation complex. To this end, we used a previously identified temperature-sensitive (ts) mutant of the yeast protein Ndc10p as a model substrate (Ravid et al., 2006). Ndc10p is an essential kinetochore protein and a key component of the CBF3 multi-subunit complex that binds to the centromere (Doheny et al., 1993). Kopski and Huffaker (1997) described an ndc10 ts mutant, termed ndc10-2, harboring a single Ala- to Thr-missense mutation at residue 914 of the protein. At the restrictive temperature of 37°C, ndcl0-2 cells assemble anaphase spindles but fail to segregate their DNA, which is consistent with a defect in the kinetochore function. This mutant phenotype was suppressed by manipulating genes encoding Ubc6p and Ubc7p, the E2-conjugating enzymes of the Doa10p ubiquitylation complex (Chen et al., 1993). We have previously demonstrated that Ndc10-2p rapidly degrades in a DOA10-dependent manner at 37°C and that the ts phenotype of ndc10-2 cells is suppressed upon DOA10 deletion (Ravid et al., 2006). Thus the degron that targets Ndc10-2p for proteolysis via Doa10p is functionally distinct from the essential protein domain(s) needed for the proper activity of Ndc10p at the kinetochore.
In this paper, we describe the identification and characterization of the Ndc10-2p degron, engaging two sets of substrates: nuclear Ndc10p and a fusion between an integral ER-membrane protein and Ndc10p serial truncations. This degron is composed of two primary determinants at the vicinity of the A914T mutation of Ndc10-2p: an amphipathic helix and a loosely structured hydrophobic C-terminal tail, both of which are required for efficient ubiquitylation and degradation. Subtle structural perturbations expose the degron, targeting the protein to proteasomal degradation. Similar combinations of hydrophobic motifs are predicted to be present in multiple proteins. Exposure of these common features may trigger quality control pathways, leading to protein ubiquitylation and degradation.
RESULTS
Ndc10-2p degradation is independent of its localization at the kinetochore
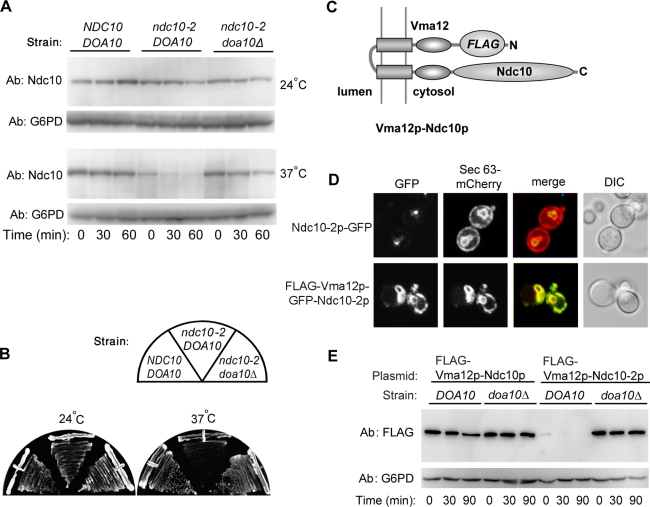
To gain new insights into the mechanism of nuclear protein quality control surveillance, we wanted to determine the specific sequences and structural determinants that mark Ndc10p as being misfolded. Indication of misfolding was based on wild-type Ndc10p and mutant ndc10-2p degradation rates after protein synthesis was blocked by the antibiotic drug cycloheximide. To this end, we set up a gene shuffle system, replacing the essential endogenous NDC10 gene with wild-type or mutated variants expressed from a plasmid (see Materials and Methods). Figure 1A shows that Ndc10p remained stable at both the permissive and restrictive incubation temperatures, while Ndc10-2p was stable at 24°C but was rapidly degraded at 37°C (comparative graphic presentations of the data appear in Figure 1A; additional cycloheximide-chase data appear in Supplemental Figure S8). In accordance with previous results using a chromosomal ndc10-2 allele (Ravid et al., 2006), deletion of DOA10 stabilized Ndc10-2p at 37°C. The decline in Ndc10-2p levels in DOA10 cells and its suppression in doa10Δ cells is in good correlation with the ts phenotype of ndc10-2 cells (Figure 1B).
FIGURE 1:
Nuclear-localized Ndc10-2p is a quality control substrate of the Doa10p E3 ligase. (A) Degradation of Ndc10p and Ndc10-2p in ndc10 Δ shuffle strains in the presence or absence of DOA10. Cells expressing NDC10 or ndc10-2 were grown at 24°C to log phase. Cells were kept at 24°C or shifted to 37°C 15 min prior to cycloheximide addition, and aliquots were taken at the indicated times. Lysates were analyzed by anti-Ndc10p and anti-G6PD immunoblotting. (B) Growth of shuffle strains described in (A). Cells were streaked on YPD plates and incubated at 24°C or 37°C for 2 d. (C) Schematic presentation of the topology of FLAG-Vma12p-Ndc10p. (D) Cellular localization of Ndc10-2p-GFP and FLAG-Vma12p-GFP-Ndc10p. Cells expressing the indicated proteins together with mCherry-Sec63p were grown to log phase and proteins were visualized by confocal fluorescence microscopy. (E) Degradation of FLAG-Vma12p-Ndc10p and FLAG-Vma12p-Ndc10-2p in DOA10 and doa10Δ cells. Cells transformed with plasmids encoding the respective proteins were assayed by cycloheximide chase and immunoblotting with anti-FLAG and anti-G6PD antibodies.
To identify other Ub-system enzymes necessary for Ndc10-2p degradation, we compared Ndc10-2p levels in a set of yeast strains, in which known or suspected ubiquitylation enzymes of protein quality control pathways were deleted. To this end, we cloned NDC10 and ndc10-2 open reading frames (ORFs) into a plasmid under a constitutive GPD1 promoter, and tagged it with an internal FLAG epitope (see Materials and Methods). Importantly, the degradation of this protein substrate was no longer temperature dependent. Temperature sensitivity appears to be a unique feature of the untagged, nuclear Ndc10p mutants.
Levels of Ndc10-2p-FLAG in yeast strains carrying deletions of the indicated Ub enzymes were estimated by immunoblotting (see Figure S1). As anticipated, of the 10 deletion strains tested, only doa10Δ and its cognate E2-Ubc7Δ significantly increased Ndc10-2p-FLAG levels, suggesting that the Doa10p complex is the main ubiquitylation apparatus for Ndc10-2p degradation. Thus, when expressed under a constitutive and a strong promoter, changes in Ndc10-2p-FLAG steady-state levels can indicate differences in Ndc10-2p degradation kinetics.
To test whether the trigger for Ndc10-2p degradation is an intrinsic property of the protein, independent of its nuclear localization, we engineered a second copy of Ndc10p targeted to the ER membrane. This was achieved by fusing NDC10 or ndc10-2 to the C-terminal region of the yeast VMA12 coding sequence, which was tagged with N-terminal FLAG epitope (Figure 1C). Vma12p is a stable, integral ER-membrane protein with cytosolic N- and C-termini (Jackson and Stevens, 1997; see also Figure S2a). Use of the Vma12p backbone has enabled evaluation within the context of the ER membrane of other Doa10p degrons derived from normally soluble proteins (Ravid et al., 2006).
To verify the localization of nuclear and ER-bound Ndc10-2p, we integrated a green fluorescent protein (GFP) into both of these substrates (see Materials and Methods). The fluorescence emission of the GFP tag showed characteristic nuclear punctate staining for Ndc10-2p-GFP, but an ER staining for Vma12p-GFP-Ndc10-2p. This was confirmed by costaining of Ndc10-2p-GFP and Vma12p-GFP-Ndc10-2p with the tagged ER protein Sec63-mCherry (Figure 1D). Cycloheximide-based protein degradation experiments revealed that, similar to the nuclear Ndc10-2p, degradation of the ER-embedded FLAG-Vma12p-Ndc10-2p is DOA10 dependent (Figure 1E). Once again, degradation of this protein substrate was no longer temperature dependent.
Identification of a C-terminal Ndc10p degron
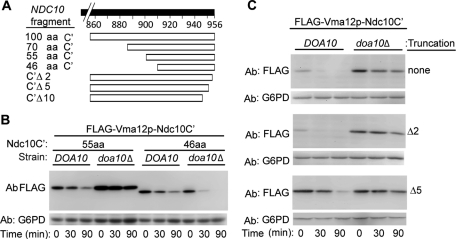
The postulated Ndc10-2p degron boundaries were defined by constructing a series of deletion mutants of FLAG-Vma12p-Ndc10p (summarized in Figure 2A). We hypothesized that the A914T missense mutation that confers instability to Ndc10-2p is part of the degron, or proximal to it. Consequently, a fragment of 100 amino acids from the NDC10 C-terminal region flanking Ala-914 was initially attached to VMA12 to obtain the fusion protein FLAG-Vma12p-Ndc10C′ (where C′ is used as an abbreviation for 100 aa of Ndc10p at the C-terminus). Importantly, intact Ndc10C′ was sufficient to confer Doa10p-dependent instability to Vma12p (Figure S2a). FLAG-Vma12p-Ndc10C′ steady-state levels were next examined in yeast strains in which known or suspected ubiquitylation enzymes of protein quality control pathways are deleted, as used in Figure S1, to test FLAG-Ndc10-2p levels. Likewise, FLAG-Vma12p-Ndc10C′ expression was increased in doa10Δ and ubc7Δ strains, but not in other tested E2s and E3 knockout strains (Figure S2b).
FIGURE 2:
Identification of the Ndc10p degron. (A) Schematic presentation of the truncation mutations shown in (B and C) and in Figure S3. (B and C) Degradation of FLAG-Vma12p-Ndc10C′ and truncated mutants in DOA10 and doa10Δ cells, assayed by cycloheximide chase and immunoblotting with anti-FLAG and anti-G6PD antibodies.
Taken together, our findings indicate that a C-terminal degron of Ndc10p is embedded within the intact protein in a latent form. Structural perturbations, such as the A914T transition or out-of-context expression in the ER, expose the degron, facilitating recognition by the Doa10p Ub-conjugation machinery and subsequent degradation of the protein.
We next tested the degradation kinetics of shortened Ndc10C′ fragments fused to Vma12p (summarized in Figure 2A). An ∼55–amino acid C-terminal peptide still retained Doa10p-dependent degradation (Figures 2B and S3), while a shorter 46–amino acid Ndc10p fragment was degraded in a DOA10-independent manner (Figure 2B). Further C-terminal truncations of FLAG-Vma12p-Ndc10C′ revealed that elimination of the last two amino acids (Δ2), had no effect on degradation, while the removal of the last five amino acids (Δ5) significantly stabilized the protein (Figure 2C). Thus the minimal Doa10p-dependent Ndc10p degron resides within the extreme 55–amino acid, C-terminal fragment of the protein, excluding the last two C-terminal amino acids. Our observation that the Ndc10p degron conferred Doa10-dependent degradation on an otherwise stable protein, Vma12p, indicates that it acts as an autonomous determinant.
A hydrophobic C-terminal segment is essential for Ndc10-2p degradation
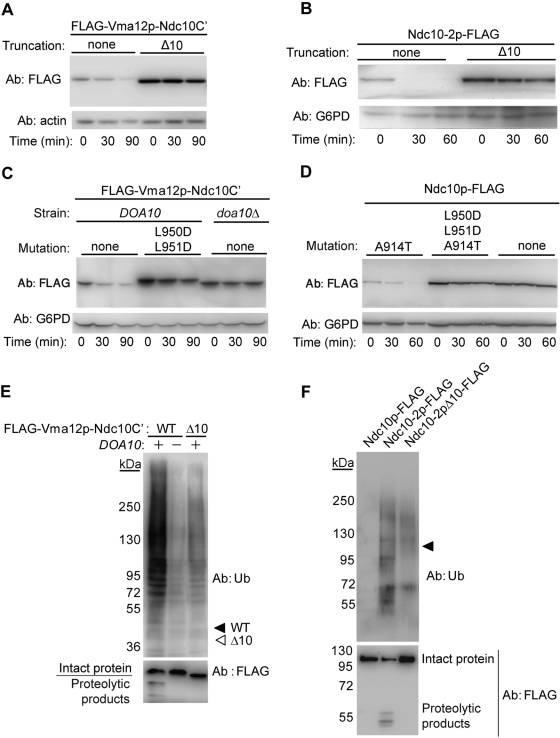
The Δ5-truncation stabilizing FLAG-Vma12p-Ndc10C′ eliminates three amino acids with hydrophobic residues that are part of a 6-mer hydrophobic segment (GLLVYL). Thus hydrophobicity may represent an essential feature of the degron. To test this theory, we examined whether removal of the entire 6-mer hydrophobic sequence, plus four flanking amino acids (Δ10), further increases FLAG-Vma12p-Ndc10C′ stability, and found that indeed it was the case (Figure 3A). We also examined the degradation of the Δ10 nuclear Ndc10-2p in the gene shuffle strain (ndc10Δ), and observed that it was similarly stabilized (Figure 3B).
FIGURE 3:
The hydrophobic C-terminal segment of Ndc10p is required for ubiquitylation. (A and B) Stabilization of C-terminally truncated (Δ10) FLAG-Vma12p-Ndc10C′ and Ndc10-2p-FLAG, assayed by cycloheximide chase and immunoblotting with anti-FLAG and anti-G6PD antibodies. (C and D) Hydrophobic-to-charge mutations (LL–DD) within the extreme C-terminal tail of FLAG-Vma12p-Ndc10C′ and Ndc10-2p-FLAG stabilized both proteins. Degradation was measured as above. (E) Impaired ubiquitylation of C-terminal–truncated FLAG-Vma12p-Ndc10C′ expressed in Rpt2-RF cells. Proteasome-inhibited cells transformed with the indicated plasmids were subjected to immunoprecipitation using anti-FLAG beads. Ubiquitylated proteins were visualized by immunoblotting with anti-Ub antibodies. Arrowheads indicate the migration distances of the unconjugated proteins: filled, FLAG-Vma12p-Ndc10C`; empty, FLAG-Vma12p-Ndc10C`Δ10. (F). Impaired ubiquitylation of C-terminal–truncated Ndc10-2p expressed in PRE6-Tet off cells. Cells expressing the indicated plasmids were treated with doxycycline (1.5 μg/ml) 24 h prior to harvesting. Lysed proteins were immunoprecipitated using anti-FLAG beads. Ubiquitylated proteins were visualized by immunoblotting with anti-Ub antibodies. Arrowhead indicates the migration distances of the unconjugated proteins.
We next disrupted the hydrophobic GLLVYL tail sequence by mutating it to GDDVYL and tested the double mutation effect on the stability of both the above described ER and nuclear substrates. This hydrophobic-to-charged residue replacement resulted in substantial stabilization of both proteins (Figure 3, C and D), indicating that hydrophobicity of the C-terminal tail is a key feature of the Ndc10p degron.
To test the functional properties of the Ndc10p C-terminal tail, we examined whether it is required for Ub conjugation. To this end, we compared Ub conjugation to FLAG-Vma12p-Ndc10C′, with or without 10 amino acids at the extreme C-terminus (Δ10), in rpt2-RF cells with impaired proteasome activity (Rubin et al., 1998). Immunoblot analysis using anti-Ub antibodies (Figure 3E) showed that FLAG-Vma12p-Ndc10C′ was ubiquitylated in a Doa10p-dependent manner. However, ubiquitylation was substantially reduced in the Δ10 derivative. Similar results were observed for the nuclear, FLAG-tagged Ndc10-2p expressed in PRE6-Tet off cells [where the α4 subunit of the proteasome Pre6p was expressed from a tetracycline titratable promoter (Tet-off; Hughes et al., 2000; Figure 3F). Therefore we concluded that an intact Ndc10p C-terminal tail is required for ubiquitylation.
Secondary structure analysis of the Ndc10p degron
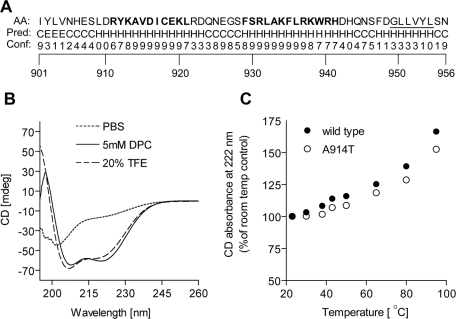
Since the extreme C′ terminal tail is necessary but insufficient to confer instability to Ndc10p, we sought to identify additional motifs within the last 55 amino acids of Ndc10p. Bioinformatics secondary-structure prediction analysis of this region, using the algorithms PSIPRED (McGuffin et al., 2000), JNET (Cuff and Barton, 2000), and SSPRO (Cheng et al., 2005), predicted the presence of two α-helices connected via a short random coil in the Ndc10p degron region (Figure 4A shows the PSIPRED prediction). These helices are followed by a C-terminal region with a lower helical propensity that contains the 6-mer hydrophobic sequence.
FIGURE 4:
Structural analysis of the Ndc10p degron. (A) Secondary structure prediction of Ndc10p C-terminal region using the PSIPRED server (McGuffin et al., 2000). Confidence is ranked from 0 to 9. H, helix; C, random coil; E, extended sheet; bold, predicted helical structures with high confidence score; underlined, C-terminal hydrophobic segment. (B and C) CD analysis. Five repetitions were taken for each measurement. (B) CD spectra of a peptide derived from aa 910–942 of Ndc10p in the indicated solvents. (C) Changes in the CD absorbance at 222 nm of Ndc10p- and Ndc10-2p-derived peptides, as a function of temperature elevation. CD absorbance at room temperature (23°C) was given the value of 100%.
Circular dichroism (CD) analysis of a synthetic peptide containing the two predicted helices of Ndc10p (aa 910–942) supported this structural model to some extent by displaying absorption peaks indicative of α-helical structure in 20% trifluoroethanol (TFE), a helix-stabilizing solvent. The helical structure was further stabilized in a detergent solution [5 mM dodecylphosphocholine (DPC)], suggesting a hydrophobic propensity of the helices (Figure 4B). Similar CD spectra characteristics were also observed using a synthetic peptide of the same length containing the A914T mutation (see Figure S4), indicating that the A914T mutation did not disturb the secondary structure. Changes in the signal at 222 nm, as a function of temperature elevation from room temperature (23°C) to 95°C, showed an increase of CD absorbance (Figures 4C and S4), indicating an apparent temperature-dependent loss of the helical secondary structure. Again, we could not detect significant absorbance differences between the wild-type and the mutant peptides, suggesting that in the context of the short synthetic peptides the A914T mutation did not cause an additional, temperature-dependent loss of the secondary structure (Figure 4C).
Disruption of predicted amphipathic helices 1 and 2 of the Ndc10p degron triggers degradation.
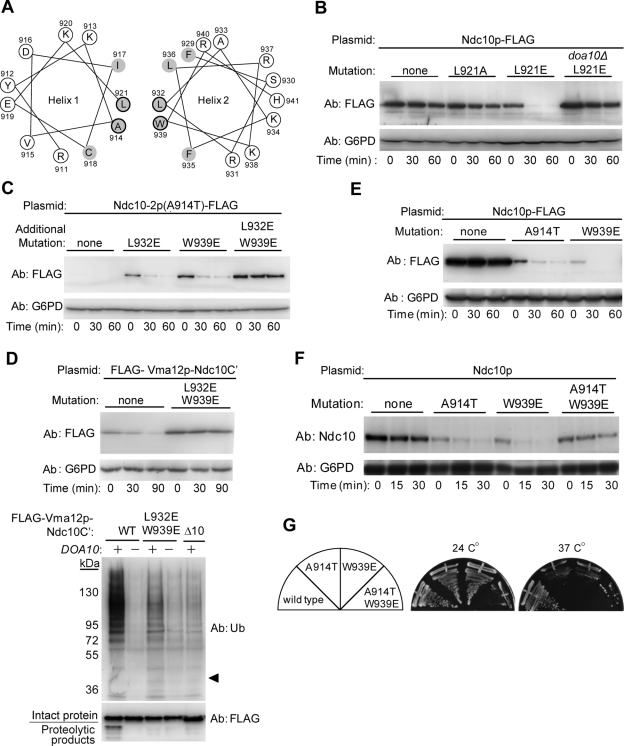
Three-dimensional structure modeling of the Ndc10p degron employing I-TASSER (Roy et al., 2010) predicts with a high degree of confidence that the postulated helix 1 (aa 910–921) and helix 2 (aa 929–941) of the Ndc10 degron are amphipathic, with their hydrophobic surfaces facing each other (Figures 5A and S7). The original mutation inducing Ndc10-2p instability (A914T) replaces a hydrophobic residue with a relatively polar residue. Thus we hypothesized that this mutation disrupts the hydrophobic interaction between the two amphipathic helices, likely increasing the aperture between them (Figure S7). To test this theory, key hydrophobic residues in Ndc10p-FLAG and Ndc10-2p-FLAG were substituted with either Ala to maintain hydrophobicity, or Glu to disrupt it. Similar to the conditional lethal A914T allele, substitution of Leu-921 (the adjacent residue in the predicted hydrophobic face of helix 1) with Glu enhanced Doa10p-dependent degradation, while substitution with Ala was inert (Figure 5B). In agreement with our initial hypothesis, two putative mechanisms may explain our findings: the first assumes that the structural change per se serves as a recognition determinant, while the second assumes that the increased aperture exposes a hydrophobic element on the opposite helix that is recognized by the Ub-ligase system.
FIGURE 5:
Disruption of amphipathic helices 1 and 2 of the Ndc10p degron triggers degradation. (A) Schematic presentation of the two predicted amphipathic helices in the C′ terminal region of Ndc10p. Interface hydrophobic residues are marked in gray. Mutated interface residues in (B to H) are circled. (B, C, and E) Degradation of FLAG-Ndc10p and the indicted mutants expressed in ndc10Δ cells, assayed by cycloheximide chase and immunoblotting with anti-FLAG and anti-G6PD antibodies. (D) Impaired ubiquitylation of FLAG-Vma12p-Ndc10C′(L932E, W939E) expressed in Rpt2-RF cells. Top, degradation of intact and mutated FLAG-Vma12p-Ndc10C′, assayed by cycloheximide-chase and immunoblotting. Bottom, the proteins in (D) expressed in proteasome-inhibited Rpt2-RF and Rpt2-RF doa10Δ cells were purified using anti-FLAG beads. Ubiquitylated proteins were detected by immunoblotting with anti-Ub antibodies. Arrowhead indicates the migration distance of the unconjugated proteins. (F) Degradation of Ndc10p and the indicated interface-residue mutants, expressed from the endogenous promoter in ndc10Δ cells. Analysis was performed as described in (D). (G) Growth of cells examined in (F). Cells were grown on nutrient-rich (YPD) agar plates and incubated at 24°C or 37°C for 2 d.
To determine whether an exposed hydrophobic surface is an essential feature of the Ndc10p degron (our second assumption), we further disrupted the hydrophobic surface of helix 2 in Ndc110-2p (A914T) through substitution of either Leu-932 or Trp-939 with Glu. Indeed, we found that these mutations increased Ndc110-2p levels, while the matching Ala substitutions in helix 2 had no effect (Figures 5C and S5). Combined substitution of both Leu-932 and Trp-939 with Glu further increased Ndc10-2p levels and greatly stabilized it, indicating a synergistic effect. These results confirm our hypothesis that an exposed hydrophobic helical surface is an essential feature of the Ndc10p degron. Therefore disruption of the hydrophobicity of both helices, as shown in Figure 5C, obliterated the degron's ability to trigger protein degradation.
Having established a role for the exposed intact helix 2 in Ndc10-2p degradation, we next investigated whether it is required for ubiquitylation, once again employing the ER-bound FLAG-Vma12p-Ndc10C′ substrate. Like the nuclear Ndc10-2p-FLAG, the double mutant FLAG-Vma12p-Ndc10C′ (L932E, W939E) was more stable than the intact fusion protein (Figure 5D, top). This result correlates with the levels of Ub-Ndc10p conjugates: The unstable FLAG-Vma12p-Ndc10C′ was highly ubiquitylated, while ubiquitylation of the stable mutant was substantially reduced, albeit not to the extent of the Δ10 mutant (Figure 5D, bottom).
Mutagenesis results combined with structure predictions imply that the disruption of a putative hydrophobic interaction between helices 1 and 2 (Figure S7) triggers ubiquitylation. Consequently, disruption of helix 2 hydrophobicity in the context of a native helix 1 should also destabilize Ndc10p. As anticipated, a single W939E mutation in the postulated helix 2 destabilized wild-type Ndc10p-FLAG (Figure 5E).
To verify the importance of amphipathy in the context of the native, untagged Ndc10p expressed under its own promoter, we mutated the native Ndc10p, cloned it onto a plasmid at the corresponding sites, and expressed it in the ndc10Δ gene shuffle strain. Degradation kinetics of the mutant Ndc10p at 37°C were similar to those obtained with the Ndc10p-FLAG constructs (Figure 5F). Moreover, similar to ndc10-2, a mutant ndc10(W939E) strain showed a ts growth phenotype (Figure 5G). Unexpectedly, while the double mutant Ndc10p (A914T, W939E) was considerably stable, this strain grew poorly at both the permissive and restrictive temperatures. It is possible that the double mutation causes a loss of essential degron recognition motifs at both helices, leading to stabilization of a deleterious Ndc10p conformation (see Discussion).
In summary, the combined effects of site-specific mutations within the postulated Ndc10p C-terminal helices 1 and 2 strongly support a model according to which a disruption of hydrophobicity of either helix serves as a degradation-promoting cue.
HSP70 chaperones are required for Ndc10-2p degradation
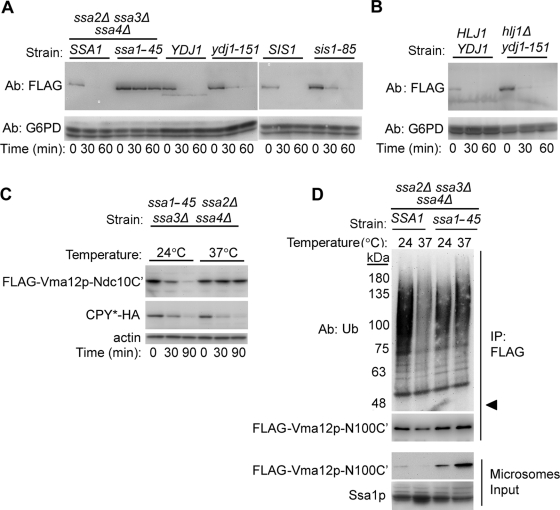
Cellular chaperones of the HSP40 and HSP70 families mediate ubiquitylation of several Doa10p substrates (Han et al., 2007; Metzger et al., 2008; Nakatsukasa et al., 2008). Therefore we wanted to identify the chaperones involved in Ndc10-2p degradation. To this end, we examined the role of the cytosolic yeast HSP70 chaperone Ssa1p, and HSP40 cochaperones Ydj1p and Hlj1p, in Ndc10-2p degradation. We also tested the effect on degradation of the cytosolic chaperone Sis1p, which is specifically required for prion propagation (Aron et al., 2007), as a negative control. Plasmids containing Ndc10-2p-FLAG were expressed in yeast strains containing ts alleles for ssa1, ydj1, or sis1, and the rate of Ndc10-2p degradation was assessed. Notably, since S. cerevisiae harbors four SSA chaperone family members with overlapping functions (Werner-Washburne et al., 1987), the role of Ssa1p in Ndc10p degradation was examined in yeast strains in which SSA2, SSA3, and SSA4 were knocked out. This manipulation had left wild-type SSA1 or the mutant ssa1-45 as the sole representatives of the SSA family. At the restrictive temperature of 37°C, Ndc10-2p-FLAG was rapidly degraded in all the parental strains, as well as in strains expressing mutant Hsp40 cochaperones (Figure 6A). However, Ndc10-2p-FLAG was substantially stabilized upon Ssa1p deactivation at 37°C. When a double mutant hlj1Δ ydj1-151 yeast strain was used, Ndc10-2p-FLAG was still rapidly degraded, suggesting that neither of the examined Hsp40 chaperones plays a significant role in the degradation pathway (Figure 6B). This result was surprising, taking into consideration the important role of Ydj1p in the degradation of other Doa10p substrates characterized so far (see Discussion).
FIGURE 6:
The HSP70 chaperone, Ssa1p, is required for Ndc10-2p degradation. (A and B) Degradation of Ndc10-2p in the indicated wild-type and mutant chaperone strains. Cells transformed with a plasmid encoding Ndc10-2p-FLAG were assayed by cycloheximide chase and immunoblotting with anti-FLAG and anti-G6PD antibodies. Cycloheximide was added 30 min after shifting the cells to 37°C, and samples were collected at the indicated times. ssa2, ssa3, and ssa4 were deleted in both SSA1 and ssa1-45 strains, to eliminate any redundant activity of the chaperones. (C) Degradation of FLAG-Vma10p-Ndc10C′ and CPY* in the ssa1-45 strain, at the permissive and restrictive temperatures. Cycloheximide-chase experiment was done as illustrated in (A and B). CPY*-HA was detected by immunoblotting with anti-HA antibodies (Roche). (D) Ubiquitylation of FLAG-Vma12p-Ndc10C′ in SSA1 and ssa1-45 strains at permissive and restrictive temperatures. Cells at log growth phase, expressing FLAG-Vma12p-Ndc10C′, were incubated for 90 min at 24°C or 37°C. Cells were then harvested, and microsomal fractions were prepared. Proteins were then subjected to immunoprecipitation, using anti-FLAG beads, which was followed by separation on an SDS–PAGE and immunoblotting with anti-Ub and anti-FLAG antibodies. Arrowhead indicates the migration distance of the unconjugated proteins.
The ssa1-45 strain propagates poorly at 37°C (unpublished data), which may indicate the accumulation of massive amounts of quality control substrates. Stabilization of Ndc10-2p in this strain background could therefore be an indirect consequence of a grossly perturbed proteostasis under stress conditions. To test it, we compared the degradation of two additional substrates: the ER-bound FLAG-Vma12p-Ndc10C′ and a mutant form of CPY (CPY*), a Hrd1p substrate (Bordallo et al., 1998). When degradation rates were tested in a ssa1-45 ts strain at 37°C, FLAG-Vma12p-Ndc10C′ was stabilized, whereas CPY* degradation was unaffected (Figure 6C). This likely excludes the possibility of a general defect in ER-associated degradation (ERAD) in ssa1-45 cells at the restrictive temperature.
To study the ubiquitylation of FLAG-Vma12p-Ndc10C′ in ssa1-45 cells, we used the protocol established by Hampton and coworkers for microsomal preparation and enrichment (Bazirgan et al., 2006). Doa10-dependant ubiquitylation of FLAG-Vma12p-Ndc10C′ in the microsomal preparations was verified by extracting the protein from wild-type and doa10Δ cells, which was followed by precipitation with anti-FLAG affinity gel and immunoblotting with anti-Ub antibodies (Figure S6). Next, SSA1 and ssa1-45 cells expressing FLAG-Vma12p-Ndc10C′ were incubated for 90 min at 24°C or 37°C. Membrane-enriched fractions were prepared and subjected to immunoprecipitation and immunoblotting, as described above (Figure 6D). At 24°C, Ub-substrate conjugates were more abundant in the wild-type SSA1 strain compared with the mutant (Figure 6D). This implies that Ssa1p is required for efficient ubiquitylation and that its function is not optimal in ssa1-45 cells, even at the permissive temperature. Notably, we did not observe any differences between the ubiquitylation pattern of FLAG-Vma12p-Ndc10C′ in ssa1-45 cells incubated at 24°C or 37°C. However, as the degradation kinetics of this substrate differ at the permissive and restrictive temperatures (Figure 6C), an additional role for Ssa1p downstream to ubiquitylation, might have been revealed (see Discussion).
DISCUSSION
In this study, we investigated the specific sequence and structural features that mark a mutant Ndc10p as misfolded. Importantly, we found that the nuclear Ndc10p is a novel quality control substrate of the ER/nuclear envelope–embedded Doa10p Ub-ligation complex, despite the existence of a quality control ubiquitylation system in the nucleus, which is mediated by the San1p E3 ligase (Gardner et al., 2005). We focused the current research on the C-terminal region of Ndc10p, since previous studies have shown that a missense mutation in this region triggered temperature-dependent degradation of the protein via the Doa10p pathway (Ravid et al., 2006).
A model for Ndc10p degradation via the Doa10p pathway
The degradation of Ndc10-2p requires the fulfillment of two conditions: the presence of a hydrophobic segment at the extreme C-terminus, and the exposure of a hydrophobic surface of a nearby amphipathic helix. This bipartite signal is obligatory for degrading both nuclear and ER-bound Ndc10p. Consistent with these results, we suggest a model for the interaction of mutant Ndc10p with the Ub system: DNA-bound Ndc10p is a relatively stable protein harboring a buried degron. Specific mutations within either of the two amphipathic helices at the C-terminus expose the cryptic degron, which is composed of a helical hydrophobic surface and a hydrophobic sequence at the extreme C′ terminus. The exposure of the degron enables the interaction with Doa10p, possibly assisted by HSP70 chaperones, as implied by the additional requirement for Ssa1 (Figure 6). These chaperones may play an additional role in degradation by escorting ubiquitylated substrates en route to the 26S proteasome.
Mild hydrophobic perturbations serve as degradation cues for protein quality control
Single-site missense mutations introducing a polar or a negative charge into the hydrophobic face of the predicted amphipathic helices of the Ndc10p degron trigger rapid degradation of the mutant protein (Figure 5). It is intriguing that these subtle mutations, which according to our in vitro and in silico data do not disrupt the overall secondary structure of Ndc10p, can so profoundly impact the protein's fate. A spatial explanation for these findings was provided by Gianni and colleagues, who studied the major states involved in the folding process of a PDZ domain (Gianni et al., 2010). They suggested that misfolded intermediates may often be quite compact, rather than substantially unstructured and highly dynamic.
The exposure of buried internal hydrophobic residues is a sign of protein misfolding that may result in irreversible protein inactivation or aggregation (Soto, 2003). There is, most likely, a thermodynamic threshold that, when crossed, reduces a protein's chance to either refold or be efficiently destroyed. Therefore the earlier an aberrant protein is detected, the lower the likelihood that it will reach this threshold and accumulate in a deleterious form. This possibility is supported by data from this study: while a single hydrophobic-to-charged mutation induced rapid ubiquitylation and degradation of mutant Ndc10p (Figure 5, B and E), a similar double mutation produced a stable protein (Figure 5, C and F) that was harmful to the cell (Figure 5G). Presumably, the singly mutated Ndc10p was recognized through the exposed hydrophobic face of the second intact helix, whereas the double mutant could not be disposed of, due to the loss of recognition determinants in both helices, and therefore accumulated in a toxic form, possibly as an aggregate.
C-terminal hydrophobic tail is a second key determinant of the Ndc10p degron
The results presented in this study show that the predicted loosely structured hydrophobic determinant at Ndc10p extreme C-terminus is an essential element of the degron. Interestingly, the presence of this tail is necessary for ubiquitylation (Figure 3), while previous studies have suggested that loosely structured sequences may serve as proteasome-degradation initiation cues, allowing the first insertion into the internal cavity of the proteasome (Prakash et al., 2004; Takeuchi et al., 2007). Our results do not exclude the possibility that the Ndc10p C-terminal tail additionally serves as a 26S proteasome initiation element. However, the requirement for an intact hydrophobic sequence (Figure 3, C and D) argues that it is not only the length and folding status but also the context that defines this C-terminal region as part of a degron element that targets Ndc10p for degradation. These conclusions are in agreement with a study by Fredrickson and colleagues, who demonstrated the important role of C-terminal hydrophobic residues for nuclear quality control in yeast (Fredrickson et al., 2011).
Ubiquitin-mediated nuclear and cytosolic quality control
Deng and Hochstrasser provided a spatial explanation for the capacity of the Doa10p complex to ubiquitylate nuclear substrates, demonstrating the localization of the E3 ligase at the cytosolic leaflet of the ER membrane, as well as at the inner nuclear envelope membrane (Deng and Hochstrasser, 2006). The best-characterized nuclear Doa10p substrate is the yeast transcriptional repressor Matα2p. This protein is ubiquitylated at the ER/nuclear envelope membrane, as well as in the nuclear matrix, by two distinct Ub-ligation systems (Wilcox and Laney, 2009; Xie et al., 2010). While Matα2p degradation serves a regulatory function during mating-type switching, the degradation of mutant Ndc10p bears the characteristics of a quality control mechanism.
In light of previous studies demonstrating the presence of a quality control ubiquitylation system in the nucleus that is mediated by the San1p E3 ligase (Gardner et al., 2005), our findings raise the question of how mutant Ndc10p evades this system. A plausible answer to this question emerged from studies by Gardner and coworkers, who demonstrated the substrate-recognition mode of the nuclear E3 ligase San1p (Fredrickson et al., 2011; Rosenbaum et al., 2011). San1p appears to recognize hydrophobic segments within its substrates via a direct interaction between intrinsic, permanently disordered, N- and C-terminal regions and misfolded regions in the substrate proteins. Whether Doa10p similarly interacts with its various substrates is yet unknown. Currently, the only structural element within Doa10p shown to partake in protein degradation is the N-terminal RING domain that binds E2 enzymes (Swanson et al., 2001). Studying the function of Doa10p is a major challenge, since its DNA coding sequence has been shown to be toxic in Escherichia coli, which greatly limits molecular and genetic approaches (Mandart et al., 1994). Nevertheless, our CD and mutagenesis results (Figures 4– 6), support a model by which mild structural perturbations trigger HSP70 chaperone–dependent Ndc10-2p interaction with the Doa10p complex. The essential role of HSP70 chaperones in the degradation of Ndc10-2p may explain why San1p does not act as an E3 ligase in the degradation of mutant Ndc10p.
The degradation of several artificial cytosolic quality control substrates is mediated by the dual action of both San1p and the cytosolic E3 ligase Ubr1p, suggesting a similar mode of substrate recognition (Heck et al., 2010; Prasad et al., 2010). It is intriguing that the degradation of Doa10p and San1p/Ubr1p cytosolic quality control substrates requires Hsp70 chaperones, while the degradation of nuclear San1p substrates is likely to be chaperone independent. This implies a mechanism whereby specific yeast quality control E3 ligases, as well as HSP70 chaperones, recognize similar structural determinants within substrate proteins.
HSP70 chaperones are required for the degradation of misfolded Doa10p substrates
Similar to other Doa10p substrates, the degradation of the nuclear Ndc10-2p requires Ssa1p, an HSP70 chaperone (Figure 6A; Huyer et al., 2004; Han et al., 2007; Metzger et al., 2008; Nakatsukasa et al., 2008). Yet knockdown of both Hsp40 cochaperones, Ydj1p and Hlj1p, did not attenuate Ndc10-2p degradation significantly (Figure 6B).
The Ydj1p cochaperone facilitates the ER localization and ubiquitylation of the cytosolic substrate Ura3-CL1, as well as the ubiquitylation of the ER-membrane proteins Ste6* and Pma1-D378S (Huyer et al., 2004; Han et al., 2007; Metzger et al., 2008; Nakatsukasa et al., 2008). We observed that Ydj1p is not essential for degrading Ndc10-2p. This emphasizes a possible partitioning between the well-studied ERAD pathway and Doa10-dependent nuclear quality control. For example: a different, yet unidentified, nuclear cochaperone(s) may cooperate with Ssa1p to target misfolded nuclear proteins to the Doa10 pathway.
What might be the role of HSP70 chaperones in the degradation of Doa10p substrates? Several previous studies have shown that the ubiquitylation of model Doa10p substrates (i.e., Ura3-CL1, Ste6*, and Pma1-D378S), is impaired in strains expressing mutant SSA1, suggesting that binding of Ssa1p to the substrate may facilitate subsequent substrate interaction with Doa10p (Metzger et al., 2008; Nakatsukasa et al., 2008). The data in Figure 6D imply that Ssa1p activity is also required for targeting ubiquitylated FLAG-Vma12-Ndc10C′ for degradation. This observation is in agreement with findings by Wolf and coworkers, who demonstrated that Ssa1p activity is essential for the degradation of ubiquitylated ΔssCPG, a cytosolic form of CPY* (Park et al., 2007). The possible role of HSP70 chaperones in targeting ubiquitylated misfolded proteins to the proteasome likely explains why ubiquitylated FLAG-Vma12p-Ndc10C′ is not degraded under conditions where mutant Ssa1p is inactivated. Thus HSP70 chaperones may function at several distinct steps in protein quality control degradation, which are not mutually exclusive: maintaining aggregation-prone proteins in a soluble form prior to ubiquitylation, targeting substrates to the E3 ligase, and assisting in delivering the ubiquitylated substrates to the 26S proteasome.
MATERIALS AND METHODS
Yeast and bacterial methods
Yeast extract–peptone–dextrose (YPD)-rich media, synthetic dextrose (SD) minimal media, and lysogeny broth bacterial media were prepared using standard protocols. Standard methods were used for genomic manipulation of yeast and for recombinant DNA work (Sambrook et al., 1989).
Yeast strains
S. cerevisiae strains used in this study are listed in Table 1. Unless indicated otherwise, the genetic background for yeast strains used in this study was that of TRY107 (a his3-Δ200, leu2-3112, ura3-52, lys2-801, trp1-1, gal2). Deletion strains constructed in this study contain the KanMX cassette that was used to disrupt the ORF of the appropriate genes, and are derived from the yeast knockout collection (Open Biosystems, Huntsville, AL).
TABLE 1:
Yeast strains used in this study.
| Yeast | Genotype | Source |
|---|---|---|
| TRY107 | a his3-Δ200, leu2-3112, ura3-52, lys2-801, trp1-1, gal2 | Chen et al., 1993 |
| TRY108 | α his3-Δ200, leu2-3112, ura3-52, lys2-801, trp1-1, gal2 | Chen et al., 1993 |
| TRY171 | α his3-Δ200 leu2-3112, ura3-52, lys2-801, trp1-1, doa10-Δ1::HIS3 | Swanson et al., 2001 |
| TRY334 | α his3-11,15, leu2-3,11, ura3-52, trp1-Δ1, lys2, SSA1, ssa2-1::LEU2, ssa3-1::TRP1, ssa4-2::LYS2 | Becker et al., 1996 |
| TRY335 | α his3-11,15, leu2-3,11, ura3-52, trp1-Δ1, lys2, ssa1ts (ssa1-45), ssa2-1::LEU2, ssa3-1::TRP1, ssa4-2::LYS2 | Becker et al., 1996 |
| TRY468 | a his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | Brachmann et al., 1998 |
| TRY 581 | α ade2-1, his3-11, leu2-3112, ura3-1, trp1-1, can 1-100 | Thomas and Rothstein, 1989 |
| TRY582 | α ade2-1, his3-11, leu2-3112, ura3-1, trp1-1, can 1-100, ydj1-2::HIS3, LEU2::ydj1-151 | Caplan et al., 1992 |
| TRY583 | a ade2-1, his3-11,15, leu2-3112, ura3-1, trpl-1, ssdl-d2, can1-100, sis1::HIS3, SIS1 on CEN/LEU2 plasmid | Luke et al., 1991 |
| TRY584 | a ade2-1, his3-11,15, leu2-3112, ura3-1, trpl-1, ssdl-d2, can1-100, sis1::HIS3, NH2–HA-tagged sis1-85 on CEN/LEU2 plasmid | Luke et al., 1991 |
| TRY621 | a leu2-3112, ura3-52, ade2-10, ndc10-2-GFP-kanMX | This study |
| TRY631 | a leu2-3112, ura3-52, ade2-10, ndc10-2-GFP-kanMX | This study |
| TRY788 | a his3-Δ200, leu2-3112, ura3-52, lys2-801, trp1-1, gal2 RPT2-RF (CEN/LEU2) | Rubin et al., 1998 |
| TRY812 | a his3-Δ200, leu2-3112, ura3-52, lys2-801, trp1-1, gal2,Doa10Δ:: KanMX, RPT2-RF (CEN/LEU2) | This study |
| TRY869 | α his3-Δ200, leu2-3112, ura3-52, lys2-801, trp1-1, gal2, NDC10Δ::kanMX, NDC10(-500-3000)9Myc on pRS316 (CEN/URA3) plasmid | This study |
| TRY879 | a his3Δ1, leu2Δ0, met15Δ0, ura3Δ0:: URA::CMV-tTA, Kan-tetO7-PRE6 | Hughes et al., 2000 |
| TRY919 | α his3-Δ200, leu2-3112, ura3-52, lys2-801, trp1-1, gal2, NDC10Δ::kanMX, DOA10::His3, NDC10(-500-3000)9Myc on pRS316 (CEN/URA3) plasmid | This study |
NDC10 gene shuffle strains were constructed by replacing the endogenous NDC10 gene with plasmids containing the wild-type or mutant NDC10 gene. Briefly, a single copy of NDC10 in diploid cells was knocked out using a PCR-amplified fragment of the KanMX6 gene with flanking ends compatible to Ndc10 locus (-500-3000). A pRS316 (centromere, CEN/URA3) plasmid containing NDC10 with a 9Myc C` terminal tag (obtained by yeast recombination using the plasmid pFa6a-9Myc-Nat), was then transformed into the diploid strain, which was followed by sporulation induction. Spores lacking the NDC10 gene but expressing NDC10 from a plasmid were separated by tetrad dissection and selected for G418 resistance and growth on SD-Ura media. To introduce mutant ndc10 plasmids and to replace the wild-type NDC10-9Myc, pRS415 (CEN/LEU2) plasmids containing several ndc10 variants, either untagged or FLAG-tagged, were transformed into ndc10Δ cells expressing pRS416-Ndc10-9Myc-Nat, which was followed by selection on SD-Leu and on 5-fluoroorotic acid–containing plates. The shift in migration distance on SDS–PAGE gels indicates the replacement of Ndc10-9Myc with Ndc10 or with Ndc10-FLAG. Deletion of DOA10 in the shuffle strain was done using yeast recombination of a PCR-amplified HIS3 ORF with flanking ends compatible to the DOA10 locus.
Plasmids
Plasmids used in this study are listed in Table 2. The plasmids were constructed using standard molecular biology techniques as described below. Primers for constructing plasmids and strains using PCR amplification techniques are available upon request.
TABLE 2:
Plasmids used in this study.
| Plasmid | Relevant markers | Source |
|---|---|---|
| pTR414 | pCPY*-HA (CEN/URA3) | Ng et al., 2000 |
| pTR425 | pFa6a-GFP-KanMX6 | Longtine et al., 1998 |
| pTR740 | pRS416-Sec63-mCherry | Kaganovich et al., 2008 |
| pTR776 | pRS414GPDp- FLAG-Vma12-6HIS-Ndc10C′-Cyc1t | This study |
| pTR777 | pRS414GPDp- FLAG-Vma12-6HIS-Ndc10-2C′-Cyc1t | This study |
| pTR781 | pRS414GPDp- FLAG-Vma12-6HIS-Ndc10-Cyc1t | This study |
| pTR782 | pRS414GPDp- FLAG-Vma12-6HIS-Ndc10-2-Cyc1t | This study |
| pTR820 | pRS414GPDp- FLAG-Vma12-Ndc10C′46aa -Cyc1t | This study |
| pTR828 | pRS410GPDp- FLAG-Vma12-6HIS-Ndc10C′-Cyc1t | This study |
| pTR834 | pRS316-Ndc10 (-500-3000) | This study |
| pTR851 | pRS415GPDp-Δ11Ndc10-FLAG-Cyc1t | This study |
| pTR852 | pRS415GPDp-Δ11NdcC10-2-FLAG-Cyc1t | This study |
| pTR853 | pRS315- Ndc10 (-500-3000) | This study |
| pTR856 | pRS414GPDp- FLAG-Vma12-Cyc1t | This study |
| pTR857 | pRS414GPDp-FLAG-Vma12-Ndc10C′70aa -Cyc1t | This study |
| pTR872 | pRS415GPDp- FLAG-Vma12-6HIS-Ndc10C′-Cyc1t | This study |
| pTR883 | pRS415GPDp- FLAG-Vma12-6HIS-Ndc10C′Δ10-CyC1t | This study |
| pTR932 | pRS415GPDp- Δ11Ndc10-2-FLAG-Δ10-CyC1t | This study |
| pTR939 | pRS414GPDp- FLAG-Vma12-Ndc10C′55aa -Cyc1t | This study |
| pTR949 | pRS416GPDp- Δ11Ndc10-2-FLAG-Cyc1t | This study |
| pTR960 | pRS415GPDp-Δ11ndc10-2 L932E-FLAG-Cyc1t | This study |
| pTR967 | pRS414 GPDp FLAG-Vma12-6HIS-Ndc10C′Δ2-Cyc1t | This study |
| pTR969 | pRS414 GPDp- FLAG-Vma12-6HIS- Ndc10C′Δ5-Cyc1t | This study |
| pTR976 | pRS414 GPDp- FLAG-Vma12-6HIS-Ndc10C′W939A-Cyc1t | This study |
| pTR982 | pR414 GPDp- FLAG-Vma12-6HIS-Ndc10C′W939E-Cyc1t | This study |
| pTR990 | Ubiquitin CUP1 promoter (CEN/LYS2) | M. Hochstrasser (unpublished) |
| pTR1023 | pRS415GPDp-Δ11ndc10 L921E-FLAG-Cyc1t | This study |
| pTR1030 | pRS414 GPDp-FLAG-Vma12-GFP-Ndc10-2-Cyc1t | This study |
| pTR1033 | pRS315-Ndc10-2 (-500-3000) | This study |
| pTR1037 | pRS315-Ndc10-2W939E (-500-3000) | This study |
| pTR1047 | pRS415-GPDp- Δ11NdcC10L921A-Cyc1t | This study |
| pTR1048 | pRS415-GPDp- Δ11NdcC10-2L932A-Cyc1t | This study |
| pTR1049 | pRS415-GPDp- Δ11NdcC10-2W939A-Cyc1t | This study |
| pTR1050 | pRS415-GPDp- Δ11NdcC10-2W939E-Cyc1t | This study |
| pTR1055 | pRS315-Ndc10W939E (-500-3000) | This study |
| pTR1058 | pRS415-GPDp- Δ11NdcC10-2L932E, W939E-Cyc1t | This study |
| pTR1060 | pRS414 GPDp- FLAG-Vma12-6HIS- Ndc10C′W939E, L932E -Cyc1t | This study |
| pTR1080 | pRS415 GPDp- Δ11NdcC10-FLAGW939E-Cyc1t | This study |
| pTR1113 | pRS415GPDp- Δ11NdcC10-2 L950D,L951D -FLAG -Cyc1t | This study |
| pTR1114 | pRS414 GPDp- FLAG-Vma12-6HIS – Ndc10C′L950D,L951D-Cyc1t | This study |
Plasmids containing Ndc10p under its endogenous promoter (500 base pairs) were generated by PCR amplification of NDC10 or ndc10-2 genes from yeast genomic preparation, which was followed by cloning into pRS316 or pRS315 (CEN/LEU2) vectors using NotI and BamHI restriction sites as 5′ and 3′ cloning sites, respectively.
Plasmids overexpressing FLAG-tagged Ndc10p were generated by digesting a PCR-amplified Ndc10 with ClaI, which removes 11 amino acids from the Ndc10 N-terminal region (Goh and Kilmartin, 1993), and cloning it into pRS415GPD-expressing vector (Mumberg et al., 1995) in restriction sites SmaI and XhoI. (We assumed that Met-12 serves as the translation initiation site for Ndcl0pΔ11.) A FLAG epitope was then inserted between amino acids 866 and 867 by PCR amplification, which was followed by three-way ligation. Briefly, two fragments of Ndc10p containing the restriction sites HincII, PsiI andPsiI, SacII, respectively, were PCR-amplified. Ligation of the two fragments after cleavage with PsiI resulted in a single product containing a single FLAG epitope at the indicated site. This fragment was cleaved with HincII and SacII, ligated into a NDC10-containing plasmid, that was cut with the same enzyme at the endogenous restriction sites to yield plasmid pRS415GPD-Ndc10-FLAG. Deletions and mutations were introduced to the plasmid by site-directed mutagenesis (Stratagene, Agilent, Santa Clara, CA). Plasmids were transformed to yeast strains using a standard LiAc transformation protocol.
Plasmids for expressing Ndc10/Ndc10-2 at the ER membrane were generated by homology recombination between a PCR-amplified 6His-NDC10 cloned from a plasmid containing glutathione S-transferase–Ndc10 (Montpetit et al., 2006) into a pHis-parallel 2 expression vector, and a plasmid containing Deg1-FLAG-Vma12-ProtA (Ravid et al., 2006), replacing protein A. FLAG-VMA12-6HIS-NDC10 was then PCR-amplified, cleaved by PstI and SalI restriction enzymes, and ligated into plasmid pRS414GPD (CEN/TRP1; Mumberg et al., 1995) at the same restriction sites to yield the plasmid pRS414GPD-FLAG-Vma12p-6His-Ndc10p (pTR781). Plasmid pTR782 was made by site-directed mutagenesis (A914T), using plasmid pTR781 as a template.
To determine the minimal Ndc10 degron, Ndc10 in plasmid pRS414GPD-FLAG-Vma12p-6His-Ndc10p was replaced with shorter C-terminal fragments that were PCR-amplified and digested with AgeI and SalI restriction enzymes. All ORF insertions were verified by sequencing. Plasmid pRS410-GPD-FLAG-Vma12p-6His-Ndc10C′, used for studying protein degradation in chaperone-knockdown strains, was obtained by replacing the TRP1 marker in plasmid pRS414-GPD-FLAG-Vma12p-6His-Ndc10C′ with KanMX by yeast recombination using a PCR fragment amplified from trp1Δ yeast strain (part of the Open Biosystems yeast knockout collection). To obtain plasmid FLAG-Vma12p-GFP-Ndc10p, a GFP fragment from plasmid pFa6a-GFP was PCR-amplified and cloned into plasmid pRS414GPD-FLAG-Vma12p-6His-Ndc10p at AgeI and StuI sites.
Cycloheximide chase and immunoblot analyses
Unless otherwise indicated, all experiments were done at 30°C. Cells were grown to log phase, then cycloheximide (0.5 mg/ml) was added, and aliquots from each time point were taken. Protein extraction was carried out by incubating cells with 0.1 N NaOH for 5 min at 23°C and spinning down the cells. The pellets were then dissolved in sample buffer and boiled at 95°C for 5 min. Samples were separated on SDS–PAGE gels (5–15% gradient), transferred to polyvinylidene fluoride membranes, and immunoblotted. The following antibodies were used: anti-Ndc10 (a gift from P. Meluh, Johns Hopkins University, Baltimore, MD), anti-glucose-6-phosphate dehydrogenase (G6PD; Sigma-Aldrich, St. Louis, MO), anti-FLAG (Sigma), anti-actin (MP Biomedicals, Solon, OH), anti-Ssa1 (a gift from E. Craig, University of Wisconsin, Madison, WI), and anti-hemagglutinin (anti-HA; Roche, Indianapolis, IN). Proteins were visualized by enhanced chemiluminescence reaction.
Fluorescence tagging and imaging
Endogenous Ndc10p was tagged with GFP using pFA6a-GFP-kanMX plasmid (Longtine et al., 1998). The construction of plasmid FLAG-Vma12-GFP-Ndc10 was as described in Plasmids. Cells were grown to log phase and images were obtained by confocal microscopy using a Bio-Rad (Hercules, CA) MRC-1024 workstation attached to a Zeiss (Jena, Germany) Axiovert 135M microscope equipped with a 63×/1.4 objective.
Ubiquitylation assay
Ubiquitylation assays were performed according to Loayza et al. (1998). Proteasome-inhibited yeast cells coexpressing the indicated Ndc10p-FLAG and FLAG-Vma12p-Ndc10C′ plasmids, together with a plasmid containing copper-induced Ub, were incubated in selective media containing 100 μM CuSO4 until late logarithmic phase. Approximately 25 A600 cell units were harvested, and then lysed by addition of 1.5 ml of 2N NaOH/1 M β-mercaptoethanol. The lysate was incubated on ice with 5% trichloroacetic acid. Proteins were separated by centrifugation at 17,000 × g for 10 min at 4°C, and the pellet was resuspended in 100 μl sample buffer. Cell extracts were diluted 30-fold with buffer supplemented with protease inhibitors (Sigma) and 5 mM N-ethylmaleimide. Extracted proteins were incubated with Anti-FLAG M2 Affinity Gel (Sigma) at 4°C for 3 h. Bead complexes were washed three times, and proteins were separated and visualized by immunoblotting, using anti-FLAG or anti-Ub antibodies.
Ubiquitylation was also tested in microsomes, which were prepared precisely as described by Bazirgan et al. (2006). Briefly, 20 A600 units of log-phase cells were harvested and resuspended in 400 μl of ice-cold membrane fractionation buffer (MFB: 20 mM Tris, pH 7.5, 0.1 M NaCl, 0.3 M sorbitol) with protease inhibitors. Glass beads were added and lysis was conducted at 4°C by three cycles of high-speed vortexing for 45 s, using FastPrep-24 Instrument (MP Biomedicals, Irvine, CA). Lysate was collected by removing supernatant from beads, washing beads twice with 400 μl of MFB, and pooling the washes and lysate. The resulting pooled lysate was cleared by repeated 10-s microcentrifuge pulses to remove unlysed cells and large debris. The cleared supernatant contained microsome membranes, which were harvested by centrifugation at 17,000 × g for 30 min.
Microsomal pellets were resuspended in SDS-sample buffer with 50 mM dithiothreitol and diluted by ∼30-fold in buffer supplemented with protease inhibitors (Sigma) and 5 mM N-ethylmaleimide. Immunoprecipitation and immunoblots were done as described above.
Peptide synthesis and CD analysis
Peptide synthesis and CD analysis were done according to Ronen et al. (2010). Ndc10p- and Ndc10-2p-derived peptides were synthesized using a Liberty microwave-assisted peptide synthesizer (CEM with a gradient of 5–60% acetonitrile in water (both containing 0.1% vol/vol trifluoroacetic acid). Peptide purity was confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and analytical high-performance liquid chromatography.
CD spectra were recorded using a J-810 spectropolarimeter (Jasco, Easton, MD) in 25 mM potassium phosphate buffer (pH 7.3), 100 mM NaCl, and 50 μM Ndc10- or Ndc10-2-derived peptides. Far-ultraviolet CD spectra were collected over a spectral range of 190–260 nm. Data were collected each 0.1 nm and averaged over 5 acquisitions. Changes in the CD spectra were monitored as a function of temperature from 23°C to 95°C.
Supplementary Material
Acknowledgments
We thank Y. Reiss, M. Hochstrasser, R. Kulka, D. Shalev, R. Cohen, and Y. Geffen for helpful discussions during this study and for critically reviewing the manuscript. We are grateful to M. Glickman, P. Hietter, M. Hochstrasser, Daniel Kaganovich, and M. Schuldiner for strains or plasmids; to P. Meluh for the Ndc10 antibodies; and to E. Craig for the Ssa1 antibodies. This work was supported by Israel Science Foundation grant 786/08, by Marie Curie International Reintegration Grant MIRG-CT-2007-205425, and by the Lejwa Fund for Biochemistry (T.R.).
Abbreviations used:
- aa
amino acid
- CD
circular dichroism
- CEN
centromere
- CFTR
cystic fibrosis transmembrane conductance regulator
- CPY
carboxypeptidase Y
- DPC
dodecylphosphocholine
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- GFP
green fluorescent protein
- HA
hemagglutinin
- HMG CoA
3-hydroxy-3-methylglutaryl CoA
- MFB
membrane fractionation buffer
- ORF
open reading frame
- SD
synthetic dextrose
- TFE
trifluoroethanol
- ts
temperature sensitive
- Ub
ubiquitin
- YPD
yeast extract–peptone–dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-05-0463) on October 12, 2011.
REFERENCES
- Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007;26:3794–3803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga MF, Wang L, Ravid T, Hochstrasser M, Canessa CM. An amphipathic helix targets serum and glucocorticoid-induced kinase 1 to the endoplasmic reticulum-associated ubiquitin-conjugation machinery. Proc Natl Acad Sci USA. 2006;103:11178–11183. doi: 10.1073/pnas.0604816103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazirgan OA, Garza RM, Hampton RY. Determinants of RING-E2 fidelity for Hrd1p, a membrane-anchored ubiquitin ligase. J Biol Chem. 2006;281:38989–39001. doi: 10.1074/jbc.M608174200. [DOI] [PubMed] [Google Scholar]
- Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:W72–W76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443:827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- de Virgilio M, Kitzmuller C, Schwaiger E, Klein M, Kreibich G, Ivessa NE. Degradation of a short-lived glycoprotein from the lumen of the endoplasmic reticulum: the role of N-linked glycans and the unfolded protein response. Mol Biol Cell. 1999;10:4059–4073. doi: 10.1091/mbc.10.12.4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson EK, Rosenbaum JC, Locke MN, Milac TI, Gardner RG. Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol Biol Cell. 2011;22:2384–2395. doi: 10.1091/mbc.E11-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Gianni S, Ivarsson Y, De Simone A, Travaglini-Allocatelli C, Brunori M, Vendruscolo M. Structural characterization of a misfolded intermediate populated during the folding process of a PDZ domain. Nat Struct Mol Biol. 2010;17:1431–1437. doi: 10.1038/nsmb.1956. [DOI] [PubMed] [Google Scholar]
- Gilon T, Chomsky O, Kulka RG. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 1998;17:2759–2766. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilon T, Chomsky O, Kulka RG. Degradation signals recognized by the Ubc6p-Ubc7p ubiquitin-conjugating enzyme pair. Mol Cell Biol. 2000;20:7214–7219. doi: 10.1128/mcb.20.19.7214-7219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh PY, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3- methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Rine J. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J Cell Biol. 1994;125:299–312. doi: 10.1083/jcb.125.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Liu Y, Chang A. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J Biol Chem. 2007;282:26140–26149. doi: 10.1074/jbc.M701969200. [DOI] [PubMed] [Google Scholar]
- Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci USA. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast a2 repressor. Cell. 1990;61:697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- Jackson DD, Stevens TH. VMA12 encodes a yeast endoplasmic reticulum protein required for vacuolar H+-ATPase assembly. J Biol Chem. 1997;272:25928–25934. doi: 10.1074/jbc.272.41.25928. [DOI] [PubMed] [Google Scholar]
- Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Johnson PR, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell. 1998;94:217–227. doi: 10.1016/s0092-8674(00)81421-x. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopski KM, Huffaker TC. Suppressors of the ndc10-2 mutation: a role for the ubiquitin system in Saccharomyces cerevisiae kinetochore function. Genet. 1997;147:409–420. doi: 10.1093/genetics/147.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H. Quality control against misfolded proteins in the cytosol: a network for cell survival. J Biochem. 2009;146:609–616. doi: 10.1093/jb/mvp139. [DOI] [PubMed] [Google Scholar]
- Loayza D, Michaelis S. Role for the ubiquitin-proteasome system in the vacuolar degradation of Ste6p, the a-factor transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:779–789. doi: 10.1128/mcb.18.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza D, Tam A, Schmidt WK, Michaelis S. Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2767–2784. doi: 10.1091/mbc.9.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Luke MM, Sutton A, Arndt KT. Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial dnaJ proteins. J Cell Biol. 1991;114:623–638. doi: 10.1083/jcb.114.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandart E, Dufour ME, Lacroute F. Inactivation of SSM4, a new Saccharomyces cerevisiae gene, suppresses mRNA instability due to rna14 mutations. Mol Gen Genet. 1994;245:323–333. doi: 10.1007/BF00290112. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Metzger MB, Maurer MJ, Dancy BM, Michaelis S. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem. 2008;283:32302–32316. doi: 10.1074/jbc.M806424200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit B, Hazbun TR, Fields S, Hieter P. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol. 2006;174:653–663. doi: 10.1083/jcb.200605019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Spear ED, Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-H, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- Prasad R, Kawaguchi S, Ng DT. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21:2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Doolman R, Avner R, Harats D, Roitelman J. The ubiquitin-proteasome pathway mediates the regulated degradation of mammalian 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 2000;275:35840–35847. doi: 10.1074/jbc.M004793200. [DOI] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–689. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen D, Rosenberg MM, Shalev DE, Rosenberg M, Rotem S, Friedler A, Ravid S. The positively charged region of the myosin IIC non-helical tailpiece promotes filament assembly. J Biol Chem. 2010;285:7079–7086. doi: 10.1074/jbc.M109.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JC, et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Maniatis T, Fritsch EF. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi J, Chen H, Coffino P. Proteasome substrate degradation requires association plus extended peptide. EMBO J. 2007;26:123–131. doi: 10.1038/sj.emboj.7601476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. Naming a targeting signal. Cell. 1991;64:13–15. doi: 10.1016/0092-8674(91)90202-a. [DOI] [PubMed] [Google Scholar]
- Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Urban J, Volkwein C, Sommer T. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J. 2001;20:3124–3131. doi: 10.1093/emboj/20.12.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M, Stone DE, Craig EA. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox AJ, Laney JD. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nat Cell Biol. 2009;11:1481–1486. doi: 10.1038/ncb1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Rubenstein EM, Matt T, Hochstrasser M. SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev. 2010;24:893–903. doi: 10.1101/gad.1906510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kaung G, Kobayashi S, Kopito RR. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.